

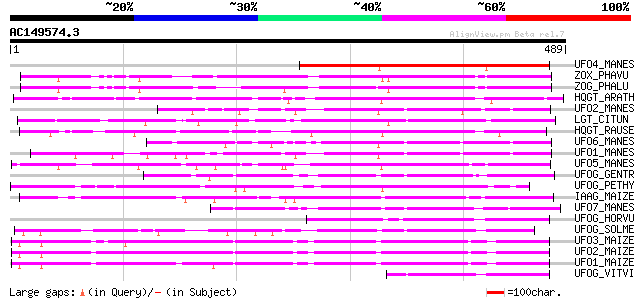
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC149574.3 + phase: 0
(489 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UFO4_MANES (Q40286) Flavonol 3-O-glucosyltransferase 4 (EC 2.4.1... 202 2e-51
ZOX_PHAVU (P56725) Zeatin O-xylosyltransferase (EC 2.4.2.40) (Ze... 195 3e-49
ZOG_PHALU (Q9ZSK5) Zeatin O-glucosyltransferase (EC 2.4.1.203) (... 192 2e-48
HQGT_ARATH (Q9M156) Probable hydroquinone glucosyltransferase (E... 186 1e-46
UFO2_MANES (Q40285) Flavonol 3-O-glucosyltransferase 2 (EC 2.4.1... 184 4e-46
LGT_CITUN (Q9MB73) Limonoid UDP-glucosyltransferase (EC 2.4.1.21... 182 1e-45
HQGT_RAUSE (Q9AR73) Hydroquinone glucosyltransferase (EC 2.4.1.2... 180 6e-45
UFO6_MANES (Q40288) Flavonol 3-O-glucosyltransferase 6 (EC 2.4.1... 177 4e-44
UFO1_MANES (Q40284) Flavonol 3-O-glucosyltransferase 1 (EC 2.4.1... 170 9e-42
UFO5_MANES (Q40287) Flavonol 3-O-glucosyltransferase 5 (EC 2.4.1... 168 3e-41
UFOG_GENTR (Q96493) Flavonol 3-O-glucosyltransferase (EC 2.4.1.9... 152 2e-36
UFOG_PETHY (Q43716) Flavonol 3-O-glucosyltransferase (EC 2.4.1.9... 141 3e-33
IAAG_MAIZE (Q41819) Indole-3-acetate beta-glucosyltransferase (E... 141 3e-33
UFO7_MANES (Q40289) Flavonol 3-O-glucosyltransferase 7 (EC 2.4.1... 138 4e-32
UFOG_HORVU (P14726) Flavonol 3-O-glucosyltransferase (EC 2.4.1.9... 132 2e-30
UFOG_SOLME (Q43641) Flavonol 3-O-glucosyltransferase (EC 2.4.1.9... 131 4e-30
UFO3_MAIZE (P16167) Flavonol 3-O-glucosyltransferase (EC 2.4.1.9... 128 4e-29
UFO2_MAIZE (P16165) Flavonol 3-O-glucosyltransferase (EC 2.4.1.9... 124 4e-28
UFO1_MAIZE (P16166) Flavonol 3-O-glucosyltransferase (EC 2.4.1.9... 123 1e-27
UFOG_VITVI (P51094) Flavonol 3-O-glucosyltransferase (EC 2.4.1.9... 92 4e-18
>UFO4_MANES (Q40286) Flavonol 3-O-glucosyltransferase 4 (EC
2.4.1.91) (UDP-glucose flavonoid 3-O-glucosyltransferase
4) (Fragment)
Length = 241
Score = 202 bits (514), Expect = 2e-51
Identities = 112/230 (48%), Positives = 152/230 (65%), Gaps = 10/230 (4%)
Query: 256 NKEEEIPSYRGKEASIDKHECLKWLDTKNINSVVYMCFGSMTHFLNSQLKEIAMGLEASG 315
NK + + RG +AS+D E LKWLD SV+Y C GS++ + QL E+ +GLE++
Sbjct: 2 NKLKLDKAERGDKASVDNTELLKWLDLWEPGSVIYACLGSISGLTSWQLAELGLGLESTN 61
Query: 316 HNFIWVVRT--QTEDGDEW-LPEGFEERTEGK-GLIIRGWSPQVMILEHEAIGAFVTHCG 371
FIWV+R ++E ++W L EG+EER + IRGWSPQV+IL H AIGAF THCG
Sbjct: 62 QPFIWVIREGEKSEGLEKWILEEGYEERKRKREDFWIRGWSPQVLILSHPAIGAFFTHCG 121
