

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC149576.2 + phase: 0
(538 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
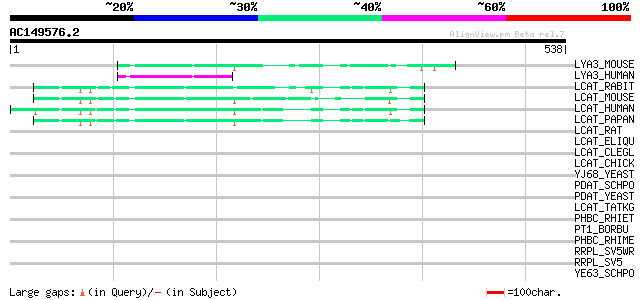
Score E
Sequences producing significant alignments: (bits) Value
LYA3_MOUSE (Q8VEB4) 1-O-acylceramide synthase precursor (EC 2.3.... 58 7e-08
LYA3_HUMAN (Q8NCC3) 1-O-acylceramide synthase precursor (EC 2.3.... 54 1e-06
LCAT_RABIT (P53761) Phosphatidylcholine-sterol acyltransferase p... 50 1e-05
LCAT_MOUSE (P16301) Phosphatidylcholine-sterol acyltransferase p... 47 1e-04
LCAT_HUMAN (P04180) Phosphatidylcholine-sterol acyltransferase p... 47 2e-04
LCAT_PAPAN (Q08758) Phosphatidylcholine-sterol acyltransferase p... 46 2e-04
LCAT_RAT (P18424) Phosphatidylcholine-sterol acyltransferase pre... 43 0.002
LCAT_ELIQU (O35573) Phosphatidylcholine-sterol acyltransferase (... 37 0.099
LCAT_CLEGL (O35502) Phosphatidylcholine-sterol acyltransferase (... 37 0.099
LCAT_CHICK (P53760) Phosphatidylcholine-sterol acyltransferase p... 37 0.099
YJ68_YEAST (P47139) Hypothetical 74.1 kDa protein in ACR1-YUH1 i... 37 0.17
PDAT_SCHPO (O94680) Phospholipid:diacylglycerol acyltransferase ... 37 0.17
PDAT_YEAST (P40345) Phospholipid:diacylglycerol acyltransferase ... 36 0.22
LCAT_TATKG (O35840) Phosphatidylcholine-sterol acyltransferase (... 36 0.29
PHBC_RHIET (Q52728) Poly-beta-hydroxybutyrate polymerase (EC 2.3... 35 0.64
PT1_BORBU (O51508) Phosphoenolpyruvate-protein phosphotransferas... 34 0.84
PHBC_RHIME (P50176) Poly-beta-hydroxybutyrate polymerase (EC 2.3... 33 1.4
RRPL_SV5WR (Q03396) RNA polymerase beta subunit (EC 2.7.7.48) (L... 32 3.2
RRPL_SV5 (Q88434) RNA polymerase beta subunit (EC 2.7.7.48) (Lar... 32 3.2
YE63_SCHPO (O14249) Hypothetical protein C6G10.03c in chromosome I 32 5.4
>LYA3_MOUSE (Q8VEB4) 1-O-acylceramide synthase precursor (EC
2.3.1.-) (ACS) (Lysosomal phospholipase A2) (LCAT-like
lysophospholipase) (LLPL)
Length = 412
Score = 57.8 bits (138), Expect = 7e-08
Identities = 84/347 (24%), Positives = 137/347 (39%), Gaps = 79/347 (22%)
Query: 105 AIDVLDPDLV-IGSEAVYYFHDMIVQMQKWGYQEGKTLFGFGYDFRQS-NRLQETMDRFA 162
+++ LDP +GS YF+ M+ + WGY G+ + G YD+R++ N
Sbjct: 123 SMEFLDPSKRNVGS----YFYTMVESLVGWGYTRGEDVRGAPYDWRRAPNENGPYFLALR 178
Query: 163 EKLELIYNAAGGKKIDLISHSMGGLLVKCFMTLHSDIF-EKYVKNWIAICAPFQG----- 216
E +E +Y GG + L++HSMG + + F+ ++ +KY+ ++++ AP+ G
Sbjct: 179 EMIEEMYQMYGGPVV-LVAHSMGNVYMLYFLQRQPQVWKDKYIHAFVSLGAPWGGVAKTL 237
Query: 217 ---APGCTNSTFLNGMSFVEGWEQNFFISKWSMHQLLIECPSIYELMACPNFHWKHVPLL 273
A G N + G + +++ + W + P
Sbjct: 238 RVLASGDNNRIPVIGPLKIREQQRSAVSTSWLL------------------------PYN 273
Query: 274 ELWRERLHEDGKSHVILESYPPRDSIEIFKQALVNNKVNHEGEELPLPFNSHIFEWANKT 333
W HE + +Y RD F+ + F F +
Sbjct: 274 HTWS---HEKVFVYTPTTNYTLRDYHRFFRD---------------IGFEDGWF--MRQD 313
