

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC149600.13 + phase: 0 /pseudo
(232 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
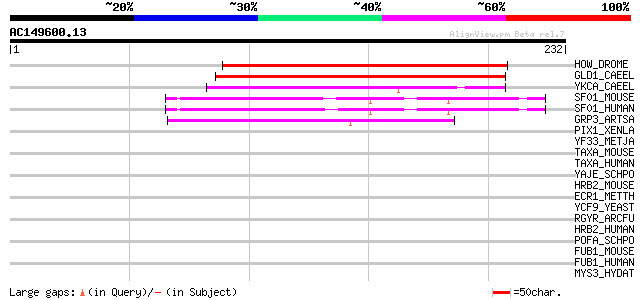
Score E
Sequences producing significant alignments: (bits) Value
HOW_DROME (O01367) Held out wings protein (KH-domain protein KH9... 120 3e-27
GLD1_CAEEL (Q17339) Female germline-specific tumor suppressor gl... 118 1e-26
YKCA_CAEEL (P42083) Hypothetical protein B0280.11 in chromosome III 87 3e-17
SF01_MOUSE (Q64213) Splicing factor 1 (Zinc finger protein 162) ... 86 6e-17
SF01_HUMAN (Q15637) Splicing factor 1 (Zinc finger protein 162) ... 86 6e-17
GRP3_ARTSA (P13230) Glycine-rich protein GRP33 70 6e-12
PIX1_XENLA (Q9W751) Pituitary homeobox 1 (X-PITX-1) (xPitx1) 35 0.15
YF33_METJA (Q58928) Hypothetical protein MJ1533 34 0.26
TAXA_MOUSE (Q6PAM1) Alpha-taxilin 34 0.33
TAXA_HUMAN (P40222) Alpha-taxilin 34 0.33
YAJE_SCHPO (Q09911) Hypothetical protein C30D11.14c in chromosome I 33 0.75
HRB2_MOUSE (Q8BGA5) HIV-1 Rev binding protein 2 homolog 32 0.97
ECR1_METTH (O26780) Probable exosome complex RNA-binding protein 1 32 0.97
YCF9_YEAST (P25586) Hypothetical 37.2 kDa protein in CHA1-PRD1 i... 32 1.3
RGYR_ARCFU (O29238) Reverse gyrase [Includes: Helicase (EC 3.6.1... 32 1.3
HRB2_HUMAN (Q13601) HIV-1 Rev binding protein 2 (Rev interacting... 32 1.3
POFA_SCHPO (Q9P7W4) F-box/WD-repeat protein pof10 (Skp1-binding ... 31 2.2
FUB1_MOUSE (Q91WJ8) Far upstream element binding protein 1 (FUSE... 31 2.2
FUB1_HUMAN (Q96AE4) Far upstream element binding protein 1 (FUSE... 31 2.2
MYS3_HYDAT (P39922) Myosin heavy chain, clone 203 (Fragment) 31 2.8
>HOW_DROME (O01367) Held out wings protein (KH-domain protein KH93F)
(Putative RNA-binding protein) (Muscle-specific protein)
(Wings held out protein) (Struthio protein)
(Quaking-related 93F)
Length = 405
Score = 120 bits (301), Expect = 3e-27
Identities = 55/119 (46%), Positives = 84/119 (70%)
Query: 90 IPHDNHPTFNFVGRLLGPRGNSLKRVEATTGCRVFIRGKGSIKDFDKEELLRGRPGFEHL 149
+P HP FNFVGR+LGPRG + K++E TGC++ +RGKGS++D KE+ RG+P +EHL
Sbjct: 143 VPVREHPDFNFVGRILGPRGMTAKQLEQETGCKIMVRGKGSMRDKKKEDANRGKPNWEHL 202
Query: 150 NEPLHILIEAELPVNVVDLRLRQAQEIIEELLKPVDESQDIYKRQQLRELAMLNSSFRE 208
++ LH+LI E N ++L QA +++LL P E +D K++QL ELA++N ++R+
