

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC119408.7 + phase: 0 /pseudo
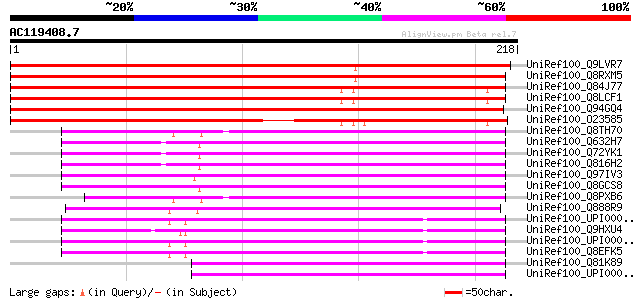
(218 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q9LVR7 Arabidopsis thaliana genomic DNA, chromosome 5,... 337 1e-91
UniRef100_Q8RXM5 Hypothetical protein At5g47420 [Arabidopsis tha... 336 2e-91
UniRef100_Q84J77 Hypothetical protein At4g17420 [Arabidopsis tha... 317 1e-85
UniRef100_Q8LCF1 Hypothetical protein [Arabidopsis thaliana] 314 1e-84
UniRef100_Q94GQ4 Hypothetical protein OJ1124_H03.5 [Oryza sativa] 280 2e-74
UniRef100_O23585 Hypothetical protein dl4745c [Arabidopsis thali... 246 2e-64
UniRef100_Q8TH70 Hypothetical protein MA4652 [Methanosarcina ace... 128 1e-28
UniRef100_Q632H7 Hypothetical protein [Bacillus cereus] 123 3e-27
UniRef100_Q72YK1 Hypothetical protein [Bacillus cereus] 122 5e-27
UniRef100_Q816H2 Hypothetical protein [Bacillus cereus] 122 7e-27
UniRef100_Q97IV3 Uncharacterized conserved protein [Clostridium ... 122 9e-27
UniRef100_Q8GCS8 Hypothetical protein [Eubacterium acidaminophilum] 120 2e-26
UniRef100_Q8PXB6 HTH DNA-binding protein [Methanosarcina mazei] 114 2e-24
UniRef100_Q888R9 Hypothetical protein [Pseudomonas syringae] 107 2e-22
UniRef100_UPI000032E9E7 UPI000032E9E7 UniRef100 entry 106 4e-22
UniRef100_Q9HXU4 Hypothetical protein [Pseudomonas aeruginosa] 106 4e-22
UniRef100_UPI00002F37B2 UPI00002F37B2 UniRef100 entry 106 5e-22
UniRef100_Q8EFK5 Hypothetical protein SO1966 [Shewanella oneiden... 106 5e-22
UniRef100_Q81K89 Hypothetical protein [Bacillus anthracis] 102 5e-21
UniRef100_UPI00003CBDBB UPI00003CBDBB UniRef100 entry 102 7e-21
>UniRef100_Q9LVR7 Arabidopsis thaliana genomic DNA, chromosome 5, P1 clone:MQL5
[Arabidopsis thaliana]
Length = 268
Score = 337 bits (864), Expect = 1e-91
Identities = 163/216 (75%), Positives = 190/216 (87%), Gaps = 1/216 (0%)
Query: 1 MAAPFFSTPFQPYVYQSQQDAIIPFQILGGESQVVQIMLKPHEKIIAKPGSMCFMSGSVE 60
MAAPFFSTPFQPYVYQSQQD I PFQILGGESQVVQIMLK EK+IAKP SMC+MSGS+E
Sbjct: 1 MAAPFFSTPFQPYVYQSQQDTITPFQILGGESQVVQIMLKSEEKVIAKPASMCYMSGSIE 60
Query: 61 MENAYLPENEVGIWQWLFGKTVTNIVVRNSGPSDGFVGIAAPYFARILPIDLATFNGEIL 120
MEN Y PE EVG+ QW+ GK+V+++V+RN+G +DGFVGIAAPY ARILPIDLA F GEIL
Sbjct: 61 MENTYTPEQEVGVLQWILGKSVSSVVLRNTGQNDGFVGIAAPYLARILPIDLAMFGGEIL 120
Query: 121 CQPDAFLCSVNDVKVSNTVDQRGRNVV-AGAEVFLRQKLSGQGLAFILGGGSVVQKILEV 179
CQPDAFLCSV+DVKV N+VDQR RN+V AGAE FLRQ+LSGQGLAFIL GGSVVQK+LEV
Sbjct: 121 CQPDAFLCSVHDVKVVNSVDQRARNIVAAGAEGFLRQRLSGQGLAFILAGGSVVQKVLEV 180
