

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC149580.1 + phase: 0 /pseudo
(260 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
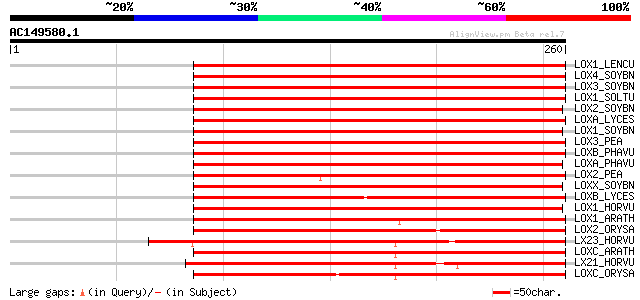
Score E
Sequences producing significant alignments: (bits) Value
LOX1_LENCU (P38414) Lipoxygenase (EC 1.13.11.12) 293 2e-79
LOX4_SOYBN (P38417) Lipoxygenase-4 (EC 1.13.11.12) (L-4) (VSP94) 273 4e-73
LOX3_SOYBN (P09186) Seed lipoxygenase-3 (EC 1.13.11.12) (L-3) 265 6e-71
LOX1_SOLTU (P37831) Lipoxygenase 1 (EC 1.13.11.12) 264 2e-70
LOX2_SOYBN (P09439) Seed lipoxygenase-2 (EC 1.13.11.12) (L-2) 263 3e-70
LOXA_LYCES (P38415) Lipoxygenase A (EC 1.13.11.12) 262 5e-70
LOX1_SOYBN (P08170) Seed lipoxygenase-1 (EC 1.13.11.12) (L-1) 262 5e-70
LOX3_PEA (P09918) Seed lipoxygenase-3 (EC 1.13.11.12) 261 1e-69
LOXB_PHAVU (P27481) Lipoxygenase (EC 1.13.11.12) (Fragment) 261 2e-69
LOXA_PHAVU (P27480) Lipoxygenase 1 (EC 1.13.11.12) 255 7e-68
LOX2_PEA (P14856) Seed lipoxygenase-2 (EC 1.13.11.12) 254 2e-67
LOXX_SOYBN (P24095) Seed lipoxygenase (EC 1.13.11.12) 250 3e-66
LOXB_LYCES (P38416) Lipoxygenase B (EC 1.13.11.12) 245 7e-65
LOX1_HORVU (P29114) Lipoxygenase 1 (EC 1.13.11.12) 237 2e-62
LOX1_ARATH (Q06327) Lipoxygenase 1 (EC 1.13.11.12) 230 3e-60
LOX2_ORYSA (P29250) Lipoxygenase L-2 (EC 1.13.11.12) 212 6e-55
LX23_HORVU (Q8GSM2) Lipoxygenase 2.3, chloroplast precursor (EC ... 207 2e-53
LOXC_ARATH (P38418) Lipoxygenase, chloroplast precursor (EC 1.13... 204 1e-52
LX21_HORVU (P93184) Lipoxygenase 2.1, chloroplast precursor (EC ... 196 4e-50
LOXC_ORYSA (P38419) Lipoxygenase, chloroplast precursor (EC 1.13... 192 9e-49
>LOX1_LENCU (P38414) Lipoxygenase (EC 1.13.11.12)
Length = 866
Score = 293 bits (751), Expect = 2e-79
Identities = 136/174 (78%), Positives = 153/174 (87%)
Query: 87 WLLAKAYAVVNDSCCHQLVSHWLKTHAVVEPFVIATNRHLSVLHPIHKLLVPHYRGTMTI 146
WLLAKA+ VVNDSC HQLVSHWL THAVVEPF+IATNRHLSV+HPIHKLL+PHYR TM I
Sbjct: 506 WLLAKAFVVVNDSCYHQLVSHWLNTHAVVEPFIIATNRHLSVVHPIHKLLLPHYRDTMNI 565
Query: 147 NARSRNILINAGGIVESTFLFEKYCMEMSAVVYKDWVFAEEGLPTDLKNRGMAVDDDSSP 206
NA +RN+L+NA GI+ESTFL+ Y MEMSAVVYKDWVF ++GLP DL RG+AV D SSP
Sbjct: 566 NALARNVLVNAEGIIESTFLWGNYAMEMSAVVYKDWVFPDQGLPNDLIKRGVAVKDPSSP 625
Query: 207 HGLRLLIEDYPYASDGLEIWAAIKSWVDEYVNFYYESDKDVKDDEELKAFWKEL 260
HG+RLLIEDYPYASDGLEIWAAIKSWV+EYVNFYY+SD + D EL+AFWKEL