Sbjct: 203 SDDLHVLITVEDTENRATVKLAQAVAEVQKLLVPQAEGEDELKKRQLMELAIINGTYRD 261
>GLD1_CAEEL (Q17339) Female germline-specific tumor suppressor gld-1
(Defective in germ line development protein 1)
Length = 463
Score = 118 bits (296), Expect = 1e-26
Identities = 53/121 (43%), Positives = 85/121 (69%)
Query: 87 RLDIPHDNHPTFNFVGRLLGPRGNSLKRVEATTGCRVFIRGKGSIKDFDKEELLRGRPGF 146
++ +P + +P +NFVGR+LGPRG + K++E TGC++ +RGKGS++D KE RG+ +
Sbjct: 208 KIYVPKNEYPDYNFVGRILGPRGMTAKQLEQDTGCKIMVRGKGSMRDKSKESAHRGKANW 267
Query: 147 EHLNEPLHILIEAELPVNVVDLRLRQAQEIIEELLKPVDESQDIYKRQQLRELAMLNSSF 206
EHL + LH+L++ E N V ++L+ A E +++LL P E D KR+QL ELA++N ++
Sbjct: 268 EHLEDDLHVLVQCEDTENRVHIKLQAALEQVKKLLIPAPEGTDELKRKQLMELAIINGTY 327
Query: 207 R 207
R
Sbjct: 328 R 328
>YKCA_CAEEL (P42083) Hypothetical protein B0280.11 in chromosome III
Length = 634
Score = 87.0 bits (214), Expect = 3e-17
Identities = 46/126 (36%), Positives = 75/126 (59%), Gaps = 4/126 (3%)
Query: 83 KKMLRLDIPHDNHPTFNFVGRLLGPRGNSLKRVEATTGCRVFIRGKGSIKDFDKEELLRG 142
+K+ ++ P + N VGRL+GPRG +++++E GC++FIRGKG KD KEE LR
Sbjct: 512 EKIDKVFFPPETANNTNPVGRLIGPRGMTIRQLEKDLGCKLFIRGKGCTKDDAKEERLRE 571
Query: 143 RPGFEHLNEPLHILIEAEL-PVNVVDLRLRQAQEIIEELLKPVDESQDIYKRQQLRELAM 201
R G+EHL EP+H++I +L +++++E L+ D KR QL +LA+
Sbjct: 572 RVGWEHLKEPIHVMISVRSDSEEAASEKLSSIKKMLQEFLEHTDSE---LKRSQLMQLAV 628
Query: 202 LNSSFR 207
+ + +
Sbjct: 629 IEGTLK 634
>SF01_MOUSE (Q64213) Splicing factor 1 (Zinc finger protein 162)
(Transcription factor ZFM1) (mZFM) (Zinc finger gene in
MEN1 locus) (Mammalian branch point binding protein
mBBP) (BBP) (CW17)
Length = 653
Score = 86.3 bits (212), Expect = 6e-17
Identities = 62/165 (37%), Positives = 89/165 (53%), Gaps = 20/165 (12%)
Query: 66 LNVDWQRAPAISNSHIVKKMLRLDIPHDNHPTFNFVGRLLGPRGNSLKRVEATTGCRVFI 125
LN D+ + PA + ++ IP D +P NFVG L+GPRGN+LK +E ++ I
Sbjct: 119 LNPDF-KPPADYKPPATRVSDKVMIPQDEYPEINFVGLLIGPRGNTLKNIEKECNAKIMI 177
Query: 126 RGKGSIKDFDKEELLRGRPGFEHL---NEPLHILIEAELPVNVVDLRLRQAQEIIEELLK 182
RGKGS+ EE GR + L +EPLH L+ A NV ++A E I +LK
Sbjct: 178 RGKGSV-----EEGKVGRKDGQMLPGEDEPLHALVTANTMENV-----KKAVEQIRNILK 227
Query: 183 ---PVDESQDIYKRQQLRELAMLNSSFREESPQLSGSLSPFTSNE 224
E Q+ ++ QLRELA LN + RE+ ++ L P+ S+E
Sbjct: 228 QGIETPEDQNDLRKMQLRELARLNGTLREDDNRI---LRPWQSSE 269