Query: 334 REILSSAKLPSGVKFYNIYGTNLATPHSICYGNADKPVSDLQELRYLQARYVCV-DGDGT 392
E L A P GV+ + +YGT + TP+S Y + P D + +C DGDGT
Sbjct: 314 TEGLVEAMTPPGVELHCLYGTGVPTPNSFYYESF--PDRDPK---------ICFGDGDGT 362
Query: 393 VPVES---AKADGFNAEERVG---IPG-EHRGILCEPHLFRILKHWL 432
V +ES +A E RV +PG EH +L LK L
Sbjct: 363 VNLESVLQCQAWQSRQEHRVSLQELPGSEHIEMLANATTLAYLKRVL 409
>LYA3_HUMAN (Q8NCC3) 1-O-acylceramide synthase precursor (EC
2.3.1.-) (ACS) (Lysosomal phospholipase A2) (LCAT-like
lysophospholipase) (LLPL) (UNQ341/PRO540)
Length = 412
Score = 53.9 bits (128), Expect = 1e-06
Identities = 33/114 (28%), Positives = 59/114 (50%), Gaps = 6/114 (5%)
Query: 105 AIDVLDPDLVIGSEAVYYFHDMIVQMQKWGYQEGKTLFGFGYDFRQS-NRLQETMDRFAE 163
+++ LDP S YFH M+ + WGY G+ + G YD+R++ N E
Sbjct: 123 SLEFLDPSK---SSVGSYFHTMVESLVGWGYTRGEDVRGAPYDWRRAPNENGPYFLALRE 179
Query: 164 KLELIYNAAGGKKIDLISHSMGGLLVKCFMTLHSDIF-EKYVKNWIAICAPFQG 216
+E +Y GG + L++HSMG + F+ + +KY++ ++++ AP+ G
Sbjct: 180 MIEEMYQLYGGPVV-LVAHSMGNMYTLYFLQRQPQAWKDKYIRAFVSLGAPWGG 232
Score = 33.5 bits (75), Expect = 1.4
Identities = 24/65 (36%), Positives = 34/65 (51%), Gaps = 12/65 (18%)
Query: 335 EILSSAKLPSGVKFYNIYGTNLATPHSICYGNADKPVSDLQELRYLQARYVCV-DGDGTV 393
E L A +P GV+ + +YGT + TP S Y + P D + +C DGDGTV
Sbjct: 315 EGLVEATMPPGVQLHCLYGTGVPTPDSFYYESF--PDRDPK---------ICFGDGDGTV 363
Query: 394 PVESA 398
++SA
Sbjct: 364 NLKSA 368
>LCAT_RABIT (P53761) Phosphatidylcholine-sterol acyltransferase
precursor (EC 2.3.1.43) (Lecithin-cholesterol
acyltransferase) (Phospholipid-cholesterol
acyltransferase)
Length = 440
Score = 50.4 bits (119), Expect = 1e-05
Identities = 91/405 (22%), Positives = 159/405 (38%), Gaps = 87/405 (21%)
Query: 24 PQAYVNPNLDPVLLVPGVGGSILNAVNESDGSQERVWVRFLSAE------------YKLK 71
P+A ++ + PV+LVPG G+ L A + D W+ + E L
Sbjct: 38 PKAELSNHTRPVILVPGCLGNQLEA--KLDKPSVVNWMCYRKTEDFFTIWLDLNMFLPLG 95
Query: 72 TKLWSR-----YDPSTGKTVTLDQKSRIVVPEDRHGLHAIDVLDPDLVIGSEAVYYFHDM 126
W Y+ S+G+ V + +I VP ++++ LD + + G Y H +
Sbjct: 96 VDCWIDNTRVVYNRSSGR-VVISPGVQIRVP-GFGKTYSVEYLDNNKLAG-----YMHTL 148
Query: 127 IVQMQKWGYQEGKTLFGFGYDFR-QSNRLQETMDRFAEKLELIYNAAGGKKIDLISHSMG 185
+ + GY +T+ YD+R + ++ +E + A +E ++ AA GK + LI HS+G
Sbjct: 149 VQNLVNNGYVRDETVRAAPYDWRLEPSQQEEYYGKLAGLVEEMH-AAYGKPVFLIGHSLG 207
Query: 186 GLLVKCFMTLHSDIF-EKYVKNWIAICAPFQGAPGCTNSTFLNGMSFVEGWEQNFFISKW 244
L + F+ + ++++ +I++ AP+ G+ + +G I
Sbjct: 208 CLHLLYFLLRQPQSWKDRFIDGFISLGAPWGGS---IKPMLVLASGDNQGIPLMSSIKLR 264
Query: 245 SMHQLLIECPSIYELMACPNFHWKHVPLLELWRERLHEDGKSHVILE----SYPPRDSIE 300
++ P ++ P +W E HV + +Y RD
Sbjct: 265 EEQRITTTSPWMF-------------PSQGVWPE-------DHVFISTPSFNYTGRDFKR 304
Query: 301 IFKQALVNNKVNHEGEELPLPFNSHIFEWANKTREILSSAKLPSGVKFYNIYGTNLATPH 360
F+ L F + W ++R++L+ P GV+ Y +YG L TPH