Query: 180 GEVLAVDVSCIVAVTSTVDIQIKYNGPARRTMFGVS 215
GEV ++DVSCI A+T ++D++IK N P RR +FG++
Sbjct: 181 GEVFSIDVSCIAALTPSIDVRIKNNAPFRRALFGLT 216
>UniRef100_Q8RXM5 Hypothetical protein At5g47420 [Arabidopsis thaliana]
Length = 282
Score = 336 bits (862), Expect = 2e-91
Identities = 163/214 (76%), Positives = 188/214 (87%), Gaps = 1/214 (0%)
Query: 1 MAAPFFSTPFQPYVYQSQQDAIIPFQILGGESQVVQIMLKPHEKIIAKPGSMCFMSGSVE 60
MAAPFFSTPFQPYVYQSQQD I PFQILGGESQVVQIMLK EK+IAKP SMC+MSGS+E
Sbjct: 1 MAAPFFSTPFQPYVYQSQQDTITPFQILGGESQVVQIMLKSEEKVIAKPASMCYMSGSIE 60
Query: 61 MENAYLPENEVGIWQWLFGKTVTNIVVRNSGPSDGFVGIAAPYFARILPIDLATFNGEIL 120
MEN Y PE EVG+ QW+ GK+V+++V+RN+G +DGFVGIAAPY ARILPIDLA F GEIL
Sbjct: 61 MENTYTPEQEVGVLQWILGKSVSSVVLRNTGQNDGFVGIAAPYLARILPIDLAMFGGEIL 120
Query: 121 CQPDAFLCSVNDVKVSNTVDQRGRNVV-AGAEVFLRQKLSGQGLAFILGGGSVVQKILEV 179
CQPDAFLCSV+DVKV N+VDQR RN+V AGAE FLRQ+LSGQGLAFIL GGSVVQK+LEV
Sbjct: 121 CQPDAFLCSVHDVKVVNSVDQRARNIVAAGAEGFLRQRLSGQGLAFILAGGSVVQKVLEV 180
Query: 180 GEVLAVDVSCIVAVTSTVDIQIKYNGPARRTMFG 213
GEV ++DVSCI A+T ++D++IK N P RR +FG
Sbjct: 181 GEVFSIDVSCIAALTPSIDVRIKNNAPFRRALFG 214
>UniRef100_Q84J77 Hypothetical protein At4g17420 [Arabidopsis thaliana]
Length = 285
Score = 317 bits (813), Expect = 1e-85
Identities = 159/217 (73%), Positives = 185/217 (84%), Gaps = 4/217 (1%)
Query: 1 MAAPFFSTPFQPYVYQSQQDAIIPFQILGGESQVVQIMLKPHEKIIAKPGSMCFMSGSVE 60
MAAPFFSTPFQPYVYQSQ+D I PFQILGGE+QVVQIMLKP EK+IAKPGSMC+MSGS+E
Sbjct: 1 MAAPFFSTPFQPYVYQSQEDTITPFQILGGEAQVVQIMLKPQEKVIAKPGSMCYMSGSIE 60
Query: 61 MENAYLPENEVGIWQWLFGKTVTNIVVRNSGPSDGFVGIAAPYFARILPIDLATFNGEIL 120
M+N Y PE EVG+ QW+ GK+V++IV+RN+G +DGFVGIAAP ARILP+DLA F G+IL
Sbjct: 61 MDNTYTPEQEVGVVQWILGKSVSSIVLRNTGQNDGFVGIAAPSLARILPLDLAMFGGDIL 120
Query: 121 CQPDAFLCSVNDVKVSNTVDQ--RGRNV-VAGAEVFLRQKLSGQGLAFILGGGSVVQKIL 177
CQPDAFLCSV+DVKV NTV Q R RN+ AGAE LRQ+LSGQGLAFI+ GGSVVQK L
Sbjct: 121 CQPDAFLCSVHDVKVVNTVYQRHRARNIAAAGAEGVLRQRLSGQGLAFIIAGGSVVQKNL 180
Query: 178 EVGEVLAVDVSCIVAVTSTVDIQIKYN-GPARRTMFG 213
EVGEVL +DVSCI A+T +++ QIKYN P RR +FG
Sbjct: 181 EVGEVLTIDVSCIAALTPSINFQIKYNAAPVRRAVFG 217
>UniRef100_Q8LCF1 Hypothetical protein [Arabidopsis thaliana]
Length = 285
Score = 314 bits (804), Expect = 1e-84
Identities = 157/217 (72%), Positives = 184/217 (84%), Gaps = 4/217 (1%)
Query: 1 MAAPFFSTPFQPYVYQSQQDAIIPFQILGGESQVVQIMLKPHEKIIAKPGSMCFMSGSVE 60
MAAPFFSTPFQPYVYQSQ+D I PFQILGGE+QVVQIMLKP EK+IAKPGSMC+MSGS+E
Sbjct: 1 MAAPFFSTPFQPYVYQSQEDTITPFQILGGEAQVVQIMLKPQEKVIAKPGSMCYMSGSIE 60