>SF01_HUMAN (Q15637) Splicing factor 1 (Zinc finger protein 162)
(Transcription factor ZFM1) (Zinc finger gene in MEN1
locus) (Mammalian branch point binding protein mBBP)
(BBP)
Length = 639
Score = 86.3 bits (212), Expect = 6e-17
Identities = 62/165 (37%), Positives = 89/165 (53%), Gaps = 20/165 (12%)
Query: 66 LNVDWQRAPAISNSHIVKKMLRLDIPHDNHPTFNFVGRLLGPRGNSLKRVEATTGCRVFI 125
LN D+ + PA + ++ IP D +P NFVG L+GPRGN+LK +E ++ I
Sbjct: 119 LNPDF-KPPADYKPPATRVSDKVMIPQDEYPEINFVGLLIGPRGNTLKNIEKECNAKIMI 177
Query: 126 RGKGSIKDFDKEELLRGRPGFEHL---NEPLHILIEAELPVNVVDLRLRQAQEIIEELLK 182
RGKGS+K E GR + L +EPLH L+ A NV ++A E I +LK
Sbjct: 178 RGKGSVK-----EGKVGRKDGQMLPGEDEPLHALVTANTMENV-----KKAVEQIRNILK 227
Query: 183 ---PVDESQDIYKRQQLRELAMLNSSFREESPQLSGSLSPFTSNE 224
E Q+ ++ QLRELA LN + RE+ ++ L P+ S+E
Sbjct: 228 QGIETPEDQNDLRKMQLRELARLNGTLREDDNRI---LRPWQSSE 269
>GRP3_ARTSA (P13230) Glycine-rich protein GRP33
Length = 308
Score = 69.7 bits (169), Expect = 6e-12
Identities = 42/122 (34%), Positives = 68/122 (55%), Gaps = 2/122 (1%)
Query: 67 NVDWQRAPAISNSHIVKKMLRLDIPHDNHPTFNFVGRLLGPRGNSLKRVEATTGCRVFIR 126
N D + N VK + R +P D P +NF+G+LLGP G+++K+++ T ++ I
Sbjct: 57 NTDGSGFMDLYNDTKVKLVSRCCLPVDQFPKYNFLGKLLGPGGSTMKQLQDETMTKISIL 116
Query: 127 GKGSIKDFDKEELLR--GRPGFEHLNEPLHILIEAELPVNVVDLRLRQAQEIIEELLKPV 184
G+GS++D +KEE LR G + HLNE LHI I + R+ A I++ + P
Sbjct: 117 GRGSMRDRNKEEELRNSGDVKYAHLNEQLHIEIISIASPAEAHARMAYALTEIKKYITPE 176
Query: 185 DE 186
++
Sbjct: 177 ED 178
>PIX1_XENLA (Q9W751) Pituitary homeobox 1 (X-PITX-1) (xPitx1)
Length = 305
Score = 35.0 bits (79), Expect = 0.15
Identities = 32/106 (30%), Positives = 49/106 (46%), Gaps = 12/106 (11%)
Query: 22 SGLMQNQGFSDYDRVQFG------STKPSLMPSLDTTSSFTGWNSLSHEGLNVDWQRAPA 75
SGLMQ YD + G +TK SL P+ +T SFT +NS+S + +
Sbjct: 154 SGLMQ-----PYDEMYAGYPYNNWATK-SLTPAPLSTKSFTFFNSMSPLSSQSMFSGPSS 207
Query: 76 ISNSHIVKKMLRLDIPHDNHPTFNFVGRLLGPRGNSLKRVEATTGC 121
IS+ + M +P + + N + L G+SL ++TGC
Sbjct: 208 ISSMSMPSSMGHSAVPGMANSSLNNINNLNNISGSSLNSAMSSTGC 253
>YF33_METJA (Q58928) Hypothetical protein MJ1533
Length = 642
Score = 34.3 bits (77), Expect = 0.26
Identities = 23/118 (19%), Positives = 51/118 (42%), Gaps = 26/118 (22%)
Query: 100 FVGRLLGPRGNSLKRVEATTGCRVFIRGKGSIKDFDKEELLRGRP--------------- 144
++G ++G G + ++E G ++ ++ K ++ D E + R