Sbjct: 305 FFED---------------LHFEEGWYMWL-QSRDLLAGLPAP-GVEVYCLYGIGLPTPH 347
Query: 361 SICYGNA---DKPVSDLQELRYLQARYVCVDGDGTVPVESAKADG 402
+ Y + PV L E DGD TV S G
Sbjct: 348 TYIYDHGFPYTDPVGVLYE-----------DGDDTVATSSTDLCG 381
>LCAT_MOUSE (P16301) Phosphatidylcholine-sterol acyltransferase
precursor (EC 2.3.1.43) (Lecithin-cholesterol
acyltransferase) (Phospholipid-cholesterol
acyltransferase)
Length = 438
Score = 47.0 bits (110), Expect = 1e-04
Identities = 98/410 (23%), Positives = 158/410 (37%), Gaps = 97/410 (23%)
Query: 24 PQAYVNPNLDPVLLVPGVGGSILNAVNESDGSQERVWVRFLSAE------------YKLK 71
P+A ++ + PV+LVPG G+ L A + D W+ + E L
Sbjct: 38 PKAELSNHTRPVILVPGCLGNRLEA--KLDKPDVVNWMCYRKTEDFFTIWLDFNLFLPLG 95
Query: 72 TKLWSR-----YDPSTGKTVTLDQKSRIVVPEDRHGLHAIDVLDPDLVIGSEAVYYFHDM 126
W Y+ S+G+ V+ +I VP +++ +D + + G Y H +
Sbjct: 96 VDCWIDNTRIVYNHSSGR-VSNAPGVQIRVPGFGK-TESVEYVDDNKLAG-----YLHTL 148
Query: 127 IVQMQKWGYQEGKTLFGFGYDFRQSNRLQ-ETMDRFAEKLELIYNAAGGKKIDLISHSMG 185
+ + GY +T+ YD+R + Q E + A +E +Y AA GK + LI HS+G
Sbjct: 149 VQNLVNNGYVRDETVRAAPYDWRLAPHQQDEYYKKLAGLVEEMY-AAYGKPVFLIGHSLG 207
Query: 186 GLLVKCFMTLHSDIF-EKYVKNWIAICAPFQG--------APGCTNS-TFLNGMSFVEGW 235
L V F+ + + ++ +I++ AP+ G A G L+ + E
Sbjct: 208 CLHVLHFLLRQPQSWKDHFIDGFISLGAPWGGSIKAMRILASGDNQGIPILSNIKLKE-- 265
Query: 236 EQNFFISKWSMHQLLIECPSIYELMACPNFHWKHVPLLELWRERLHEDGKSHVILESYPP 295
EQ + M P + ++ PNF++ V E + LH + H+ L+S
Sbjct: 266 EQRITTTSPWMLPAPHVWPEDHVFISTPNFNYT-VQDFERFFTDLHFEEGWHMFLQS--- 321
Query: 296 RDSIEIFKQALVNNKVNHEGEELPLPFNSHIFEWANKTREILSSAKLPSGVKFYNIYGTN 355
RD + E LP P GV+ Y +YG
Sbjct: 322 RDLL----------------ERLPAP-----------------------GVEVYCLYGVG 342
Query: 356 LATPHSICYGN---ADKPVSDLQELRYLQARYVCVDGDGTVPVESAKADG 402
TPH+ Y + PV+ L E DGD TV S + G
Sbjct: 343 RPTPHTYIYDHNFPYKDPVAALYE-----------DGDDTVATRSTELCG 381
>LCAT_HUMAN (P04180) Phosphatidylcholine-sterol acyltransferase
precursor (EC 2.3.1.43) (Lecithin-cholesterol
acyltransferase) (Phospholipid-cholesterol
acyltransferase)
Length = 440
Score = 46.6 bits (109), Expect = 2e-04
Identities = 98/435 (22%), Positives = 163/435 (36%), Gaps = 97/435 (22%)
Query: 1 MTMLLEDVLR-SVELWLRLIKKPQ---PQAYVNPNLDPVLLVPGVGGSILNAVNESDGSQ 56
+T+LL +L + WL + P P+A ++ + PV+LVPG G+ L A + D
Sbjct: 11 VTLLLGLLLPPAAPFWLLNVLFPPHTTPKAELSNHTRPVILVPGCLGNQLEA--KLDKPD 68
Query: 57 ERVWVRFLSAE------------YKLKTKLWSR-----YDPSTGKTVTLDQKSRIVVPED 99
W+ + E L W Y+ S+G V+ +I VP
Sbjct: 69 VVNWMCYRKTEDFFTIWLDLNMFLPLGVDCWIDNTRVVYNRSSG-LVSNAPGVQIRVPGF 127
Query: 100 RHGLHAIDVLDPDLVIGSEAVYYFHDMIVQMQKWGYQEGKTLFGFGYDFRQSNRLQETMD 159
++++ LD + G Y H ++ + GY +T+ YD+R QE
Sbjct: 128 GK-TYSVEYLDSSKLAG-----YLHTLVQNLVNNGYVRDETVRAAPYDWRLEPGQQEEYY 181
Query: 160 RFAEKLELIYNAAGGKKIDLISHSMGGLLVKCFMTLHSDIF-EKYVKNWIAICAPFQG-- 216
R L +AA GK + LI HS+G L + F+ + ++++ +I++ AP+ G