Query: 61 MENAYLPENEVGIWQWLFGKTVTNIVVRNSGPSDGFVGIAAPYFARILPIDLATFNGEIL 120
M+N Y PE EVG+ QW+ GK+V++IV+RN+G +DGFVGIAAP ARILP+DLA F G+IL
Sbjct: 61 MDNTYTPEQEVGVVQWILGKSVSSIVLRNTGQNDGFVGIAAPSLARILPLDLAMFGGDIL 120
Query: 121 CQPDAFLCSVNDVKVSNTVDQ--RGRNV-VAGAEVFLRQKLSGQGLAFILGGGSVVQKIL 177
CQPDAFLCSV+DVKV N+V Q R RN+ AGAE LRQ+LSGQGLAFI+ GGSVVQK L
Sbjct: 121 CQPDAFLCSVHDVKVVNSVYQRHRARNIAAAGAEGVLRQRLSGQGLAFIIAGGSVVQKNL 180
Query: 178 EVGEVLAVDVSCIVAVTSTVDIQIKYN-GPARRTMFG 213
EVGE L +DVSCI A+T +++ QIKYN P RR +FG
Sbjct: 181 EVGEFLTIDVSCIAALTPSINFQIKYNAAPVRRAVFG 217
>UniRef100_Q94GQ4 Hypothetical protein OJ1124_H03.5 [Oryza sativa]
Length = 282
Score = 280 bits (716), Expect = 2e-74
Identities = 128/212 (60%), Positives = 171/212 (80%)
Query: 1 MAAPFFSTPFQPYVYQSQQDAIIPFQILGGESQVVQIMLKPHEKIIAKPGSMCFMSGSVE 60
MAAPFFSTPFQPYVYQSQ+ ++ FQI GG+ QV+Q+M+K EK+ KPG+MC+MSG+++
Sbjct: 1 MAAPFFSTPFQPYVYQSQEGSVTAFQISGGDVQVLQVMVKSQEKLTVKPGTMCYMSGNIQ 60
Query: 61 MENAYLPENEVGIWQWLFGKTVTNIVVRNSGPSDGFVGIAAPYFARILPIDLATFNGEIL 120
+N YLPEN+ G+WQW+FGK++++ V N G DG+VGI+AP+ RILP+DLA F GE+L
Sbjct: 61 TDNNYLPENDGGVWQWIFGKSISSSVFFNPGSDDGYVGISAPFPGRILPMDLANFGGELL 120
Query: 121 CQPDAFLCSVNDVKVSNTVDQRGRNVVAGAEVFLRQKLSGQGLAFILGGGSVVQKILEVG 180
CQ DAFLCSVNDV V++TV+QR RN+ GAEV L+QKL GQG+AF++GGGSV+QKIL
Sbjct: 121 CQADAFLCSVNDVSVTSTVEQRPRNIEIGAEVILKQKLRGQGMAFLVGGGSVMQKILAPR 180
Query: 181 EVLAVDVSCIVAVTSTVDIQIKYNGPARRTMF 212
EV+ VD +CIVA+T+T++ Q+K RR +F
Sbjct: 181 EVITVDAACIVAMTTTINFQLKTPNQPRRVVF 212
>UniRef100_O23585 Hypothetical protein dl4745c [Arabidopsis thaliana]
Length = 270
Score = 246 bits (629), Expect = 2e-64
Identities = 138/236 (58%), Positives = 164/236 (69%), Gaps = 35/236 (14%)
Query: 1 MAAPFFSTPFQPYVYQSQQDAIIPFQILGGESQVVQIMLKPHEKIIAKPGSMCFMSGSVE 60
MAAPFFSTPFQPYVYQSQ+D I PFQILGGE+QVVQIMLKP EK+IAKPGSMC+MSGS+E
Sbjct: 1 MAAPFFSTPFQPYVYQSQEDTITPFQILGGEAQVVQIMLKPQEKVIAKPGSMCYMSGSIE 60
Query: 61 MENAYLPENEVGIWQWLFGKTVTNIVVRNSGPSDGFVGIAAPYFARILPIDLATFNGEIL 120
M+N Y PE EVG+ QW+ GK+V++IV+RN+G +DGFVGIAAP ARILP
Sbjct: 61 MDNTYTPEQEVGVVQWILGKSVSSIVLRNTGQNDGFVGIAAPSLARILP----------- 109
Query: 121 CQPDAFLCSVNDVKVSNTVDQ--RGRNV-VAGAE------------------VFLRQKLS 159
PDAFLCSV+DVKV NTV Q R RN+ AGAE + R +L
Sbjct: 110 --PDAFLCSVHDVKVVNTVYQRHRARNIAAAGAERVVVVGGSETKAFWSGSCFYYRWRLW 167
Query: 160 GQGLAFILGGGSVVQKILEVGEVLAVDVSCIVAVTSTVDIQIKYN-GPARRTMFGV 214
L + VVQK LEVGEVL +DVSCI A+T +++ QIKYN P RR +FG+
Sbjct: 168 PFKLILLHLLHKVVQKNLEVGEVLTIDVSCIAALTPSINFQIKYNAAPVRRAVFGL 223
>UniRef100_Q8TH70 Hypothetical protein MA4652 [Methanosarcina acetivorans]