Sbjct: 521 YIGAIIGKGGKEISKLEDMLGLKISVKEKEKEEEKDMERIYRKYEYVNELESTRIYETDK 580
Query: 145 ------GFEHLNEPLHILIEAELPVNVV-----DLRLRQAQEIIEELLKPVDESQDIY 191
G + E + I I+ +L V +R+ + ++ +E+L+ +DE +DIY
Sbjct: 581 YVVVDVGEDFAGENIRIYIDGKLLTTVTVRNDGTVRINKKTKVGKEILEAIDEGRDIY 638
>TAXA_MOUSE (Q6PAM1) Alpha-taxilin
Length = 554
Score = 33.9 bits (76), Expect = 0.33
Identities = 32/103 (31%), Positives = 51/103 (49%), Gaps = 22/103 (21%)
Query: 147 EHLNEPLHILIEAELPVNVVDLRLRQAQEIIEE-----------LLKPVDESQ---DIYK 192
EH+++ + +L +VD +L+QAQE+++E LLK ESQ ++ K
Sbjct: 321 EHIDK---VFKHKDLQQQLVDAKLQQAQEMLKEAEERHQREKEFLLKEAVESQRMCELMK 377
Query: 193 RQQL---RELAMLNSSFREESPQLSGSLSPFTS--NEMIKRAK 230
+Q+ ++LA+ F E LS S FT+ EM K K
Sbjct: 378 QQETHLKQQLALYTEKFEEFQNTLSKSSEVFTTFKQEMEKMTK 420
>TAXA_HUMAN (P40222) Alpha-taxilin
Length = 546
Score = 33.9 bits (76), Expect = 0.33
Identities = 32/103 (31%), Positives = 51/103 (49%), Gaps = 22/103 (21%)
Query: 147 EHLNEPLHILIEAELPVNVVDLRLRQAQEIIEE-----------LLKPVDESQ---DIYK 192
EH+++ + +L +VD +L+QAQE+++E LLK ESQ ++ K
Sbjct: 321 EHIDK---VFKHKDLQQQLVDAKLQQAQEMLKEAEERHQREKDFLLKEAVESQRMCELMK 377
Query: 193 RQQL---RELAMLNSSFREESPQLSGSLSPFTS--NEMIKRAK 230
+Q+ ++LA+ F E LS S FT+ EM K K
Sbjct: 378 QQETHLKQQLALYTEKFEEFQNTLSKSSEVFTTFKQEMEKMTK 420
>YAJE_SCHPO (Q09911) Hypothetical protein C30D11.14c in chromosome I
Length = 534
Score = 32.7 bits (73), Expect = 0.75
Identities = 25/97 (25%), Positives = 48/97 (48%), Gaps = 13/97 (13%)
Query: 98 FNFVGRLLGPRGNSLKRVEATTGCRVFIRGKGSIKDFDKEELLRGRPGFEHLNEPLHILI 157
F+ ++GP+G +K ++ T RV I+G+GS F + R +EP+H+ I
Sbjct: 303 FHLRQAIVGPQGAYVKHIQQETRTRVQIKGQGSA--FIEPSTNR------ESDEPIHLCI 354
Query: 158 EAELPVNVVDLRLRQAQEIIEELLKPVDESQDIYKRQ 194
+ P +++A+ + E+L+ V + +K Q
Sbjct: 355 MSHDP-----NAIQRAKVLCEDLIASVHQQYKAWKSQ 386
>HRB2_MOUSE (Q8BGA5) HIV-1 Rev binding protein 2 homolog
Length = 380
Score = 32.3 bits (72), Expect = 0.97
Identities = 12/25 (48%), Positives = 19/25 (76%)
Query: 103 RLLGPRGNSLKRVEATTGCRVFIRG 127
RL+GP+G++LK +E T C V ++G
Sbjct: 162 RLIGPKGSTLKALELLTNCYVMVQG 186
>ECR1_METTH (O26780) Probable exosome complex RNA-binding protein 1
Length = 311
Score = 32.3 bits (72), Expect = 0.97
Identities = 18/72 (25%), Positives = 36/72 (50%), Gaps = 7/72 (9%)
Query: 101 VGRLLGPRGNSLKRVEATTGCRVFIRGKGSIKDFDKEELLRGRPGFEHLNEPLHILIEAE 160