Query: 372 WNSVLEGVVAGVPMITWPVAAEQFYNEKLVTEVLKTGVPVGVKKWVM-----KVGDNVEW 426
WNS LEG+ AGVP++ P+ AEQFYNEKLV EVL GV VGV+ V K G ++
Sbjct: 122 WNSTLEGISAGVPIVACPLFAEQFYNEKLVVEVLGIGVSVGVEAAVTWGLEDKCGAVMKK 181
Query: 427 DAVEKAVKRVME-GEEAYEMRNKAKMLAEMAKKAVEEDGSSYSQLNALIE 475
+ V+KA++ VM+ G+E E R +A+ + EMAK+ +EE GSSY + LI+
Sbjct: 182 EQVKKAIEIVMDKGKEGEERRRRAREIGEMAKRTIEEGGSSYLDMEMLIQ 231
>ZOX_PHAVU (P56725) Zeatin O-xylosyltransferase (EC 2.4.2.40)
(Zeatin O-beta-D-xylosyltransferase)
Length = 454
Score = 195 bits (495), Expect = 3e-49
Identities = 147/484 (30%), Positives = 229/484 (46%), Gaps = 58/484 (11%)
Query: 10 ILVFPFMGHGHTIPTIDMAKLFASKGVRVTIV--TTPLNKPPISKALEQSKIHFNNIDIQ 67
+L+ PF GH P + ++ L A++ + V V T + + + S IHF+ ++
Sbjct: 11 VLLLPFPVQGHLNPFLQLSHLIAAQNIAVHYVGTVTHIRQAKLRYHNATSNIHFHAFEVP 70
Query: 68 TIKFPCVEAGLPEGCENVDSIPSVSFVPAFFAAIRLLQQPFEELLL----QQKPHCVVAD 123
P P E D PS +P+F A+ L ++P +LL Q K ++ D
Sbjct: 71 PYVSP------PPNPE--DDFPS-HLIPSFEASAHL-REPVGKLLQSLSSQAKRVVLIND 120
Query: 124 MFFPWATDSAAKFG-IPRIVFHGTSFFSLCASQCMKKYQPYKNVSSDTDLFEITDLPGNI 182
AA F + R F Q + +++ D +E P
Sbjct: 121 SLMASVAQDAANFSNVERYCF-----------------QVFSALNTAGDFWEQMGKP--- 160
Query: 183 KMTRLQLPNTLTENDPISQSFAKLFEEIKDSEVRSYGVIVNSFYELENVYADYYREVLGI 242
+ P+ + IS F + + G I N+ +E Y + G
Sbjct: 161 PLADFHFPDIPSLQGCISAQFTDFLTAQNEFRKFNNGDIYNTSRVIEGPYVELLERFNGG 220
Query: 243 KE-WHIGPFSIHNRNKEEEIPSYRGKEASIDKHECLKWLDTKNINSVVYMCFGSMTHFLN 301
KE W +GPF+ K++ I H C++WLD + +SV+Y+ FG+ T +
Sbjct: 221 KEVWALGPFTPLAVEKKDSIGF---------SHPCMEWLDKQEPSSVIYVSFGTTTALRD 271
Query: 302 SQLKEIAMGLEASGHNFIWVVRTQTE----DGDEW----LPEGFEERTEGKGLIIRGWSP 353
Q++E+A GLE S FIWV+R + DG E LPEGFEER EG GL++R W+P
Sbjct: 272 EQIQELATGLEQSKQKFIWVLRDADKGDIFDGSEAKRYELPEGFEERVEGMGLVVRDWAP 331
Query: 354 QVMILEHEAIGAFVTHCGWNSVLEGVVAGVPMITWPVAAEQFYNEKLVTEVLKTGVPVGV 413
Q+ IL H + G F++HCGWNS LE + GVPM TW + ++Q N LVT+VLK G+ V
Sbjct: 332 QMEILSHSSTGGFMSHCGWNSCLESLTRGVPMATWAMHSDQPRNAVLVTDVLKVGLI--V 389
Query: 414 KKWVMKVGDNVEWDAVEKAVKRVMEGEEAYEMRNKAKMLAEMAKKAVEEDGSSYSQLNAL 473
K W + V +E AV+R+ME +E E+R +A L + ++++E G S ++ +
Sbjct: 390 KDWEQR-KSLVSASVIENAVRRLMETKEGDEIRKRAVKLKDEIHRSMDEGGVSRMEMASF 448
Query: 474 IEEL 477
I +
Sbjct: 449 IAHI 452
>ZOG_PHALU (Q9ZSK5) Zeatin O-glucosyltransferase (EC 2.4.1.203)
(Trans-zeatin O-beta-D-glucosyltransferase)
Length = 459