Sbjct: 626 HGVRLLIEDYPYASDGLEIWAAIKSWVEEYVNFYYKSDAAIAQDAELQAFWKEL 679
>LOX4_SOYBN (P38417) Lipoxygenase-4 (EC 1.13.11.12) (L-4) (VSP94)
Length = 853
Score = 273 bits (697), Expect = 4e-73
Identities = 123/174 (70%), Positives = 149/174 (84%)
Query: 87 WLLAKAYAVVNDSCCHQLVSHWLKTHAVVEPFVIATNRHLSVLHPIHKLLVPHYRGTMTI 146
WLLAKAY VVND+C HQ++SHWL THA+VEPFVIATNR LSV+HPI+KLL PHYR TM I
Sbjct: 493 WLLAKAYVVVNDACYHQIISHWLSTHAIVEPFVIATNRQLSVVHPIYKLLFPHYRDTMNI 552
Query: 147 NARSRNILINAGGIVESTFLFEKYCMEMSAVVYKDWVFAEEGLPTDLKNRGMAVDDDSSP 206
N+ +R L+NA GI+E TFL+ +Y MEMSAV+YKDWVF ++ LP DL RG+AV D S+P
Sbjct: 553 NSLARKALVNADGIIEKTFLWGRYSMEMSAVIYKDWVFTDQALPNDLVKRGVAVKDPSAP 612
Query: 207 HGLRLLIEDYPYASDGLEIWAAIKSWVDEYVNFYYESDKDVKDDEELKAFWKEL 260
HG+RLLIEDYPYASDGLEIW AIKSWV EYV+FYY+SD++++ D EL+A+WKEL
Sbjct: 613 HGVRLLIEDYPYASDGLEIWDAIKSWVQEYVSFYYKSDEELQKDPELQAWWKEL 666
>LOX3_SOYBN (P09186) Seed lipoxygenase-3 (EC 1.13.11.12) (L-3)
Length = 857
Score = 265 bits (678), Expect = 6e-71
Identities = 122/174 (70%), Positives = 144/174 (82%)
Query: 87 WLLAKAYAVVNDSCCHQLVSHWLKTHAVVEPFVIATNRHLSVLHPIHKLLVPHYRGTMTI 146
WLLAKAY VVNDSC HQLVSHWL THAVVEPF+IATNRHLSV+HPI+KLL PHYR TM I
Sbjct: 498 WLLAKAYVVVNDSCYHQLVSHWLNTHAVVEPFIIATNRHLSVVHPIYKLLHPHYRDTMNI 557
Query: 147 NARSRNILINAGGIVESTFLFEKYCMEMSAVVYKDWVFAEEGLPTDLKNRGMAVDDDSSP 206
N +R L+N GG++E TFL+ +Y +EMSAVVYKDWVF ++ LP DL RGMA++D S P
Sbjct: 558 NGLARLSLVNDGGVIEQTFLWGRYSVEMSAVVYKDWVFTDQALPADLIKRGMAIEDPSCP 617
Query: 207 HGLRLLIEDYPYASDGLEIWAAIKSWVDEYVNFYYESDKDVKDDEELKAFWKEL 260
HG+RL+IEDYPYA DGLEIW AIK+WV EYV YY+SD +++D EL+A WKEL
Sbjct: 618 HGIRLVIEDYPYAVDGLEIWDAIKTWVHEYVFLYYKSDDTLREDPELQACWKEL 671
>LOX1_SOLTU (P37831) Lipoxygenase 1 (EC 1.13.11.12)
Length = 861
Score = 264 bits (674), Expect = 2e-70
Identities = 121/174 (69%), Positives = 143/174 (81%)
Query: 87 WLLAKAYAVVNDSCCHQLVSHWLKTHAVVEPFVIATNRHLSVLHPIHKLLVPHYRGTMTI 146
W LAKAY VND HQL+SHWL THAV+EPFVIATNR LSVLHPIHKLL PH+R TM I
Sbjct: 502 WQLAKAYVAVNDVGVHQLISHWLNTHAVIEPFVIATNRQLSVLHPIHKLLYPHFRDTMNI 561
Query: 147 NARSRNILINAGGIVESTFLFEKYCMEMSAVVYKDWVFAEEGLPTDLKNRGMAVDDDSSP 206
NA +R +L+NAGG++EST K+ MEMSAVVYKDWVF ++ LP DL RG+AV+D SSP
Sbjct: 562 NASARQLLVNAGGVLESTVFQSKFAMEMSAVVYKDWVFPDQALPADLVKRGVAVEDSSSP 621
Query: 207 HGLRLLIEDYPYASDGLEIWAAIKSWVDEYVNFYYESDKDVKDDEELKAFWKEL 260