V RL+G RG+ + V+ T C + + G + ++G P E + E + ++I+ E
Sbjct: 151 VPRLIGKRGSMINMVKEKTHCDIVVGQNGVV-------WIKGEPDMERIAEKVVLMIDRE 203
Query: 161 LPVNVVDLRLRQ 172
+ + R+R+
Sbjct: 204 AHTSGLTDRVRE 215
>YCF9_YEAST (P25586) Hypothetical 37.2 kDa protein in CHA1-PRD1
intergenic region
Length = 316
Score = 32.0 bits (71), Expect = 1.3
Identities = 12/25 (48%), Positives = 18/25 (72%)
Query: 103 RLLGPRGNSLKRVEATTGCRVFIRG 127
RL+GP GN+LK +E T C + ++G
Sbjct: 144 RLVGPNGNTLKALELLTKCYILVQG 168
>RGYR_ARCFU (O29238) Reverse gyrase [Includes: Helicase (EC
3.6.1.-); Topoisomerase (EC 5.99.1.3)]
Length = 1054
Score = 32.0 bits (71), Expect = 1.3
Identities = 30/108 (27%), Positives = 50/108 (45%), Gaps = 6/108 (5%)
Query: 122 RVFIRGKGSIKDFDKEELLRGRPGF-EHLNEPLHILIEAELPVNVVDLRLRQAQEI-IEE 179
R +I+G G L +G E +E L IE + D+ + E+ E+
Sbjct: 427 RTYIQGSGRTSRLFAGGLTKGASFLLEDDSELLSAFIER---AKLYDIEFKSIDEVDFEK 483
Query: 180 LLKPVDESQDIYKRQQLRELAMLNSSFREESPQLSGSLSPFTSNEMIK 227
L + +DES+D Y+R+Q +L + + F ESP + +S F +K
Sbjct: 484 LSRELDESRDRYRRRQEFDL-IKPALFIVESPTKARQISRFFGKPSVK 530
>HRB2_HUMAN (Q13601) HIV-1 Rev binding protein 2 (Rev interacting
protein 1) (Rip-1)
Length = 381
Score = 32.0 bits (71), Expect = 1.3
Identities = 11/25 (44%), Positives = 19/25 (76%)
Query: 103 RLLGPRGNSLKRVEATTGCRVFIRG 127
RL+GP+G++LK +E T C + ++G
Sbjct: 163 RLIGPKGSTLKALELLTNCYIMVQG 187
>POFA_SCHPO (Q9P7W4) F-box/WD-repeat protein pof10 (Skp1-binding
protein 2)
Length = 662
Score = 31.2 bits (69), Expect = 2.2
Identities = 25/97 (25%), Positives = 44/97 (44%), Gaps = 2/97 (2%)
Query: 135 DKEELLRGRPGFE-HLNEPLHILIEAELPVNVVDLRLRQAQEIIEELLKPVDESQDIYKR 193
++ E + R FE H E L L E E+ V L + + + +L VD+ ++ +K
Sbjct: 558 ERREKMEARQKFEQHFGEGLVGLSEEEIIAYVTMLSQEEEAKRMVQLSMDVDKIEEDFKE 617
Query: 194 QQLRELAMLNS-SFREESPQLSGSLSPFTSNEMIKRA 229
+ + LN+ S E PQ +++ E I+ A
Sbjct: 618 NDEQATSSLNALSSNHEPPQEQANVAELNEQEQIELA 654
>FUB1_MOUSE (Q91WJ8) Far upstream element binding protein 1 (FUSE
binding protein 1) (FBP)
Length = 651
Score = 31.2 bits (69), Expect = 2.2
Identities = 31/141 (21%), Positives = 56/141 (38%), Gaps = 40/141 (28%)
Query: 101 VGRLLGPRGNSLKRVEATTGCRVFIRGKG------------------SIKDFDKEELLRG 142
VG ++G G + R++ +GC++ I S K + + +G
Sbjct: 108 VGFIIGRGGEQISRIQQESGCKIQIAPDSGGLPERSCMLTGTPESVQSAKRLLDQIVEKG 167