Score = 192 bits (487), Expect = 2e-48
Identities = 146/489 (29%), Positives = 224/489 (44%), Gaps = 68/489 (13%)
Query: 10 ILVFPFMGHGHTIPTIDMAKLFASKGVRVTIV--TTPLNKPPISKALEQSKIHFNNIDIQ 67
+L+ PF GH + +++L ++ + V V T + + + S IHF+ +
Sbjct: 16 VLLIPFPAQGHLNQFLHLSRLIVAQNIPVHYVGTVTHIRQATLRYNNPTSNIHFHAFQVP 75
Query: 68 TIKFPCVEAGLPEGCENVDSIPSVSFVPAFFAAIRLLQQPFEELLL----QQKPHCVVAD 123
P P E D PS +P+F A+ L ++P +LL Q K V+ D
Sbjct: 76 PFVSP------PPNPE--DDFPS-HLIPSFEASAHL-REPVGKLLQSLSSQAKRVVVIND 125
Query: 124 MFFPWATDSAAKFG-IPRIVFHGTSFFSLCASQCMKKYQPYKNVSSDTDLFEITDLPGNI 182
AA + FH S F+ + +P D E L G I
Sbjct: 126 SLMASVAQDAANISNVENYTFHSFSAFNTSGDFWEEMGKP---PVGDFHFPEFPSLEGCI 182
Query: 183 KMTRLQLPNTLTENDPISQSFAKLFEEIKDSEVRSYGVIVNSFYELENVYADYYREVL-- 240
A F+ + ++ Y V Y E+L
Sbjct: 183 ---------------------AAQFKGFRTAQYEFRKFNNGDIYNTSRVIEGPYVELLEL 221
Query: 241 ---GIKEWHIGPFSIHNRNKEEEIPSYRGKEASIDKHECLKWLDTKNINSVVYMCFGSMT 297
G K W +GPF+ K++ I +H C++WLD + +SV+Y+ FG+ T
Sbjct: 222 FNGGKKVWALGPFNPLAVEKKDSIGF---------RHPCMEWLDKQEPSSVIYISFGTTT 272
Query: 298 HFLNSQLKEIAMGLEASGHNFIWVVRTQTEDGDEW---------LPEGFEERTEGKGLII 348
+ Q+++IA GLE S FIWV+R + + GD + LP+GFEER EG GL++
Sbjct: 273 ALRDEQIQQIATGLEQSKQKFIWVLR-EADKGDIFAGSEAKRYELPKGFEERVEGMGLVV 331
Query: 349 RGWSPQVMILEHEAIGAFVTHCGWNSVLEGVVAGVPMITWPVAAEQFYNEKLVTEVLKTG 408
R W+PQ+ IL H + G F++HCGWNS LE + GVP+ TWP+ ++Q N LVTEVLK G
Sbjct: 332 RDWAPQLEILSHSSTGGFMSHCGWNSCLESITMGVPIATWPMHSDQPRNAVLVTEVLKVG 391
Query: 409 VPVGVKKWVMKVGDNVEWDAVEKAVKRVMEGEEAYEMRNKAKMLAEMAKKAVEEDGSSYS 468
+ VK W + V VE V+R+ME +E EMR +A L ++++E G S+
Sbjct: 392 LV--VKDWAQR-NSLVSASVVENGVRRLMETKEGDEMRQRAVRLKNAIHRSMDEGGVSHM 448
Query: 469 QLNALIEEL 477
++ + I +
Sbjct: 449 EMGSFIAHI 457
>HQGT_ARATH (Q9M156) Probable hydroquinone glucosyltransferase (EC
2.4.1.218) (Arbutin synthase)
Length = 480
Score = 186 bits (472), Expect = 1e-46
Identities = 154/506 (30%), Positives = 249/506 (48%), Gaps = 53/506 (10%)
Query: 4 QSNPLHILVFPFMGHGHTIPTIDMAK-LFASKGVRVTIVTTPLNKPPISKALEQSKIHFN 62
+S H+ + P G GH IP ++ AK L G+ VT V + P SKA +++ +
Sbjct: 3 ESKTPHVAIIPSPGMGHLIPLVEFAKRLVHLHGLTVTFVIA--GEGPPSKA-QRTVLDSL 59
Query: 63 NIDIQTIKFPCVEAGLPEGCENVDSIPSVSFVPAFFAAIRLLQQPFEELLLQQKPHCVVA 122
I ++ P V+ ++S S++ V +R + F E + P +V
Sbjct: 60 PSSISSVFLPPVDLTDLSSSTRIESRISLT-VTRSNPELRKVFDSFVEG--GRLPTALVV 116
Query: 123 DMFFPWATDSAAKFGIPRIVFHGTSFFSLCASQCMKKYQPYKNVSSD-TDLFEITDLPGN 181