HG+RLLIEDYPYA DGLEIW+AIKSWV +Y +FYY SD+++ D EL+A+WKEL
Sbjct: 622 HGVRLLIEDYPYAVDGLEIWSAIKSWVTDYCSFYYGSDEEILKDNELQAWWKEL 675
>LOX2_SOYBN (P09439) Seed lipoxygenase-2 (EC 1.13.11.12) (L-2)
Length = 865
Score = 263 bits (672), Expect = 3e-70
Identities = 121/173 (69%), Positives = 141/173 (80%)
Query: 87 WLLAKAYAVVNDSCCHQLVSHWLKTHAVVEPFVIATNRHLSVLHPIHKLLVPHYRGTMTI 146
WLLAKAY VVNDSC HQL+SHWL THAV+EPF+IATNRHLS LHPI+KLL PHYR TM I
Sbjct: 507 WLLAKAYVVVNDSCYHQLMSHWLNTHAVIEPFIIATNRHLSALHPIYKLLTPHYRDTMNI 566
Query: 147 NARSRNILINAGGIVESTFLFEKYCMEMSAVVYKDWVFAEEGLPTDLKNRGMAVDDDSSP 206
NA +R LINA GI+E +FL K+ +EMS+ VYK+WVF ++ LP DL RG+A+ D S+P
Sbjct: 567 NALARQSLINADGIIEKSFLPSKHSVEMSSAVYKNWVFTDQALPADLIKRGVAIKDPSAP 626
Query: 207 HGLRLLIEDYPYASDGLEIWAAIKSWVDEYVNFYYESDKDVKDDEELKAFWKE 259
HGLRLLIEDYPYA DGLEIWAAIK+WV EYV+ YY D DVK D EL+ +WKE
Sbjct: 627 HGLRLLIEDYPYAVDGLEIWAAIKTWVQEYVSLYYARDDDVKPDSELQQWWKE 679
>LOXA_LYCES (P38415) Lipoxygenase A (EC 1.13.11.12)
Length = 860
Score = 262 bits (670), Expect = 5e-70
Identities = 121/174 (69%), Positives = 144/174 (82%)
Query: 87 WLLAKAYAVVNDSCCHQLVSHWLKTHAVVEPFVIATNRHLSVLHPIHKLLVPHYRGTMTI 146
W LAKAY VNDS HQL+SHWL THAV+EPFVIATNR LSVLHPIHKLL PH+R TM I
Sbjct: 501 WQLAKAYVAVNDSGVHQLISHWLNTHAVIEPFVIATNRQLSVLHPIHKLLYPHFRDTMNI 560
Query: 147 NARSRNILINAGGIVESTFLFEKYCMEMSAVVYKDWVFAEEGLPTDLKNRGMAVDDDSSP 206
NA +R ILINAGG++EST K+ MEMSAVVYKDWVF ++ LP DL RG+AV+D SSP
Sbjct: 561 NALARQILINAGGVLESTVFPSKFAMEMSAVVYKDWVFPDQALPADLVKRGVAVEDSSSP 620
Query: 207 HGLRLLIEDYPYASDGLEIWAAIKSWVDEYVNFYYESDKDVKDDEELKAFWKEL 260
HG+RLLI+DYPYA DGLEIW+AIKSWV +Y +FYY S++++ D EL+A+WKE+
Sbjct: 621 HGVRLLIDDYPYAVDGLEIWSAIKSWVTDYCSFYYGSNEEILKDNELQAWWKEV 674
>LOX1_SOYBN (P08170) Seed lipoxygenase-1 (EC 1.13.11.12) (L-1)
Length = 839
Score = 262 bits (670), Expect = 5e-70
Identities = 120/173 (69%), Positives = 142/173 (81%)
Query: 87 WLLAKAYAVVNDSCCHQLVSHWLKTHAVVEPFVIATNRHLSVLHPIHKLLVPHYRGTMTI 146
WLLAKAY +VNDSC HQL+SHWL THA +EPFVIAT+RHLSVLHPI+KLL PHYR M I
Sbjct: 479 WLLAKAYVIVNDSCYHQLMSHWLNTHAAMEPFVIATHRHLSVLHPIYKLLTPHYRNNMNI 538
Query: 147 NARSRNILINAGGIVESTFLFEKYCMEMSAVVYKDWVFAEEGLPTDLKNRGMAVDDDSSP 206
NA +R LINA GI+E+TFL KY +EMS+ VYK+WVF ++ LP DL RG+A+ D S+P
Sbjct: 539 NALARQSLINANGIIETTFLPSKYSVEMSSAVYKNWVFTDQALPADLIKRGVAIKDPSTP 598