Length = 263
Score = 128 bits (321), Expect = 1e-28
Identities = 73/200 (36%), Positives = 113/200 (56%), Gaps = 11/200 (5%)
Query: 23 IPFQILGGESQVVQIMLKPHEKIIAKPGSMCFMSGSVEMENAYLPEN-------EVGIWQ 75
I ++I+G + Q+V+I L P E + A+ G+M +M ++M+ + E + G+ +
Sbjct: 5 IDYEIIGNDMQIVEIELDPGEAVQAEAGAMAYMGPGIQMQTSMGSEGGGLLGGLKKGLKR 64
Query: 76 WLFGKT--VTNIVVRNSGPSDGFVGIAAPYFARILPIDLATFNGEILCQPDAFLCSVNDV 133
L G++ +TN + + SG G V AAPY +I+P+DL+ F G ILCQ DAFLC+ V
Sbjct: 65 ALTGESFFITNFIHKGSGK--GHVAFAAPYPGKIIPLDLSKFGGSILCQKDAFLCAARGV 122
Query: 134 KVSNTVDQRGRNVVAGAEVFLRQKLSGQGLAFILGGGSVVQKILEVGEVLAVDVSCIVAV 193
V +R + G E F+ Q+L G G AF+ GG+VV+K L GE VD C+ A
Sbjct: 123 DVEVAFTRRIGTGLFGGEGFILQRLRGNGFAFVHIGGTVVRKDLAPGETYHVDTGCVAAF 182
Query: 194 TSTVDIQIKYNGPARRTMFG 213
T TV+ I ++ + +FG
Sbjct: 183 TETVNYDITWSKDFKNALFG 202
>UniRef100_Q632H7 Hypothetical protein [Bacillus cereus]
Length = 263
Score = 123 bits (309), Expect = 3e-27
Identities = 69/203 (33%), Positives = 109/203 (52%), Gaps = 14/203 (6%)
Query: 23 IPFQILGGESQVVQIMLKPHEKIIAKPGSMCFMSGSVEMENAYLPENEVGIWQWLFGK-- 80
I +++ G + Q V+I L P E +IA+ G+M M +EME + + G LFGK
Sbjct: 9 IEYKLYGDDMQFVEIELDPEESVIAEAGAMMMMEDYIEMETIF--GDGSGPSGGLFGKLM 66
Query: 81 ----------TVTNIVVRNSGPSDGFVGIAAPYFARILPIDLATFNGEILCQPDAFLCSV 130
++ V N+G V AAPY +I+P+DL + G+++CQ DAFLC+
Sbjct: 67 GAGKRLVTGESMFMTVFTNTGHGKRHVSFAAPYPGKIIPVDLTEYQGKVVCQKDAFLCAA 126
Query: 131 NDVKVSNTVDQRGRNVVAGAEVFLRQKLSGQGLAFILGGGSVVQKILEVGEVLAVDVSCI 190
V + ++ G E F+ QKL G GLAF+ GG+V ++ L+ GE L +D C+
Sbjct: 127 KGVSIGIEFTKKIGTGFFGGEGFIMQKLEGDGLAFMHAGGTVYKRELKPGEKLRIDTGCL 186
Query: 191 VAVTSTVDIQIKYNGPARRTMFG 213
VA+T V+ +++ G + +FG
Sbjct: 187 VAMTKDVNYDVEFVGKVKTALFG 209
>UniRef100_Q72YK1 Hypothetical protein [Bacillus cereus]
Length = 260
Score = 122 bits (307), Expect = 5e-27
Identities = 68/203 (33%), Positives = 109/203 (53%), Gaps = 14/203 (6%)
Query: 23 IPFQILGGESQVVQIMLKPHEKIIAKPGSMCFMSGSVEMENAYLPENEVGIWQWLFGK-- 80
I +++ G + Q V+I L P E +IA+ G+M M +EME + + G LFGK
Sbjct: 6 IEYKLYGDDMQFVEIELDPEESVIAEAGAMMMMEDYIEMETIF--GDGSGPSGGLFGKLM 63
Query: 81 ----------TVTNIVVRNSGPSDGFVGIAAPYFARILPIDLATFNGEILCQPDAFLCSV 130
++ V N+G V AAPY +I+P+DL + G+++CQ DAFLC+
Sbjct: 64 GAGKRLVTGESMFMTVFTNTGHGKRHVSFAAPYPGKIIPVDLTEYQGKVVCQKDAFLCAA 123
Query: 131 NDVKVSNTVDQRGRNVVAGAEVFLRQKLSGQGLAFILGGGSVVQKILEVGEVLAVDVSCI 190
V + ++ G E F+ QKL G GLAF+ GG+V ++ L+ GE L +D C+
Sbjct: 124 KGVSIGIEFTKKIGTGFFGGEGFIMQKLEGDGLAFMHAGGTVYKRELKHGEKLRIDTGCL 183