D+F A D A +F +P +F+ T+ L + K + VS + +L E LPG
Sbjct: 117 DLFGTDAFDVAVEFHVPPYIFYPTTANVLSFFLHLPKLD--ETVSCEFRELTEPLMLPGC 174
Query: 182 IKMTRLQ-LPNTLTENDPISQSFAKLFEEIKDSEVRSYGVIVNSFYELENVYADYYREVL 240
+ + L D + + K++E G++VN+F+ELE +E
Sbjct: 175 VPVAGKDFLDPAQDRKDDAYKWLLHNTKRYKEAE----GILVNTFFELEPNAIKALQEP- 229
Query: 241 GIKE---WHIGPFSIHNRNKEEEIPSYRGKEASIDKHECLKWLDTKNINSVVYMCFGSMT 297
G+ + + +GP + N K+E ++ ECLKWLD + + SV+Y+ FGS
Sbjct: 230 GLDKPPVYPVGP--LVNIGKQEA--------KQTEESECLKWLDNQPLGSVLYVSFGSGG 279
Query: 298 HFLNSQLKEIAMGLEASGHNFIWVVRTQT-------------EDGDEWLPEGFEERTEGK 344
QL E+A+GL S F+WV+R+ + D +LP GF ERT+ +
Sbjct: 280 TLTCEQLNELALGLADSEQRFLWVIRSPSGIANSSYFDSHSQTDPLTFLPPGFLERTKKR 339
Query: 345 GLIIRGWSPQVMILEHEAIGAFVTHCGWNSVLEGVVAGVPMITWPVAAEQFYNEKLVTEV 404
G +I W+PQ +L H + G F+THCGWNS LE VV+G+P+I WP+ AEQ N L++E
Sbjct: 340 GFVIPFWAPQAQVLAHPSTGGFLTHCGWNSTLESVVSGIPLIAWPLYAEQKMNAVLLSED 399
Query: 405 LKTGVPVGVKKWVMKVGDN--VEWDAVEKAVKRVMEGEEAYEMRNKAKMLAEMAKKAVEE 462
++ + + GD+ V + V + VK +MEGEE +RNK K L E A + +++
Sbjct: 400 IRAALR-------PRAGDDGLVRREEVARVVKGLMEGEEGKGVRNKMKELKEAACRVLKD 452
Query: 463 DGSSYSQLNALIEELRSLSHHQHISQ 488
DG+S L+ + L+ +H + + Q
Sbjct: 453 DGTSTKALS--LVALKWKAHKKELEQ 476
>UFO2_MANES (Q40285) Flavonol 3-O-glucosyltransferase 2 (EC
2.4.1.91) (UDP-glucose flavonoid 3-O-glucosyltransferase
2) (Fragment)
Length = 346
Score = 184 bits (467), Expect = 4e-46
Identities = 134/370 (36%), Positives = 192/370 (51%), Gaps = 58/370 (15%)
Query: 131 DSAAKFGIPRIVFHGTSFFSLCASQCMKK------YQPYKNVSSDTDLFEITDLPGNIKM 184
D A +FGIP +F + L ++K + P + SDT+L I N
Sbjct: 7 DLADEFGIPSYIFFASGGGFLGFMLYVQKIHDEENFNPIEFKDSDTEL--IVPSLVNPFP 64
Query: 185 TRLQLPNTLTENDPISQ--SFAKLFEEIKDSEVRSYGVIVNSFYELENVYADYYREVLGI 242
TR+ LP+++ + Q + AK F + K G+IVN+F ELE+ + ++
Sbjct: 65 TRI-LPSSILNKERFGQLLAIAKKFRQAK-------GIIVNTFLELESRAIESFKVP--- 113
Query: 243 KEWHIGPF---SIHNRNKEEEIPSYRGKEASIDKHECLKWLDTKNINSVVYMCFGSMTHF 299
+H+GP RN EI ++WLD + SVV++CFGSM F
Sbjct: 114 PLYHVGPILDVKSDGRNTHPEI---------------MQWLDDQPEGSVVFLCFGSMGSF 158
Query: 300 LNSQLKEIAMGLEASGHNFIWVVR-----------TQTEDGDEWLPEGFEERTEGKGLII 348
QLKEIA LE SGH F+W +R T ED + LPEGF ERT G +I
Sbjct: 159 SEDQLKEIAYALENSGHRFLWSIRRPPPPDKIASPTDYEDPRDVLPEGFLERTVAVGKVI 218
Query: 349 RGWSPQVMILEHEAIGAFVTHCGWNSVLEGVVAGVPMITWPVAAEQFYN--EKLVTEVLK 406
GW+PQV +L H AIG FV+HCGWNSVLE + GVP+ TWP+ AEQ +N E +V L