Query: 207 HGLRLLIEDYPYASDGLEIWAAIKSWVDEYVNFYYESDKDVKDDEELKAFWKE 259
HG+RLLIEDYPYA+DGLEIWAAIK+WV EYV YY D DVK+D EL+ +WKE
Sbjct: 599 HGVRLLIEDYPYAADGLEIWAAIKTWVQEYVPLYYARDDDVKNDSELQHWWKE 651
>LOX3_PEA (P09918) Seed lipoxygenase-3 (EC 1.13.11.12)
Length = 861
Score = 261 bits (667), Expect = 1e-69
Identities = 119/174 (68%), Positives = 144/174 (82%)
Query: 87 WLLAKAYAVVNDSCCHQLVSHWLKTHAVVEPFVIATNRHLSVLHPIHKLLVPHYRGTMTI 146
WLLAKAY +VNDSC HQLVSHWL THAVVEPFVIATNRHLS LHPI+KLL PHYR TM I
Sbjct: 502 WLLAKAYVIVNDSCYHQLVSHWLNTHAVVEPFVIATNRHLSCLHPIYKLLYPHYRDTMNI 561
Query: 147 NARSRNILINAGGIVESTFLFEKYCMEMSAVVYKDWVFAEEGLPTDLKNRGMAVDDDSSP 206
N+ +R L+N GGI+E TFL+ +Y MEMS+ VYK+WVF E+ LP DL RGMA++D SSP
Sbjct: 562 NSLARLSLVNDGGIIEKTFLWGRYSMEMSSKVYKNWVFTEQALPADLIKRGMAIEDPSSP 621
Query: 207 HGLRLLIEDYPYASDGLEIWAAIKSWVDEYVNFYYESDKDVKDDEELKAFWKEL 260
G++L++EDYPYA DGLEIWA IK+WV +YV+ YY SD+ ++ D EL+A+WKEL
Sbjct: 622 CGVKLVVEDYPYAVDGLEIWAIIKTWVQDYVSLYYTSDEKLRQDSELQAWWKEL 675
>LOXB_PHAVU (P27481) Lipoxygenase (EC 1.13.11.12) (Fragment)
Length = 741
Score = 261 bits (666), Expect = 2e-69
Identities = 119/174 (68%), Positives = 145/174 (82%)
Query: 87 WLLAKAYAVVNDSCCHQLVSHWLKTHAVVEPFVIATNRHLSVLHPIHKLLVPHYRGTMTI 146
WLLAKAY VVNDSC HQLVSHWL THAVVEPFV+ATNR LSV+HP++KLL PHYR TM I
Sbjct: 387 WLLAKAYVVVNDSCYHQLVSHWLNTHAVVEPFVLATNRQLSVVHPVYKLLFPHYRDTMNI 446
Query: 147 NARSRNILINAGGIVESTFLFEKYCMEMSAVVYKDWVFAEEGLPTDLKNRGMAVDDDSSP 206
N+ +R L+NA GI+E TFL+ +Y +E+SAV+YKDW ++ LP DL RG+AV D S+P
Sbjct: 447 NSLARKSLVNADGIIEKTFLWGRYALELSAVIYKDWSLHDQALPNDLVKRGVAVKDPSAP 506
Query: 207 HGLRLLIEDYPYASDGLEIWAAIKSWVDEYVNFYYESDKDVKDDEELKAFWKEL 260
HG++L+IEDYPYASDGLEIW AIKSWV EYV FYY+SD+ ++ D EL+A+WKEL
Sbjct: 507 HGVKLVIEDYPYASDGLEIWDAIKSWVVEYVAFYYKSDEVLQQDSELQAWWKEL 560
>LOXA_PHAVU (P27480) Lipoxygenase 1 (EC 1.13.11.12)
Length = 862
Score = 255 bits (652), Expect = 7e-68
Identities = 117/173 (67%), Positives = 143/173 (82%)
Query: 87 WLLAKAYAVVNDSCCHQLVSHWLKTHAVVEPFVIATNRHLSVLHPIHKLLVPHYRGTMTI 146
WLLAKAY VVNDSC HQL+SHWL THAV+EPFVIATNRHLSVLHPI+KLL+PHYR TM I
Sbjct: 502 WLLAKAYVVVNDSCYHQLMSHWLNTHAVMEPFVIATNRHLSVLHPIYKLLLPHYRDTMNI 561
Query: 147 NARSRNILINAGGIVESTFLFEKYCMEMSAVVYKDWVFAEEGLPTDLKNRGMAVDDDSSP 206
NA +R LINAGG++E +FL ++ +EMS+ VYK WVF ++ LP DL RGMAV+D SSP