Query: 191 VAVTSTVDIQIKYNGPARRTMFG 213
VA+T ++ +++ G + +FG
Sbjct: 184 VAMTKDINYDVEFVGKVKTALFG 206
>UniRef100_Q816H2 Hypothetical protein [Bacillus cereus]
Length = 260
Score = 122 bits (306), Expect = 7e-27
Identities = 68/203 (33%), Positives = 109/203 (53%), Gaps = 14/203 (6%)
Query: 23 IPFQILGGESQVVQIMLKPHEKIIAKPGSMCFMSGSVEMENAYLPENEVGIWQWLFGK-- 80
I +++ G + Q V+I L P E +IA+ G+M M +EME + + G LFGK
Sbjct: 6 IEYKLYGDDMQFVEIELDPEESVIAEAGAMMMMEDYIEMETIF--GDGSGPSGGLFGKLM 63
Query: 81 ----------TVTNIVVRNSGPSDGFVGIAAPYFARILPIDLATFNGEILCQPDAFLCSV 130
++ V N+G V AAPY +I+P+DL + G+++CQ DAFLC+
Sbjct: 64 GAGKRLVTGESMFMTVFTNTGHGKRHVSFAAPYPGKIIPVDLTEYQGKVVCQKDAFLCAA 123
Query: 131 NDVKVSNTVDQRGRNVVAGAEVFLRQKLSGQGLAFILGGGSVVQKILEVGEVLAVDVSCI 190
V + ++ G E F+ QKL G GLAF+ GG+V ++ L+ GE L +D C+
Sbjct: 124 KGVSLGIEFTKKIGTGFFGGEGFIMQKLEGDGLAFMHAGGTVYKRELKPGEKLRIDTGCL 183
Query: 191 VAVTSTVDIQIKYNGPARRTMFG 213
VA+T ++ +++ G + +FG
Sbjct: 184 VAMTKDINYDVEFVGKVKTALFG 206
>UniRef100_Q97IV3 Uncharacterized conserved protein [Clostridium acetobutylicum]
Length = 260
Score = 122 bits (305), Expect = 9e-27
Identities = 67/203 (33%), Positives = 110/203 (54%), Gaps = 12/203 (5%)
Query: 23 IPFQILGGESQVVQIMLKPHEKIIAKPGSMCFMSGSVEMENAYLPENEVGIWQWLF---- 78
I ++I+G E Q+V++ L P+E ++A+ G+M +M S+EME + ++ G L
Sbjct: 5 IDYKIIGSEMQIVEVELDPYESVVAEAGAMMYMDSSIEMETIFGDGSDKGSSGGLVSKLM 64
Query: 79 --------GKTVTNIVVRNSGPSDGFVGIAAPYFARILPIDLATFNGEILCQPDAFLCSV 130
G+++ + N G V AAPY +I+P+DL++FN ++CQ D FLC+
Sbjct: 65 GAGKRLVTGESLFMTIFTNRGVGKQKVAFAAPYPGKIIPMDLSSFNNYMICQKDCFLCAA 124
Query: 131 NDVKVSNTVDQRGRNVVAGAEVFLRQKLSGQGLAFILGGGSVVQKILEVGEVLAVDVSCI 190
V + ++ + G E F+ QKL G GL F+ GG++VQ+ L EV+ VD C+
Sbjct: 125 KGVSIGVEFTRKIGVGLFGGEGFMLQKLEGDGLTFVHSGGTIVQRELLPKEVIKVDTGCL 184
Query: 191 VAVTSTVDIQIKYNGPARRTMFG 213
VA T V+ I+ + +FG
Sbjct: 185 VAFTRDVNYDIEMVKGIKSAIFG 207
>UniRef100_Q8GCS8 Hypothetical protein [Eubacterium acidaminophilum]
Length = 268
Score = 120 bits (301), Expect = 2e-26
Identities = 68/203 (33%), Positives = 105/203 (51%), Gaps = 12/203 (5%)
Query: 23 IPFQILGGESQVVQIMLKPHEKIIAKPGSMCFMSGSVEMENAYLPENEVGIWQWLFGK-- 80
I + + G + Q+V+I L P E +IA+ G+M +M ++ME + G L GK
Sbjct: 7 IDYTLHGDDLQLVEIELDPGESVIAEAGAMLYMENGIQMEAVLGDASGKGEGTGLMGKLL 66
Query: 81 ----------TVTNIVVRNSGPSDGFVGIAAPYFARILPIDLATFNGEILCQPDAFLCSV 130
++ + N G V AAPY +I+P DL + G I+CQ DAFLC+
Sbjct: 67 GAGKRVIMGESLFMTLFTNKGSQKQKVAFAAPYPGKIVPFDLNAYGGRIICQKDAFLCAA 126
Query: 131 NDVKVSNTVDQRGRNVVAGAEVFLRQKLSGQGLAFILGGGSVVQKILEVGEVLAVDVSCI 190