Sbjct: 219 -GWAPQVAVLAHPAIGGFVSHCGWNSVLESLWFGVPIATWPMYAEQQFNAFEMVVELGLG 277
Query: 407 TGVPVGVKKWVMKVGDNVEWDAVEKAVKRVMEGEEAYEMRNKAKMLAEMAKKAVEEDGSS 466
+ +G +K + G V D +E+A++++ME + E R K K + E +K A+ + GSS
Sbjct: 278 VEIDMGYRK---ESGIIVNSDKIERAIRKLMENSD--EKRKKVKEMREKSKMALIDGGSS 332
Query: 467 YSQLNALIEE 476
+ L I++
Sbjct: 333 FISLGDFIKD 342
>LGT_CITUN (Q9MB73) Limonoid UDP-glucosyltransferase (EC 2.4.1.210)
(Limonoid glucosyltransferase) (Limonoid GTase) (LGTase)
Length = 511
Score = 182 bits (463), Expect = 1e-45
Identities = 133/490 (27%), Positives = 234/490 (47%), Gaps = 44/490 (8%)
Query: 8 LHILVFPFMGHGHTIPTIDMAKLFASKGVRVTIVTTPLNKPPISKALEQSKIHFNNIDIQ 67
+H+L+ F GHGH P + + +L ASKG +T+ TTP + + +
Sbjct: 7 VHVLLVSFPGHGHVNPLLRLGRLLASKGFFLTL-TTPESFGKQMRKAGNFTYEPTPVGDG 65
Query: 68 TIKFPCVEAGLPEGCENVDSIPSVSFVPAFFAAIRLLQQPFEELLLQQKPH------CVV 121
I+F E G E P + + A + L+ + ++++ C++
Sbjct: 66 FIRFEFFEDGWDE------DDPRREDLDQYMAQLELIGKQVIPKIIKKSAEEYRPVSCLI 119
Query: 122 ADMFFPWATDSAAKFGIPRIVFHGTSFFSLCASQCMKKYQPYKN----VSSDTDLFEITD 177
+ F PW +D A G+P + S CA C Y Y + S+ +
Sbjct: 120 NNPFIPWVSDVAESLGLPSAMLWVQS----CA--CFAAYYHYFHGLVPFPSEKEPEIDVQ 173
Query: 178 LPGNIKMTRLQLPNTLTENDP---ISQSFAKLFEEIKDSEVRSYGVIVNSFYELENVYAD 234
LP + ++P+ L + P + ++ +E + + + +++++FYELE D
Sbjct: 174 LPCMPLLKHDEMPSFLHPSTPYPFLRRAILGQYENLG----KPFCILLDTFYELEKEIID 229
Query: 235 YYREVLGIKEWHIGPFSIHNRNKEEEIPSYRGKEASIDKHECLKWLDTKNINSVVYMCFG 294
Y ++ IK +GP K + P+ ++ + EC+ WLD K +SVVY+ FG
Sbjct: 230 YMAKICPIKP--VGPLF-----KNPKAPTLTVRDDCMKPDECIDWLDKKPPSSVVYISFG 282
Query: 295 SMTHFLNSQLKEIAMGLEASGHNFIWVVRTQTEDGDEW---LPEGFEERTEGKGLIIRGW 351
++ + Q++EI L SG +F+WV++ ED LP+GF E+ KG +++ W
Sbjct: 283 TVVYLKQEQVEEIGYALLNSGISFLWVMKPPPEDSGVKIVDLPDGFLEKVGDKGKVVQ-W 341
Query: 352 SPQVMILEHEAIGAFVTHCGWNSVLEGVVAGVPMITWPVAAEQFYNEKLVTEVLKTGVPV 411
SPQ +L H ++ FVTHCGWNS +E + +GVP+IT+P +Q + + +V KTG+ +
Sbjct: 342 SPQEKVLAHPSVACFVTHCGWNSTMESLASGVPVITFPQWGDQVTDAMYLCDVFKTGLRL 401
Query: 412 GVKKWVMKVGDNVEWDAVEKAVKRVMEGEEAYEMRNKAKMLAEMAKKAVEEDGSSYSQLN 471
+ ++ + D VEK + G +A + A + A++AV + GSS +
Sbjct: 402 CRGEAENRI---ISRDEVEKCLLEATAGPKAVALEENALKWKKEAEEAVADGGSSDRNIQ 458
Query: 472 ALIEELRSLS 481
A ++E+R S
Sbjct: 459 AFVDEVRRTS 468
>HQGT_RAUSE (Q9AR73) Hydroquinone glucosyltransferase (EC 2.4.1.218)
(Arbutin synthase)
Length = 470
Score = 180 bits (457), Expect = 6e-45