Sbjct: 562 NALARQSLINAGGVIERSFLPGEFAVEMSSAVYKSWVFTDQALPADLIKRGMAVEDPSSP 621
Query: 207 HGLRLLIEDYPYASDGLEIWAAIKSWVDEYVNFYYESDKDVKDDEELKAFWKE 259
+GLRL++EDYPYA DGLEIW I++WV +YV+ YY ++ VK D EL+A+WKE
Sbjct: 622 YGLRLVVEDYPYAVDGLEIWDTIQTWVKDYVSLYYPTNDAVKKDTELQAWWKE 674
>LOX2_PEA (P14856) Seed lipoxygenase-2 (EC 1.13.11.12)
Length = 864
Score = 254 bits (648), Expect = 2e-67
Identities = 119/175 (68%), Positives = 142/175 (81%), Gaps = 1/175 (0%)
Query: 87 WLLAKAYAVVNDSCCHQLVSHWLKTHAVVEPFVIATNRHLSVLHPIHKLLVPHYRGTM-T 145
WLLAKAY VVNDSC HQL+SHWL THAV+EPFVIATNR LSV+HPI+KLL PHYR TM
Sbjct: 504 WLLAKAYVVVNDSCYHQLMSHWLNTHAVIEPFVIATNRQLSVVHPINKLLAPHYRDTMMN 563
Query: 146 INARSRNILINAGGIVESTFLFEKYCMEMSAVVYKDWVFAEEGLPTDLKNRGMAVDDDSS 205
INA +R+ LINA G++E +FL KY +EMS+ VYK WVF ++ LP DL R MAV D SS
Sbjct: 564 INALARDSLINANGLIERSFLPSKYAVEMSSAVYKYWVFTDQALPNDLIKRNMAVKDSSS 623
Query: 206 PHGLRLLIEDYPYASDGLEIWAAIKSWVDEYVNFYYESDKDVKDDEELKAFWKEL 260
P+GLRLLIEDYPYA DGLEIW AIK+WV +YV+ YY +D D+K+D EL+ +WKE+
Sbjct: 624 PYGLRLLIEDYPYAVDGLEIWTAIKTWVQDYVSLYYATDNDIKNDSELQHWWKEV 678
>LOXX_SOYBN (P24095) Seed lipoxygenase (EC 1.13.11.12)
Length = 864
Score = 250 bits (638), Expect = 3e-66
Identities = 116/173 (67%), Positives = 140/173 (80%)
Query: 87 WLLAKAYAVVNDSCCHQLVSHWLKTHAVVEPFVIATNRHLSVLHPIHKLLVPHYRGTMTI 146
WLLAKA+ +VNDS HQLVSHWL THAV+EPF IATNRHLSVLHPI+KLL PHYR T+ I
Sbjct: 505 WLLAKAHVIVNDSGYHQLVSHWLNTHAVMEPFAIATNRHLSVLHPIYKLLYPHYRDTINI 564
Query: 147 NARSRNILINAGGIVESTFLFEKYCMEMSAVVYKDWVFAEEGLPTDLKNRGMAVDDDSSP 206
N +R LINA GI+E +FL KY +EMS+ VYK+WVF ++ LP DL RG+A++D S+P
Sbjct: 565 NGLARQSLINADGIIEKSFLPGKYSIEMSSSVYKNWVFTDQALPADLVKRGLAIEDPSAP 624
Query: 207 HGLRLLIEDYPYASDGLEIWAAIKSWVDEYVNFYYESDKDVKDDEELKAFWKE 259
HGLRL+IEDYPYA DGLEIW AIK+WV EYV+ YY +D V+ D EL+A+WKE
Sbjct: 625 HGLRLVIEDYPYAVDGLEIWDAIKTWVHEYVSLYYPTDAAVQQDTELQAWWKE 677
>LOXB_LYCES (P38416) Lipoxygenase B (EC 1.13.11.12)
Length = 859
Score = 245 bits (626), Expect = 7e-65
Identities = 119/174 (68%), Positives = 133/174 (76%), Gaps = 1/174 (0%)
Query: 87 WLLAKAYAVVNDSCCHQLVSHWLKTHAVVEPFVIATNRHLSVLHPIHKLLVPHYRGTMTI 146
W AKAY VND HQL+SHWL THAV+EPFV+ATNRHLSVLHPIHKLL PH+R TM I
Sbjct: 501 WQFAKAYVAVNDMGIHQLISHWLNTHAVIEPFVVATNRHLSVLHPIHKLLHPHFRNTMNI 560
Query: 147 NARSRNILINAGGIVESTFLFEKYCMEMSAVVYKDWVFAEEGLPTDLKNRGMAVDDDSSP 206