Sbjct: 182 RKLAGLVEEMHAAYGKPVFLIGHSLGCLHLLYFLLRQPQAWKDRFIDGFISLGAPWGGSI 241
Query: 217 ------APGCTNSTFLNGMSFVEGWEQNFFISKWSMHQLLIECPSIYELMACPNFHWKHV 270
A G + ++ ++ S W M + P + ++ P+F
Sbjct: 242 KPMLVLASGDNQGIPIMSSIKLKEEQRITTTSPW-MFPSRMAWPEDHVFISTPSF----- 295
Query: 271 PLLELWRERLHEDGKSHVILESYPPRDSIEIFKQALVNNKVNHEGEELPLPFNSHIFEWA 330
+Y RD F L F + W
Sbjct: 296 ---------------------NYTGRDFQRFFAD---------------LHFEEGWYMWL 319
Query: 331 NKTREILSSAKLPSGVKFYNIYGTNLATPHSICYGNA---DKPVSDLQELRYLQARYVCV 387
++R++L+ P GV+ Y +YG L TP + Y + PV L E
Sbjct: 320 -QSRDLLAGLPAP-GVEVYCLYGVGLPTPRTYIYDHGFPYTDPVGVLYE----------- 366
Query: 388 DGDGTVPVESAKADG 402
DGD TV S + G
Sbjct: 367 DGDDTVATRSTELCG 381
>LCAT_PAPAN (Q08758) Phosphatidylcholine-sterol acyltransferase
precursor (EC 2.3.1.43) (Lecithin-cholesterol
acyltransferase) (Phospholipid-cholesterol
acyltransferase)
Length = 440
Score = 46.2 bits (108), Expect = 2e-04
Identities = 89/406 (21%), Positives = 158/406 (37%), Gaps = 89/406 (21%)
Query: 24 PQAYVNPNLDPVLLVPGVGGSILNAVNESDGSQERVWVRFLSAE------------YKLK 71
P+A ++ + PV+LVPG G+ L A + D W+ + E L
Sbjct: 38 PKAELSNHTRPVILVPGCLGNQLEA--KLDKPDVVNWMCYRKTEDFFTIWLDLNMFLPLG 95
Query: 72 TKLWSR-----YDPSTGKTVTLDQKSRIVVPEDRHGLHAIDVLDPDLVIGSEAVYYFHDM 126
W Y+ S+G V+ +I VP ++++ LD + G Y H +
Sbjct: 96 VDCWIDNTRVVYNRSSG-LVSNAPGVQIRVPGFGK-TYSVEYLDSSKLAG-----YLHTL 148
Query: 127 IVQMQKWGYQEGKTLFGFGYDFR-QSNRLQETMDRFAEKLELIYNAAGGKKIDLISHSMG 185
+ + GY +T+ YD+R + + +E + A +E ++ AA GK + LI HS+G
Sbjct: 149 VQNLVNNGYVRDETVRAAPYDWRLEPGQQEEYYHKLAGLVEEMH-AAYGKPVFLIGHSLG 207
Query: 186 GLLVKCFMTLHSDIF-EKYVKNWIAICAPFQG--------APGCTNSTFLNGMSFVEGWE 236
L + F+ + ++++ +I++ AP+ G A G + ++ +
Sbjct: 208 CLHLLYFLLRQPQAWKDRFIDGFISLGAPWGGSIKPMLVLASGDNQGIPIMSSIKLKEEQ 267
Query: 237 QNFFISKWSMHQLLIECPSIYELMACPNFHWKHVPLLELWRERLHEDGKSHVILESYPPR 296
+ S W M + P + ++ P+F +Y R
Sbjct: 268 RITTTSPW-MFPSRLAWPEDHVFISTPSF--------------------------NYTGR 300
Query: 297 DSIEIFKQALVNNKVNHEGEELPLPFNSHIFEWANKTREILSSAKLPSGVKFYNIYGTNL 356
D F L F + W ++R++L+ P GV+ Y +YG L
Sbjct: 301 DFQRFFAD---------------LHFEEGWYMWL-QSRDLLAGLPAP-GVEVYCLYGVGL 343
Query: 357 ATPHSICYGNADKPVSDLQELRYLQARYVCVDGDGTVPVESAKADG 402
TP + Y + P +D ++ Y DGD TV S + G
Sbjct: 344 PTPRTYIYDHG-FPYTDPVDVLY-------EDGDDTVATRSTELCG 381
>LCAT_RAT (P18424) Phosphatidylcholine-sterol acyltransferase
precursor (EC 2.3.1.43) (Lecithin-cholesterol
acyltransferase) (Phospholipid-cholesterol
acyltransferase)
Length = 440
Score = 42.7 bits (99), Expect = 0.002
Identities = 91/426 (21%), Positives = 160/426 (37%), Gaps = 100/426 (23%)
Query: 11 SVELWLRLIKKPQ---PQAYVNPNLDPVLLVPGVGGSILNAVNESDGSQE---------- 57
+ WL + P P+A ++ + PV+LVPG G+ L A +
Sbjct: 22 ATSFWLLNVLFPPHTTPKAELSNHTRPVILVPGCMGNRLEAKLDKPNVVNWLCYRKTEDF 81