Identities = 151/497 (30%), Positives = 228/497 (45%), Gaps = 75/497 (15%)
Query: 9 HILVFPFMGHGHTIPTIDMAKLFASK---GVRVTIVTTPLNKPPISKALEQSKIHFNNID 65
HI + P G GH IP ++ AK + GV I T P+ KA ++S +
Sbjct: 6 HIAMVPTPGMGHLIPLVEFAKRLVLRHNFGVTFIIPTDG----PLPKA-QKSFLDALPAG 60
Query: 66 IQTIKFPCVEAGLPEGCENVDSIPSVSFVPAFFAAIRLLQQPF-----EELLLQQKPHCV 120
+ + P V + D +P+ + PF + LL K +
Sbjct: 61 VNYVLLPPV---------SFDDLPADVRIETRICLTITRSLPFVRDAVKTLLATTKLAAL 111
Query: 121 VADMFFPWATDSAAKFGIPRIVFHGTSFFSLCASQCMKKYQPYKNVSSD-TDLFEITDLP 179
V D+F A D A +F + +F+ T+ ++C S + + VS + D+ E +P
Sbjct: 112 VVDLFGTDAFDVAIEFKVSPYIFYPTT--AMCLSLFFHLPKLDQMVSCEYRDVPEPLQIP 169
Query: 180 GNIKMTRLQLPNTLTENDPISQSFAKLFEEIKDSEVRSYGVIVNSFYELENVYADYYREV 239
G I + + D + ++ L + K + G++VN+F +LE
Sbjct: 170 GCIPIHGKDFLDPA--QDRKNDAYKCLLHQAKRYRLAE-GIMVNTFNDLEP--------- 217
Query: 240 LGIKEWHIGPFSIHNRNKEEEIPSY-------RGKEASIDKHECLKWLDTKNINSVVYMC 292
GP + + P Y + +D ECLKWLD + SV+++
Sbjct: 218 --------GPLKALQEEDQGKPPVYPIGPLIRADSSSKVDDCECLKWLDDQPRGSVLFIS 269
Query: 293 FGSMTHFLNSQLKEIAMGLEASGHNFIWVVRTQTE--------------DGDEWLPEGFE 338
FGS ++Q E+A+GLE S F+WVVR+ + D +LPEGF
Sbjct: 270 FGSGGAVSHNQFIELALGLEMSEQRFLWVVRSPNDKIANATYFSIQNQNDALAYLPEGFL 329
Query: 339 ERTEGKGLIIRGWSPQVMILEHEAIGAFVTHCGWNSVLEGVVAGVPMITWPVAAEQFYNE 398
ERT+G+ L++ W+PQ IL H + G F+THCGWNS+LE VV GVP+I WP+ AEQ N
Sbjct: 330 ERTKGRCLLVPSWAPQTEILSHGSTGGFLTHCGWNSILESVVNGVPLIAWPLYAEQKMNA 389
Query: 399 KLVTEVLKTGVPVGVKKWVMKVGDNVEWDAVE--KAVKRVMEGEEAYEMRNKAKMLAEMA 456
++TE LK + K G+N VE AVK +MEGEE + R+ K L + A
Sbjct: 390 VMLTEGLKVALR-------PKAGENGLIGRVEIANAVKGLMEGEEGKKFRSTMKDLKDAA 442
Query: 457 KKAVEEDGSSYSQLNAL 473
+A+ +DGSS L L
Sbjct: 443 SRALSDDGSSTKALAEL 459
>UFO6_MANES (Q40288) Flavonol 3-O-glucosyltransferase 6 (EC
2.4.1.91) (UDP-glucose flavonoid 3-O-glucosyltransferase
6) (Fragment)
Length = 394
Score = 177 bits (450), Expect = 4e-44
Identities = 131/380 (34%), Positives = 197/380 (51%), Gaps = 48/380 (12%)
Query: 121 VADMFFPWATDSAAKFGIPRIVFHGTSFFSLCASQCMKKYQPYKNVSSDTDLFEITDLPG 180
V DMF D A + G+P +F F S A Y + D DL + D
Sbjct: 35 VLDMFCTSMIDVAKELGVPYYIF----FTSGAAFLGFLFYVQLIHDEQDADLTQFKDSDA 90
Query: 181 NIKMTRLQ-------LPNTLTENDPISQSFAKLFEEIKDSEVRSYGVIVNSFYELE---- 229
+ + L LP ++ D +F ++ +++++ G++VN+F ELE
Sbjct: 91 ELSVPSLANSLPARVLPASMLVKDRF-YAFIRIIRGLREAK----GIMVNTFMELESHAL 145
Query: 230 NVYADYYREVLGIKEWHIGPFSIHNRNKEEEIPSYRGKEASIDKHECLKWLDTKNINSVV 289