Query: 143 RP--GFEHLNEPLHILIEAELPVNVVDLRLRQAQEIIEELL------------------- 181
RP GF H + P + + E +P + L + + E I++L
Sbjct: 168 RPAPGFHHGDGPGNAVQEIMIPASKAGLVIGKGGETIKQLQERAGVKMVMIQDGPQNTGA 227
Query: 182 -KPVDESQDIYKRQQLRELAM 201
KP+ + D YK QQ +E+ +
Sbjct: 228 DKPLRITGDPYKVQQAKEMVL 248
>FUB1_HUMAN (Q96AE4) Far upstream element binding protein 1 (FUSE
binding protein 1) (FBP) (DNA helicase V) (HDH V)
Length = 643
Score = 31.2 bits (69), Expect = 2.2
Identities = 31/141 (21%), Positives = 56/141 (38%), Gaps = 40/141 (28%)
Query: 101 VGRLLGPRGNSLKRVEATTGCRVFIRGKG------------------SIKDFDKEELLRG 142
VG ++G G + R++ +GC++ I S K + + +G
Sbjct: 111 VGFIIGRGGEQISRIQQESGCKIQIAPDSGGLPERSCMLTGTPESVQSAKRLLDQIVEKG 170
Query: 143 RP--GFEHLNEPLHILIEAELPVNVVDLRLRQAQEIIEELL------------------- 181
RP GF H + P + + E +P + L + + E I++L
Sbjct: 171 RPAPGFHHGDGPGNAVQEIMIPASKAGLVIGKGGETIKQLQERAGVKMVMIQDGPQNTGA 230
Query: 182 -KPVDESQDIYKRQQLRELAM 201
KP+ + D YK QQ +E+ +
Sbjct: 231 DKPLRITGDPYKVQQAKEMVL 251
>MYS3_HYDAT (P39922) Myosin heavy chain, clone 203 (Fragment)
Length = 539
Score = 30.8 bits (68), Expect = 2.8
Identities = 20/72 (27%), Positives = 39/72 (53%), Gaps = 6/72 (8%)
Query: 149 LNEPLHILIEAELPVNVVDLRLRQAQEIIEELLKPVDESQ------DIYKRQQLRELAML 202
LNE L L E V V++ ++++A+E I+EL + +E Q + K+ + +++ L
Sbjct: 122 LNEALEKLDGEEHSVLVLEEKIQEAEEKIDELTEKTEELQSNISRLETEKQNRDKQIDTL 181
Query: 203 NSSFREESPQLS 214
N R++ +S
Sbjct: 182 NEDIRKQDETIS 193
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.317 0.135 0.389
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 27,479,956
Number of Sequences: 164201
Number of extensions: 1204769
Number of successful extensions: 3019
Number of sequences better than 10.0: 30
Number of HSP's better than 10.0 without gapping: 15
Number of HSP's successfully gapped in prelim test: 15
Number of HSP's that attempted gapping in prelim test: 2997
Number of HSP's gapped (non-prelim): 33
length of query: 232
length of database: 59,974,054
effective HSP length: 107
effective length of query: 125
effective length of database: 42,404,547
effective search space: 5300568375
effective search space used: 5300568375
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 64 (29.3 bits)
Medicago: description of AC149600.13