Query: 58 -RVWVRFLSAEYKLKTKLWSR-----YDPSTGKTVTLDQKSRIVVPEDRHG-LHAIDVLD 110
+W+ F + L W Y+ S+G + + + G ++++ LD
Sbjct: 82 FTIWLDF-NMFLPLGVDCWIDNTRVVYNRSSGH---MSNAPGVQIRVPGFGKTYSVEYLD 137
Query: 111 PDLVIGSEAVYYFHDMIVQMQKWGYQEGKTLFGFGYDFRQSNRLQ-ETMDRFAEKLELIY 169
+ + G Y + ++ + GY +T+ YD+R + R Q E + A +E +Y
Sbjct: 138 DNKLAG-----YLNTLVQNLVNNGYVRDETVRAAPYDWRLAPRQQDEYYQKLAGLVEEMY 192
Query: 170 NAAGGKKIDLISHSMGGLLVKCFMTLHSDIF-EKYVKNWIAICAPFQG--------APGC 220
AA GK + LI HS+G L V F+ + + ++ +I++ AP+ G A G
Sbjct: 193 -AAYGKPVFLIGHSLGCLHVLHFLLRQPQSWKDHFIDGFISLGAPWGGSIKPMRILASGD 251
Query: 221 TNS-TFLNGMSFVEGWEQNFFISKWSMHQLLIECPSIYELMACPNFHWKHVPLLELWRER 279
++ + E EQ + M P + ++ PNF++ + +
Sbjct: 252 NQGIPIMSNIKLRE--EQRITTTSPWMFPAHHVWPEDHVFISTPNFNYTGQDFERFFAD- 308
Query: 280 LHEDGKSHVILESYPPRDSIEIFKQALVNNKVNHEGEELPLPFNSHIFEWANKTREILSS 339
LH + H+ L+S R++L+
Sbjct: 309 LHFEEGWHMFLQS-----------------------------------------RDLLAG 327
Query: 340 AKLPSGVKFYNIYGTNLATPHSICYGN---ADKPVSDLQELRYLQARYVCVDGDGTVPVE 396
P GV+ Y +YG + T H+ Y + PV+ L E DGD TV
Sbjct: 328 LPAP-GVEVYCLYGVGMPTAHTYIYDHNFPYKDPVAALYE-----------DGDDTVATR 375
Query: 397 SAKADG 402
S + G
Sbjct: 376 STELCG 381
>LCAT_ELIQU (O35573) Phosphatidylcholine-sterol acyltransferase (EC
2.3.1.43) (Lecithin-cholesterol acyltransferase)
(Phospholipid-cholesterol acyltransferase) (Fragment)
Length = 299
Score = 37.4 bits (85), Expect = 0.099
Identities = 68/298 (22%), Positives = 109/298 (35%), Gaps = 89/298 (29%)
Query: 104 HAIDVLDPDLVIGSEAVYYFHDMIVQMQKWGYQEGKTLFGFGYDFRQSNRLQETMD-RFA 162
++I+ LD + + G Y H ++ + Y +T+ YD+R R QE + A
Sbjct: 53 YSIEYLDDNKLAG-----YMHTLVQNLVNNAYVRDETVRAPPYDWRLEPRHQEEYYLKLA 107
Query: 163 EKLELIYNAAGGKKIDLISHSMGGLLVKCFMTLHSDIFEKYVKNWIAICAPFQGAPGCTN 222
+E +Y A GK + LI HS+G + F+ L QG P
Sbjct: 108 GLVEEMY-ATYGKPVFLIGHSLGFCHLLYFLLLQP-----------------QGIP---- 145
Query: 223 STFLNGMSFVEGWEQNFFI-SKWSM--HQLLIECPSIYELMACPNFHWKHVPLLELWRER 279
++ + VE EQ S W HQ+ P + ++ PNF++ + +
Sbjct: 146 --IMSSIKLVE--EQRITTTSPWMFPSHQVW---PEDHVFISTPNFNYTFSDFQRFFADL 198
Query: 280 LHEDGKSHVILESYPPRDSIEIFKQALVNNKVNHEGEELPLPFNSHIFEWANKTREILSS 339
EDG + W ++R++L+
Sbjct: 199 HFEDGW-----------------------------------------YMWL-QSRDLLAG 216
Query: 340 AKLPSGVKFYNIYGTNLATPHSICYGNADKPVSDLQELRYLQARYVCVDGDGTVPVES 397
P GV+ Y +YG L TPH+ Y + P +D + Y DGD TV S
Sbjct: 217 LPAP-GVEVYCLYGVGLPTPHTYMYDHG-FPYTDPVGIIY-------EDGDDTVTTHS 265
>LCAT_CLEGL (O35502) Phosphatidylcholine-sterol acyltransferase (EC
2.3.1.43) (Lecithin-cholesterol acyltransferase)
(Phospholipid-cholesterol acyltransferase) (Fragment)
Length = 291
Score = 37.4 bits (85), Expect = 0.099
Identities = 26/90 (28%), Positives = 48/90 (52%), Gaps = 7/90 (7%)
Query: 104 HAIDVLDPDLVIGSEAVYYFHDMIVQMQKWGYQEGKTLFGFGYDFR-QSNRLQETMDRFA 162