N D ++ I + +GP + N+E ++ G E S E ++WLD + +SVV
Sbjct: 146 NSLKDDQSKIPPI--YPVGPI-LKLSNQENDV----GPEGS----EIIEWLDDQPPSSVV 194
Query: 290 YMCFGSMTHFLNSQLKEIAMGLEASGHNFIWVVR-----------TQTEDGDEWLPEGFE 338
++CFGSM F Q KEIA LE S H F+W +R T E+ E LP GF
Sbjct: 195 FLCFGSMGGFDMDQAKEIACALEQSRHRFLWSLRRPPPKGKIETSTDYENLQEILPVGFS 254
Query: 339 ERTEGKGLIIRGWSPQVMILEHEAIGAFVTHCGWNSVLEGVVAGVPMITWPVAAEQFYNE 398
ERT G G ++ GW+PQV ILEH AIG FV+HCGWNS+LE + VP+ TWP+ AEQ +N
Sbjct: 255 ERTAGMGKVV-GWAPQVAILEHPAIGGFVSHCGWNSILESIWFSVPIATWPLYAEQQFN- 312
Query: 399 KLVTEVLKTGVPVGVKKWVMKVGDNV-EWDAVEKAVKRVMEGEEAYEMRNKAKMLAEMAK 457
T V + G+ V +K K + + D +E+ +K VME E+R + K +++ ++
Sbjct: 313 -AFTMVTELGLAVEIKMDYKKESEIILSADDIERGIKCVMEHHS--EIRKRVKEMSDKSR 369
Query: 458 KAVEEDGSSYSQLNALIEEL 477
KA+ +D SS L+ LIE++
Sbjct: 370 KALMDDESSSFWLDRLIEDV 389
>UFO1_MANES (Q40284) Flavonol 3-O-glucosyltransferase 1 (EC
2.4.1.91) (UDP-glucose flavonoid 3-O-glucosyltransferase
1)
Length = 449
Score = 170 bits (430), Expect = 9e-42
Identities = 138/490 (28%), Positives = 234/490 (47%), Gaps = 75/490 (15%)
Query: 19 GHTIPTIDMAKLFASKGVRVTIVTTPLNKPPISKALEQ---SKIHFNNIDIQTIKFPCVE 75
GH + ++ AKL S+ ++I N ++ + S+I ++ ++ I P E
Sbjct: 2 GHLVSAVETAKLLLSRCHSLSITVLIFNNSVVTSKVHNYVDSQIASSSNRLRFIYLPRDE 61
Query: 76 AGLPEGCENVDSIP-----SVSFVPAFFAAIRLLQQPFEELLLQQKPHCV--VADMFFPW 128
G+ ++ SV + F +++ + P V + DMF
Sbjct: 62 TGISSFSSLIEKQKPHVKESVMKITEFGSSV-------------ESPRLVGFIVDMFCTA 108
Query: 129 ATDSAAKFGIPRIVFH--GTSFFSLCAS----QCMKKYQPYKNVSSDTDLFEITDLPGNI 182
D A +FG+P +F+ G +F + + + P + +SD +L +PG +
Sbjct: 109 MIDVANEFGVPSYIFYTSGAAFLNFMLHVQKIHDEENFNPTEFNASDGEL----QVPGLV 164
Query: 183 KMTRLQLPNTLTENDPISQSFAKLFEEIKDSEVRSYGVIVNSFYELENVYADYYREVLGI 242
P+ +S+ + E + GVI+N+F+ELE+ + +++
Sbjct: 165 N----SFPSKAMPTAILSKQWFPPLLENTRRYGEAKGVIINTFFELESHAIESFKDP--- 217
Query: 243 KEWHIGPF---SIHNRNKEEEIPSYRGKEASIDKHECLKWLDTKNINSVVYMCFGSMTHF 299
+ +GP + RN +EI ++WLD + +SVV++CFGS F
Sbjct: 218 PIYPVGPILDVRSNGRNTNQEI---------------MQWLDDQPPSSVVFLCFGSNGSF 262
Query: 300 LNSQLKEIAMGLEASGHNFIWVVR-----------TQTEDGDEWLPEGFEERTEGKGLII 348
Q+KEIA LE SGH F+W + + ED E LPEGF ERT G +I
Sbjct: 263 SKDQVKEIACALEDSGHRFLWSLADHRAPGFLESPSDYEDLQEVLPEGFLERTSGIEKVI 322
Query: 349 RGWSPQVMILEHEAIGAFVTHCGWNSVLEGVVAGVPMITWPVAAEQFYNEKLVTEVLKTG 408
GW+PQV +L H A G V+H GWNS+LE + GVP+ TWP+ AEQ +N V++ G
Sbjct: 323 -GWAPQVAVLAHPATGGLVSHSGWNSILESIWFGVPVATWPMYAEQQFN--AFQMVIELG 379