+ V ++ + G E F+ ++L G G AF+ GG++V+K L E+L VD C+
Sbjct: 127 KGISVQMEFQKKIGVGLFGGEGFIMERLEGDGFAFLHAGGAIVEKELSQAELLKVDTGCL 186
Query: 191 VAVTSTVDIQIKYNGPARRTMFG 213
VA TS VD I++ G + +FG
Sbjct: 187 VAFTSGVDYDIQFMGDIKSAIFG 209
>UniRef100_Q8PXB6 HTH DNA-binding protein [Methanosarcina mazei]
Length = 250
Score = 114 bits (284), Expect = 2e-24
Identities = 68/190 (35%), Positives = 104/190 (53%), Gaps = 11/190 (5%)
Query: 33 QVVQIMLKPHEKIIAKPGSMCFMSGSVEMENAYLPEN-------EVGIWQWLFGKT--VT 83
Q+V+I L P E + A+ G+M +M + M+ E + G+ + L G++ +T
Sbjct: 2 QIVEIELDPGEAVQAEAGAMAYMGPGILMQTGMGNEGGGLFGGLKKGLKRALTGESFFIT 61
Query: 84 NIVVRNSGPSDGFVGIAAPYFARILPIDLATFNGEILCQPDAFLCSVNDVKVSNTVDQRG 143
+ + + SG G V AAPY +ILP+DL+ F G ILCQ DAFLC+ ++V ++
Sbjct: 62 SFIHKGSGK--GHVAFAAPYPGKILPLDLSKFGGSILCQKDAFLCAAKGIEVELAFTRKL 119
Query: 144 RNVVAGAEVFLRQKLSGQGLAFILGGGSVVQKILEVGEVLAVDVSCIVAVTSTVDIQIKY 203
+ G E F+ Q+L G GLAFI GG+V++K L GE VD C+ A T V I +
Sbjct: 120 GAGLFGGEGFILQRLRGDGLAFIHIGGTVIRKDLAPGETYKVDTGCVAAFTENVTYDITW 179
Query: 204 NGPARRTMFG 213
+ + +FG
Sbjct: 180 SRDFKNALFG 189
>UniRef100_Q888R9 Hypothetical protein [Pseudomonas syringae]
Length = 244
Score = 107 bits (268), Expect = 2e-22
Identities = 65/197 (32%), Positives = 100/197 (49%), Gaps = 10/197 (5%)
Query: 25 FQILGGESQVVQIMLKPHEKIIAKPGSMCFMSGSVEMENAYLP--ENEVGIWQWLFG--- 79
F++ G E+Q V++ L P E +A+ G+M + + V+ME + G+ LFG
Sbjct: 3 FKLYGAETQFVELELDPGESAVAEAGAMMYKTCDVQMETIFGDGSNQSSGLLGSLFGAGK 62
Query: 80 -----KTVTNIVVRNSGPSDGFVGIAAPYFARILPIDLATFNGEILCQPDAFLCSVNDVK 134
+++ V G G V AAPY ILP++L F G+++CQ D+FL V
Sbjct: 63 RMLTGESLFTTVFSQQGSGKGRVAFAAPYPGTILPLNLRDFGGKLICQKDSFLAGAKGVS 122
Query: 135 VSNTVDQRGRNVVAGAEVFLRQKLSGQGLAFILGGGSVVQKILEVGEVLAVDVSCIVAVT 194
+ ++ + G E F+ QKL G G F+ GG+V + L GE L VD C+ A+T
Sbjct: 123 IGIQFQKKILTGLFGGEGFVLQKLEGDGWVFVHMGGTVRKIELAAGEALDVDTGCLAAMT 182
Query: 195 STVDIQIKYNGPARRTM 211
TVD I+ G ++M
Sbjct: 183 QTVDYDIRMVGGGIKSM 199
>UniRef100_UPI000032E9E7 UPI000032E9E7 UniRef100 entry
Length = 264
Score = 106 bits (265), Expect = 4e-22
Identities = 67/201 (33%), Positives = 105/201 (51%), Gaps = 11/201 (5%)
Query: 23 IPFQILGGESQVVQIMLKPHEKIIAKPGSMCFMSGSVEMENAYLP--ENEVGIW------ 74
+ ++I+G Q+V++ L P E +IA+ G+M ++ + E E E G +
Sbjct: 8 VDYEIIGHSMQLVEVELDPQETVIAEAGAMNYLEQDISFEAKMGDGSEPESGFFGKLMGA 67
Query: 75 --QWLFGKTVTNIVVRNSGPSDGFVGIAAPYFARILPIDLATFNGEILCQPDAFLCSVND 132
+ L G+++ N G V AAPY IL IDLA + GE++CQ D+FL +
Sbjct: 68 GKRVLTGESIFMTHFTNLGHQKRKVAFAAPYPGTILAIDLAQYGGELICQKDSFLAAALG 127