++++ LD + + G Y H ++ + GY +T+ YD+R + ++ +E + A
Sbjct: 51 YSVEYLDDNKLAG-----YMHTLVQNLVNNGYVRDETVLAAPYDWRLEPSQQEEYYQKLA 105
Query: 163 EKLELIYNAAGGKKIDLISHSMGGLLVKCF 192
+E ++ AA GK + LI HS+G L V F
Sbjct: 106 GLVEEMH-AAYGKPVFLIGHSVGCLHVLYF 134
Score = 33.5 bits (75), Expect = 1.4
Identities = 29/99 (29%), Positives = 44/99 (44%), Gaps = 21/99 (21%)
Query: 312 NHEGEELPLPFNSHIFE-----WANKTREILSSAKLPSGVKFYNIYGTNLATPHSICYGN 366
N+ G++ F FE W ++R++L+ P GV+ Y +YG L TP + Y +
Sbjct: 179 NYTGQDFKRFFEDLYFEEGWYMWL-QSRDLLAGLPAP-GVEVYCLYGVGLPTPSTYIYDH 236
Query: 367 A---DKPVSDLQELRYLQARYVCVDGDGTVPVESAKADG 402
+ PV+ L E DGD TV S + G
Sbjct: 237 SFPYKDPVAALYE-----------DGDDTVATRSTELCG 264
>LCAT_CHICK (P53760) Phosphatidylcholine-sterol acyltransferase
precursor (EC 2.3.1.43) (Lecithin-cholesterol
acyltransferase) (Phospholipid-cholesterol
acyltransferase) (Fragment)
Length = 413
Score = 37.4 bits (85), Expect = 0.099
Identities = 28/119 (23%), Positives = 57/119 (47%), Gaps = 14/119 (11%)
Query: 104 HAIDVLDPDLVIGSEAVYYFHDMIVQMQKWGYQEGKTLFGFGYDFRQSNRLQ----ETMD 159
++++ LD + G Y H ++ + GY +T+ YD+R + Q + +
Sbjct: 129 YSVEYLDQSKLAG-----YLHTLVQNLVNNGYVRDQTVRAAPYDWRVGPQEQPEYFQNLK 183
Query: 160 RFAEKLELIYNAAGGKKIDLISHSMGGLLVKCFMTLHSDIF-EKYVKNWIAICAPFQGA 217
E++ Y +++ LI HSMG L V F+ + ++Y+ +I++ AP+ G+
Sbjct: 184 ALIEEMHDEYQ----QRVFLIGHSMGNLNVLYFLLQQKQAWKDQYIGGFISLGAPWGGS 238
>YJ68_YEAST (P47139) Hypothetical 74.1 kDa protein in ACR1-YUH1
intergenic region
Length = 655
Score = 36.6 bits (83), Expect = 0.17
Identities = 23/46 (50%), Positives = 28/46 (60%), Gaps = 3/46 (6%)
Query: 144 FGYDFRQSNRLQETMDRFAEKLELIYNAAGGKK-IDLISHSMGGLL 188
FGYD+R S L + KLE IYN KK I +I+HSMGGL+
Sbjct: 325 FGYDWRLS--LDISAKHLTTKLEEIYNKQKNKKGIYIIAHSMGGLV 368
>PDAT_SCHPO (O94680) Phospholipid:diacylglycerol acyltransferase (EC
2.3.1.158) (PDAT) (Pombe LRO1 homolog 1)
Length = 623
Score = 36.6 bits (83), Expect = 0.17
Identities = 49/171 (28%), Positives = 74/171 (42%), Gaps = 27/171 (15%)
Query: 34 PVLLVPGVGGSILNAVNESDGS----QERVWVRF--LSAEYKLKTKLWSRY---DPSTGK 84
PV++VPGV S L + + ++ S ++R+W + L A + L + W + D TG
Sbjct: 147 PVIMVPGVISSGLESWSFNNCSIPYFRKRLWGSWSMLKAMF-LDKQCWLEHLMLDKKTG- 204
Query: 85 TVTLDQKSRIVVPEDRHGLHAIDVLDPDLVIGSEAVYYFHDMIVQMQKWGYQEGKTLFGF 144
LD K + G A D I S+ +I + GY E +
Sbjct: 205 ---LDPKG--IKLRAAQGFEAADFFITGYWIWSK-------VIENLAAIGY-EPNNMLSA 251
Query: 145 GYDFRQSNRLQETMDRFAEKLELIY---NAAGGKKIDLISHSMGGLLVKCF 192
YD+R S E D++ KL++ N KK+ LISHSMG + F
Sbjct: 252 SYDWRLSYANLEERDKYFSKLKMFIEYSNIVHKKKVVLISHSMGSQVTYYF 302
>PDAT_YEAST (P40345) Phospholipid:diacylglycerol acyltransferase (EC
2.3.1.158) (PDAT)
Length = 661
Score = 36.2 bits (82), Expect = 0.22
Identities = 46/176 (26%), Positives = 79/176 (44%), Gaps = 31/176 (17%)
Query: 34 PVLLVPGVGGSILNAV-----NESDGS---QERVWVRFLSAEYKLKTKL-WSRY---DPS 81