NA +R L GG S F KY MEMSA YKDWVF E+ LP DL RG+AV+D SSP
Sbjct: 561 NALARETLTYDGGFETSLFP-AKYSMEMSAAAYKDWVFPEQALPADLLKRGVAVEDLSSP 619
Query: 207 HGLRLLIEDYPYASDGLEIWAAIKSWVDEYVNFYYESDKDVKDDEELKAFWKEL 260
HG+RLLI DYPYA DGLEIWAAIKSWV EY FYY+SD+ V+ D EL+A+WKEL
Sbjct: 620 HGIRLLILDYPYAVDGLEIWAAIKSWVTEYCKFYYKSDETVEKDTELQAWWKEL 673
>LOX1_HORVU (P29114) Lipoxygenase 1 (EC 1.13.11.12)
Length = 862
Score = 237 bits (604), Expect = 2e-62
Identities = 113/173 (65%), Positives = 134/173 (77%)
Query: 87 WLLAKAYAVVNDSCCHQLVSHWLKTHAVVEPFVIATNRHLSVLHPIHKLLVPHYRGTMTI 146
W LAKAY VNDS HQLVSHWL THAV+EPFVI+TNRHLSV HP+HKLL PHYR TMTI
Sbjct: 497 WELAKAYVAVNDSGWHQLVSHWLNTHAVMEPFVISTNRHLSVTHPVHKLLSPHYRDTMTI 556
Query: 147 NARSRNILINAGGIVESTFLFEKYCMEMSAVVYKDWVFAEEGLPTDLKNRGMAVDDDSSP 206
NA +R LINAGGI E T K+ + MSAVVYKDW F E+GLP DL RGMAV+D SSP
Sbjct: 557 NALARQTLINAGGIFEMTVFPGKFALGMSAVVYKDWKFTEQGLPDDLIKRGMAVEDPSSP 616
Query: 207 HGLRLLIEDYPYASDGLEIWAAIKSWVDEYVNFYYESDKDVKDDEELKAFWKE 259
+ +RLL+ DYPYA+DGL IW AI+ +V EY+ YY +D ++ D E++A+WKE
Sbjct: 617 YKVRLLVSDYPYAADGLAIWHAIEQYVSEYLAIYYPNDGVLQGDTEVQAWWKE 669
>LOX1_ARATH (Q06327) Lipoxygenase 1 (EC 1.13.11.12)
Length = 859
Score = 230 bits (586), Expect = 3e-60
Identities = 106/175 (60%), Positives = 136/175 (77%), Gaps = 1/175 (0%)
Query: 87 WLLAKAYAVVNDSCCHQLVSHWLKTHAVVEPFVIATNRHLSVLHPIHKLLVPHYRGTMTI 146
W LAKA+ VNDS HQL+SHW++THA +EPFVIATNR LSVLHP+ KLL PH+R TM I
Sbjct: 499 WQLAKAFVGVNDSGNHQLISHWMQTHASIEPFVIATNRQLSVLHPVFKLLEPHFRDTMNI 558
Query: 147 NARSRNILINAGGIVESTFLFEKYCMEMSAVVYKD-WVFAEEGLPTDLKNRGMAVDDDSS 205
NA +R ILIN GGI E T KY MEMS+ +YK+ W F ++ LP +LK RGMAV+D +
Sbjct: 559 NALARQILINGGGIFEITVFPSKYAMEMSSFIYKNHWTFPDQALPAELKKRGMAVEDPEA 618
Query: 206 PHGLRLLIEDYPYASDGLEIWAAIKSWVDEYVNFYYESDKDVKDDEELKAFWKEL 260
PHGLRL I+DYPYA DGLE+W AI+SWV +Y+ +Y+ ++D++ D EL+A+WKE+
Sbjct: 619 PHGLRLRIKDYPYAVDGLEVWYAIESWVRDYIFLFYKIEEDIQTDTELQAWWKEV 673
>LOX2_ORYSA (P29250) Lipoxygenase L-2 (EC 1.13.11.12)
Length = 865
Score = 212 bits (540), Expect = 6e-55
Identities = 101/174 (58%), Positives = 124/174 (71%), Gaps = 1/174 (0%)
Query: 87 WLLAKAYAVVNDSCCHQLVSHWLKTHAVVEPFVIATNRHLSVLHPIHKLLVPHYRGTMTI 146
W LAKAY VND C HQL+SHWL THAV+EPFVIATNR LSV HP+HKLL+PHYR TMTI
Sbjct: 501 WQLAKAYVNVNDYCWHQLISHWLNTHAVMEPFVIATNRQLSVAHPVHKLLLPHYRDTMTI 560