Query: 133 VKVSNTVDQRGRNVVAGAEVFLRQKLSGQGLAFILGGGSVVQKILEVGEVLAVDVSCIVA 192
+VS ++R G E F+ Q L G G+AFI GG++++K L+ GE L VD C+V
Sbjct: 128 TRVSMKFNRRLGTGFFGGEGFILQSLLGDGMAFIHAGGTLIKKELK-GETLRVDTGCLVG 186
Query: 193 VTSTVDIQIKYNGPARRTMFG 213
T +D I+ G + +FG
Sbjct: 187 FTPGIDYDIERAGSLKSMVFG 207
>UniRef100_Q9HXU4 Hypothetical protein [Pseudomonas aeruginosa]
Length = 248
Score = 106 bits (265), Expect = 4e-22
Identities = 65/200 (32%), Positives = 102/200 (50%), Gaps = 11/200 (5%)
Query: 23 IPFQILGGESQVVQIMLKPHEKIIAKPGSMCFMSGSVEMENAYLPENEVG-----IW--- 74
+ ++ILG Q V+I L P E +IA+ G+M +M+G + A + + G +W
Sbjct: 6 LDYRILGESMQTVEIELDPGETVIAEAGAMNYMTGDIRF-TARMGDGSDGSLLGKLWSAG 64
Query: 75 -QWLFGKTVTNIVVRNSGPSDGFVGIAAPYFARILPIDLATFNGEILCQPDAFLCSVNDV 133
+ L G++V N G V AAPY ++ +DL G + CQ D+FLC+
Sbjct: 65 KRKLGGESVFMTHFTNEGQGKQHVAFAAPYPGSVVAVDLDDVGGRLFCQKDSFLCAAYGT 124
Query: 134 KVSNTVDQRGRNVVAGAEVFLRQKLSGQGLAFILGGGSVVQKILEVGEVLAVDVSCIVAV 193
+V +R G E F+ QKL G GL F+ GG+++++ L GE L VD C+VA
Sbjct: 125 RVGIAFTKRLGAGFFGGEGFILQKLEGDGLVFVHAGGTLIRRQLN-GETLRVDTGCLVAF 183
Query: 194 TSTVDIQIKYNGPARRTMFG 213
T +D ++ G + +FG
Sbjct: 184 TDGIDYDVQLAGGLKSMLFG 203
>UniRef100_UPI00002F37B2 UPI00002F37B2 UniRef100 entry
Length = 264
Score = 106 bits (264), Expect = 5e-22
Identities = 66/201 (32%), Positives = 105/201 (51%), Gaps = 11/201 (5%)
Query: 23 IPFQILGGESQVVQIMLKPHEKIIAKPGSMCFMSGSVEMENAYLP--ENEVGIW------ 74
+ ++I+G Q+V++ L P+E +IA+ G+M ++ + E E + G +
Sbjct: 8 VDYEIIGHSMQLVEVELDPNETVIAEAGAMNYLEQDISFEAKMGDGSEPDSGFFGKLMGA 67
Query: 75 --QWLFGKTVTNIVVRNSGPSDGFVGIAAPYFARILPIDLATFNGEILCQPDAFLCSVND 132
+ L G+++ N G V AAPY IL IDLA + GE++CQ D+FL +
Sbjct: 68 GKRALTGESIFMTHFTNLGHQKRKVAFAAPYPGTILAIDLAQYGGELICQKDSFLAAALG 127
Query: 133 VKVSNTVDQRGRNVVAGAEVFLRQKLSGQGLAFILGGGSVVQKILEVGEVLAVDVSCIVA 192
+VS ++R G E F+ Q L G G+AFI GG++++K L+ GE L VD C+V
Sbjct: 128 TRVSMKFNRRLGTGFFGGEGFILQSLQGDGMAFIHAGGTLIKKELK-GETLRVDTGCLVG 186
Query: 193 VTSTVDIQIKYNGPARRTMFG 213
T +D I+ G + FG
Sbjct: 187 FTPGIDYDIERAGSLKSMFFG 207
>UniRef100_Q8EFK5 Hypothetical protein SO1966 [Shewanella oneidensis]
Length = 264
Score = 106 bits (264), Expect = 5e-22
Identities = 67/201 (33%), Positives = 105/201 (51%), Gaps = 11/201 (5%)
Query: 23 IPFQILGGESQVVQIMLKPHEKIIAKPGSMCFMSGSVEMENAYLP--ENEVGIW------ 74
+ ++I+G Q+V++ L P+E +IA+ G+M ++ + E E + G +
Sbjct: 8 VDYEIIGHSMQLVEVELDPNETVIAEAGAMNYLEQDISFEAKMGDGSEPDSGFFGKLMGA 67
Query: 75 --QWLFGKTVTNIVVRNSGPSDGFVGIAAPYFARILPIDLATFNGEILCQPDAFLCSVND 132