PV++VPGV + + + +E D S ++R+W F + K+ W ++ DP
Sbjct: 174 PVVMVPGVISTGIESWGVIGDDECDSSAHFRKRLWGSFYMLRTMVMDKVCWLKHVMLDPE 233
Query: 82 TGKTVTLDQKSRIVVPEDRHGLHAIDVLDPDLVIGSEAVYYFHDMIVQ-MQKWGYQEGKT 140
TG LD + + G + D A Y+ + + Q + GY+ K
Sbjct: 234 TG----LDPPNFTL--RAAQGFESTDYFI--------AGYWIWNKVFQNLGVIGYEPNK- 278
Query: 141 LFGFGYDFRQSNRLQETMDRFAEKLEL---IYNAAGGKKIDLISHSMGGLLVKCFM 193
+ YD+R + E DR+ KL+ +++ G+K+ LI HSMG ++ FM
Sbjct: 279 MTSAAYDWRLAYLDLERRDRYFTKLKEQIELFHQLSGEKVCLIGHSMGSQIIFYFM 334
>LCAT_TATKG (O35840) Phosphatidylcholine-sterol acyltransferase (EC
2.3.1.43) (Lecithin-cholesterol acyltransferase)
(Phospholipid-cholesterol acyltransferase) (Fragment)
Length = 293
Score = 35.8 bits (81), Expect = 0.29
Identities = 23/73 (31%), Positives = 39/73 (52%), Gaps = 2/73 (2%)
Query: 122 YFHDMIVQMQKWGYQEGKTLFGFGYDFR-QSNRLQETMDRFAEKLELIYNAAGGKKIDLI 180
Y H ++ + GY +T+ YD+R + ++ + + A +E +Y AA GK + LI
Sbjct: 62 YMHTLVQNLVNNGYVRDETVRAAPYDWRLEPSQQDDYYQKLAGLIEEMY-AAYGKPVFLI 120
Query: 181 SHSMGGLLVKCFM 193
HS+G L V F+
Sbjct: 121 GHSLGCLHVLYFL 133
Score = 35.0 bits (79), Expect = 0.49
Identities = 33/118 (27%), Positives = 50/118 (41%), Gaps = 21/118 (17%)
Query: 293 YPPRDSIEIFKQALVNNKVNHEGEELPLPFNSHIFE-----WANKTREILSSAKLPSGVK 347
+P RD + + N+ G++ F+ FE W ++R++L+ P GV
Sbjct: 160 FPDRDVWPEDHVFISTPEFNYTGQDFERFFSDLHFEEGWYMWL-QSRDLLAGLPAP-GVD 217
Query: 348 FYNIYGTNLATPHSICYGN---ADKPVSDLQELRYLQARYVCVDGDGTVPVESAKADG 402
Y +YG L TPH+ Y + PV+ L E DGD TV S + G
Sbjct: 218 VYCLYGVGLPTPHTYIYDHNFPYKDPVAALYE-----------DGDDTVATRSTELCG 264
>PHBC_RHIET (Q52728) Poly-beta-hydroxybutyrate polymerase (EC
2.3.1.-) (Poly(3-hydroxybutyrate) polymerase) (PHB
polymerase) (PHB synthase) (Poly(3-hydroxyalkanoate)
polymerase) (PHA polymerase) (PHA synthase)
(Polyhydroxyalkanoic acid synthase)
Length = 636
Score = 34.7 bits (78), Expect = 0.64
Identities = 22/92 (23%), Positives = 42/92 (44%), Gaps = 7/92 (7%)
Query: 121 YYFHDMIVQMQ--KWGYQEGKTLFGFGYDFRQSNRLQETMDRFAEK-----LELIYNAAG 173
+Y D+ Q KW +G+T+F + ++ +A + LE I A G
Sbjct: 304 FYILDLNPQKSFIKWCVDQGQTVFVISWVNPDGRHAEKDWAAYAREGIDFALETIEKATG 363
Query: 174 GKKIDLISHSMGGLLVKCFMTLHSDIFEKYVK 205
K+++ + + +GG L+ + LH+ K +K
Sbjct: 364 EKEVNAVGYCVGGTLLAATLALHAKEKNKRIK 395
>PT1_BORBU (O51508) Phosphoenolpyruvate-protein phosphotransferase
(EC 2.7.3.9) (Phosphotransferase system, enzyme I)
Length = 573
Score = 34.3 bits (77), Expect = 0.84
Identities = 24/93 (25%), Positives = 44/93 (46%), Gaps = 1/93 (1%)
Query: 284 GKSHVILESYPPRDSIEIFKQALVNNKVNHEGEELPLPFNSHIFEWANKTREILSSAKLP 343
GK V++ + IE + + N K+N + LP N + L S+KL
Sbjct: 386 GKIRVMVPMLTIYEEIETIEYFVNNAKINLKSRGLPFDENLEVGCMIEVPSAALISSKLA 445
Query: 344 SGVKFYNIYGTNLATPHSICYGNADKPVSDLQE 376
+ +KF++I GTN T + + ++ +S+L +