Query: 147 NARSRNILINAGGIVESTFLFEKYCMEMSAVVYKDWVFAEEGLPTDLKNRGMAVDDDSSP 206
N +R LIN GGI E T + MS+ YKDW FA++ LP DL RG+ D +SP
Sbjct: 561 NGLARQTLINGGGIFEMTVFPRNDALAMSSAFYKDWSFADQALPDDLVKRGVRT-DPASP 619
Query: 207 HGLRLLIEDYPYASDGLEIWAAIKSWVDEYVNFYYESDKDVKDDEELKAFWKEL 260
+ +RLLIEDYPYA+DGL + I+ W EY+ YY +D ++ D EL+A+WKE+
Sbjct: 620 YKVRLLIEDYPYANDGLAVCTPIEQWATEYLAIYYPNDGVLQGDAELQAWWKEV 673
>LX23_HORVU (Q8GSM2) Lipoxygenase 2.3, chloroplast precursor (EC
1.13.11.12) (LOX2:Hv:3)
Length = 896
Score = 207 bits (527), Expect = 2e-53
Identities = 106/201 (52%), Positives = 136/201 (66%), Gaps = 8/201 (3%)
Query: 66 TLPYPPPGPPNSTAYHAGT-----FNWLLAKAYAVVNDSCCHQLVSHWLKTHAVVEPFVI 120
T P P P A+ G+ + W LAKA+ + +D+ HQLVSHWL+THA VEP++I
Sbjct: 510 TRPQSPTKPQWKRAFTHGSDATESWLWKLAKAHVLTHDTGYHQLVSHWLRTHACVEPYII 569
Query: 121 ATNRHLSVLHPIHKLLVPHYRGTMTINARSRNILINAGGIVESTFLFEKYCMEMSAVVY- 179
ATNR LS +HP+++LL PH+R TM INA +R LINA GI+E FL KY +E+S+V Y
Sbjct: 570 ATNRQLSRMHPVYRLLHPHFRYTMEINALAREALINADGIIEEAFLAGKYSIELSSVAYG 629
Query: 180 KDWVFAEEGLPTDLKNRGMAVDDDSSPHGLRLLIEDYPYASDGLEIWAAIKSWVDEYVNF 239
W F E LP DL NRG+AV D L L I+DYPYA DGL IW +IK W +YV+F
Sbjct: 630 AAWQFNTEALPEDLINRGLAVRRDDGE--LELAIKDYPYADDGLLIWGSIKQWASDYVDF 687
Query: 240 YYESDKDVKDDEELKAFWKEL 260
YY+SD DV DEEL+A+W+E+
Sbjct: 688 YYKSDGDVAGDEELRAWWEEV 708
>LOXC_ARATH (P38418) Lipoxygenase, chloroplast precursor (EC
1.13.11.12)
Length = 896
Score = 204 bits (520), Expect = 1e-52
Identities = 93/175 (53%), Positives = 128/175 (73%), Gaps = 1/175 (0%)
Query: 87 WLLAKAYAVVNDSCCHQLVSHWLKTHAVVEPFVIATNRHLSVLHPIHKLLVPHYRGTMTI 146
W LAK +A+ +D+ HQL+SHWL+THA EP++IA NR LS +HPI++LL PH+R TM I
Sbjct: 534 WNLAKTHAISHDAGYHQLISHWLRTHACTEPYIIAANRQLSAMHPIYRLLHPHFRYTMEI 593
Query: 147 NARSRNILINAGGIVESTFLFEKYCMEMSAVVY-KDWVFAEEGLPTDLKNRGMAVDDDSS 205
NAR+R L+N GGI+E+ F KY +E+S+ VY K W F +EGLP DL RG+A +D ++
Sbjct: 594 NARARQSLVNGGGIIETCFWPGKYALELSSAVYGKLWRFDQEGLPADLIKRGLAEEDKTA 653
Query: 206 PHGLRLLIEDYPYASDGLEIWAAIKSWVDEYVNFYYESDKDVKDDEELKAFWKEL 260
HG+RL I DYP+A+DGL +W AIK WV +YV YY ++ + DEEL+ +W E+
Sbjct: 654 EHGVRLTIPDYPFANDGLILWDAIKEWVTDYVKHYYPDEELITSDEELQGWWSEV 708
>LX21_HORVU (P93184) Lipoxygenase 2.1, chloroplast precursor (EC
1.13.11.12) (LOX-100) (LOX2:Hv:1)
Length = 936
Score = 196 bits (499), Expect = 4e-50