+ L G+++ N G V AAPY IL IDLA + GE++CQ D+FL +
Sbjct: 68 GKRALTGESIFMTHFTNLGHQKCKVAFAAPYPGTILAIDLAQYGGELICQKDSFLAAALG 127
Query: 133 VKVSNTVDQRGRNVVAGAEVFLRQKLSGQGLAFILGGGSVVQKILEVGEVLAVDVSCIVA 192
+VS ++R G E F+ Q L G G+AFI GG++++K L+ GE L VD C+V
Sbjct: 128 TRVSMKFNRRLGTGFFGGEGFILQSLQGDGMAFIHAGGTLIKKELK-GETLRVDTGCLVG 186
Query: 193 VTSTVDIQIKYNGPARRTMFG 213
T VD I+ G + FG
Sbjct: 187 FTPGVDYDIERAGSLKSMFFG 207
>UniRef100_Q81K89 Hypothetical protein [Bacillus anthracis]
Length = 226
Score = 102 bits (255), Expect = 5e-21
Identities = 49/135 (36%), Positives = 79/135 (58%)
Query: 79 GKTVTNIVVRNSGPSDGFVGIAAPYFARILPIDLATFNGEILCQPDAFLCSVNDVKVSNT 138
G+++ V N+G V AAPY +I+P+DL + G+++CQ DAFLC+ V +
Sbjct: 38 GESMFMTVFTNTGHGKRHVSFAAPYPGKIIPVDLTEYQGKVVCQKDAFLCAAKGVSIGIE 97
Query: 139 VDQRGRNVVAGAEVFLRQKLSGQGLAFILGGGSVVQKILEVGEVLAVDVSCIVAVTSTVD 198
++ G E F+ QKL G GLAF+ GG+V ++ L+ GE L +D C+VA+T V+
Sbjct: 98 FTKKIGTGFFGGEGFIMQKLEGDGLAFMHAGGTVYKRELKPGEKLRIDTGCLVAMTKDVN 157
Query: 199 IQIKYNGPARRTMFG 213
+++ G + +FG
Sbjct: 158 YDVEFVGKVKTALFG 172
>UniRef100_UPI00003CBDBB UPI00003CBDBB UniRef100 entry
Length = 226
Score = 102 bits (254), Expect = 7e-21
Identities = 48/135 (35%), Positives = 79/135 (57%)
Query: 79 GKTVTNIVVRNSGPSDGFVGIAAPYFARILPIDLATFNGEILCQPDAFLCSVNDVKVSNT 138
G+++ V N+G V AAPY +I+P+DL + G+++CQ DAFLC+ V +
Sbjct: 38 GESMFMTVFTNTGHGKRHVSFAAPYPGKIIPVDLTEYQGKVVCQKDAFLCAAKGVSIGIE 97
Query: 139 VDQRGRNVVAGAEVFLRQKLSGQGLAFILGGGSVVQKILEVGEVLAVDVSCIVAVTSTVD 198
++ G E F+ QKL G GLAF+ GG+V ++ L+ GE L +D C+VA+T ++
Sbjct: 98 FTKKIGTGFFGGEGFIMQKLEGDGLAFMHAGGTVYKRELKPGEKLRIDTGCLVAMTKDIN 157
Query: 199 IQIKYNGPARRTMFG 213
+++ G + +FG
Sbjct: 158 YDVEFVGKVKTALFG 172
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.322 0.139 0.412
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 356,126,333
Number of Sequences: 2790947
Number of extensions: 14078432
Number of successful extensions: 26739
Number of sequences better than 10.0: 58
Number of HSP's better than 10.0 without gapping: 37
Number of HSP's successfully gapped in prelim test: 21
Number of HSP's that attempted gapping in prelim test: 26656
Number of HSP's gapped (non-prelim): 59
length of query: 218
length of database: 848,049,833
effective HSP length: 122
effective length of query: 96
effective length of database: 507,554,299
effective search space: 48725212704
effective search space used: 48725212704
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 72 (32.3 bits)
Medicago: description of AC119408.7