Sbjct: 446 NKLKFFSI-GTNDLTQYVLAVDRGNQKISNLYD 477
>PHBC_RHIME (P50176) Poly-beta-hydroxybutyrate polymerase (EC
2.3.1.-) (Poly(3-hydroxybutyrate) polymerase) (PHB
polymerase) (PHB synthase) (Poly(3-hydroxyalkanoate)
polymerase) (PHA polymerase) (PHA synthase)
(Polyhydroxyalkanoic acid synthase)
Length = 611
Score = 33.5 bits (75), Expect = 1.4
Identities = 16/71 (22%), Positives = 35/71 (48%), Gaps = 5/71 (7%)
Query: 132 KWGYQEGKTLFGFGYDFRQSNRLQETMDRFAEK-----LELIYNAAGGKKIDLISHSMGG 186
KW +G+T+F + + + +A + L++I A G ++++ I + +GG
Sbjct: 293 KWAVDQGQTVFVISWVNPDERHASKDWEAYAREGIGFALDIIEQATGEREVNSIGYCVGG 352
Query: 187 LLVKCFMTLHS 197
L+ + LH+
Sbjct: 353 TLLAATLALHA 363
>RRPL_SV5WR (Q03396) RNA polymerase beta subunit (EC 2.7.7.48)
(Large structural protein) (L protein)
Length = 2255
Score = 32.3 bits (72), Expect = 3.2
Identities = 20/68 (29%), Positives = 32/68 (46%), Gaps = 9/68 (13%)
Query: 266 HWKHVPLLELWR-------ERLHEDGKSHVILESYPPRDSIEIFKQALVNNKVNHEGEEL 318
HWK V L++ + E L K I S P +D + +F+++L+ + H L
Sbjct: 449 HWKEVSLIKFKKCFDADAGEELSIFMKDKAI--SAPKQDWMSVFRRSLIKQRHQHHQVPL 506
Query: 319 PLPFNSHI 326
P PFN +
Sbjct: 507 PNPFNRRL 514
>RRPL_SV5 (Q88434) RNA polymerase beta subunit (EC 2.7.7.48) (Large
structural protein) (L protein)
Length = 2255
Score = 32.3 bits (72), Expect = 3.2
Identities = 20/68 (29%), Positives = 32/68 (46%), Gaps = 9/68 (13%)
Query: 266 HWKHVPLLELWR-------ERLHEDGKSHVILESYPPRDSIEIFKQALVNNKVNHEGEEL 318
HWK V L++ + E L K I S P +D + +F+++L+ + H L
Sbjct: 449 HWKEVSLIKFKKCFDADAGEELSIFMKDKAI--SAPKQDWMSVFRRSLIKQRHQHHQVPL 506
Query: 319 PLPFNSHI 326
P PFN +
Sbjct: 507 PNPFNRRL 514
>YE63_SCHPO (O14249) Hypothetical protein C6G10.03c in chromosome I
Length = 428
Score = 31.6 bits (70), Expect = 5.4
Identities = 18/52 (34%), Positives = 27/52 (51%)
Query: 151 SNRLQETMDRFAEKLELIYNAAGGKKIDLISHSMGGLLVKCFMTLHSDIFEK 202
S +++ET F E LE G +K+ L+ HSMGG L + + + EK
Sbjct: 148 SEKVEETERFFTESLETWRIGHGIEKMILVGHSMGGYLSAVYAMQYPERVEK 199
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.318 0.137 0.419
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 70,840,709
Number of Sequences: 164201
Number of extensions: 3268295
Number of successful extensions: 7350
Number of sequences better than 10.0: 30
Number of HSP's better than 10.0 without gapping: 4
Number of HSP's successfully gapped in prelim test: 26
Number of HSP's that attempted gapping in prelim test: 7316
Number of HSP's gapped (non-prelim): 52
length of query: 538
length of database: 59,974,054
effective HSP length: 115
effective length of query: 423
effective length of database: 41,090,939
effective search space: 17381467197
effective search space used: 17381467197
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 68 (30.8 bits)
Medicago: description of AC149576.2