Identities = 97/180 (53%), Positives = 130/180 (71%), Gaps = 5/180 (2%)
Query: 83 GTFNWLLAKAYAVVNDSCCHQLVSHWLKTHAVVEPFVIATNRHLSVLHPIHKLLVPHYRG 142
G++ W LAKA+ + +D+ HQLVSHWL+THA EP++IA NR LS +HP+++LL PH+R
Sbjct: 563 GSWLWQLAKAHILAHDAGVHQLVSHWLRTHACTEPYIIAANRQLSQMHPVYRLLHPHFRF 622
Query: 143 TMTINARSRNILINAGGIVESTFLFEKYCMEMSAVVY-KDWVFAEEGLPTDLKNRGMAVD 201
TM INA++R +LINAGGI+E +F+ +Y +E+S+V Y + W F E LP DL RGMAV
Sbjct: 623 TMEINAQARAMLINAGGIIEGSFVPGEYSLELSSVAYDQQWRFDMEALPEDLIRRGMAV- 681
Query: 202 DDSSPHG-LRLLIEDYPYASDGLEIWAAIKSWVDEYVNFYYESDKDVKDDEELKAFWKEL 260
+P+G L L IEDYPYA+DGL +W AIK W YV YY D+ DDEEL+A+W E+
Sbjct: 682 --RNPNGELELAIEDYPYANDGLLVWDAIKQWALTYVQHYYPCAADIVDDEELQAWWTEV 739
>LOXC_ORYSA (P38419) Lipoxygenase, chloroplast precursor (EC
1.13.11.12)
Length = 923
Score = 192 bits (487), Expect = 9e-49
Identities = 93/175 (53%), Positives = 125/175 (71%), Gaps = 2/175 (1%)
Query: 87 WLLAKAYAVVNDSCCHQLVSHWLKTHAVVEPFVIATNRHLSVLHPIHKLLVPHYRGTMTI 146
W +AKA+ +D+ H+L++HWL+TH VEP++IA NR LS +HPI++LL PH+R TM I
Sbjct: 561 WRMAKAHVRAHDAGHHELITHWLRTHCAVEPYIIAANRQLSEMHPIYQLLRPHFRYTMRI 620
Query: 147 NARSRNILINAGGIVESTFLFEKYCMEMSAVVY-KDWVFAEEGLPTDLKNRGMAVDDDSS 205
NAR+ + I+AGGI+E +F +KY ME+S+V Y K W F E LP DL RGMA +D ++
Sbjct: 621 NARAARV-ISAGGIIERSFSPQKYSMELSSVAYDKLWRFDTEALPADLVRRGMAEEDPTA 679
Query: 206 PHGLRLLIEDYPYASDGLEIWAAIKSWVDEYVNFYYESDKDVKDDEELKAFWKEL 260
GL+L IEDYP+A+DGL IW AIK+WV YV +Y V DEEL+AFW E+
Sbjct: 680 EQGLKLAIEDYPFANDGLLIWDAIKTWVQAYVARFYPDADSVAGDEELQAFWTEV 734
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.316 0.137 0.435
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 38,843,737
Number of Sequences: 164201
Number of extensions: 1983185
Number of successful extensions: 12670
Number of sequences better than 10.0: 379
Number of HSP's better than 10.0 without gapping: 117
Number of HSP's successfully gapped in prelim test: 270
Number of HSP's that attempted gapping in prelim test: 10106
Number of HSP's gapped (non-prelim): 2052
length of query: 260
length of database: 59,974,054
effective HSP length: 108
effective length of query: 152
effective length of database: 42,240,346
effective search space: 6420532592
effective search space used: 6420532592
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 64 (29.3 bits)
Medicago: description of AC149580.1