Query: 409 VPVGVK-KWVMKVGDNVEWDAVEKAVKRVMEGEEAYEMRNKAKMLAEMAKKAVEEDGSSY 467
+ V +K + G+ V+ D +E+ ++ +M+ + + R K K ++E ++ A+ E GSSY
Sbjct: 380 LAVEIKMDYRNDSGEIVKCDQIERGIRCLMKHDS--DRRKKVKEMSEKSRGALMEGGSSY 437
Query: 468 SQLNALIEEL 477
L+ LI+++
Sbjct: 438 CWLDNLIKDM 447
>UFO5_MANES (Q40287) Flavonol 3-O-glucosyltransferase 5 (EC
2.4.1.91) (UDP-glucose flavonoid 3-O-glucosyltransferase
5)
Length = 487
Score = 168 bits (426), Expect = 3e-41
Identities = 149/508 (29%), Positives = 234/508 (45%), Gaps = 73/508 (14%)
Query: 2 DSQSNPLHILVFPFMGHGHTIPTIDMAKLFASK-GVRVTIV----TTPLNKPPISKALEQ 56
D S P HI++ G GH IP +++ K + VTI T +P + ++
Sbjct: 5 DLNSKP-HIVLLSSPGLGHLIPVLELGKRIVTLCNFDVTIFMVGSDTSAAEPQVLRSAMT 63
Query: 57 SKIHFNNIDIQTIKFPCVEAGLPEGCENVDSIPSVSFVPAFFAAIRLLQQPFEELL--LQ 114
K+ C LP + P + F +R ++ F + L+
Sbjct: 64 PKL-------------CEIIQLPPPNISCLIDPEATVCTRLFVLMREIRPAFRAAVSALK 110
Query: 115 QKPHCVVADMFFPWATDSAAKFGIPRIVFHGTSFFSLCASQCMKKYQPY--KNVSSDTDL 172
+P ++ D+F + + A + GI + V+ ++ + L + Y P K V + L
Sbjct: 111 FRPAAIIVDLFGTESLEVAKELGIAKYVYIASNAWFL----ALTIYVPILDKEVEGEFVL 166
Query: 173 -FEITDLPG--NIKMTRLQLPNTLTENDPISQSFAKLFEEIKDSEVRSYGVIVNSFYELE 229
E +PG ++ + P N S+ F +L EI ++ G+++N++ LE
Sbjct: 167 QKEPMKIPGCRPVRTEEVVDPMLDRTNQQYSEYF-RLGIEIPTAD----GILMNTWEALE 221
Query: 230 NVYADYYREV--LG----IKEWHIGPFSIHNRNKEEEIPSYRGKEASIDKHECLKWLDTK 283
R+V LG + + IGP R E L WLD +
Sbjct: 222 PTTFGALRDVKFLGRVAKVPVFPIGPLR-------------RQAGPCGSNCELLDWLDQQ 268
Query: 284 NINSVVYMCFGSMTHFLNSQLKEIAMGLEASGHNFIWVVRTQT---------------ED 328
SVVY+ FGS Q+ E+A GLE S FIWVVR T +D
Sbjct: 269 PKESVVYVSFGSGGTLSLEQMIELAWGLERSQQRFIWVVRQPTVKTGDAAFFTQGDGADD 328
Query: 329 GDEWLPEGFEERTEGKGLIIRGWSPQVMILEHEAIGAFVTHCGWNSVLEGVVAGVPMITW 388
+ PEGF R + GL++ WSPQ+ I+ H ++G F++HCGWNSVLE + AGVP+I W
Sbjct: 329 MSGYFPEGFLTRIQNVGLVVPQWSPQIHIMSHPSVGVFLSHCGWNSVLESITAGVPIIAW 388
Query: 389 PVAAEQFYNEKLVTEVLKTGVPVGVKKWVMKVGDNVEWDAVEKAVKRVMEGEEAYEMRNK 448
P+ AEQ N L+TE L GV V K K + V+ + +E+ ++R+M EE E+R +
Sbjct: 389 PIYAEQRMNATLLTEEL--GVAVRPKNLPAK--EVVKREEIERMIRRIMVDEEGSEIRKR 444
Query: 449 AKMLAEMAKKAVEEDGSSYSQLNALIEE 476
+ L + +KA+ E GSS++ ++AL E
Sbjct: 445 VRELKDSGEKALNEGGSSFNYMSALGNE 472
>UFOG_GENTR (Q96493) Flavonol 3-O-glucosyltransferase (EC 2.4.1.91)
(UDP-glucose flavonoid 3-O-glucosyltransferase)
Length = 453
Score = 152 bits (383), Expect = 2e-36
Identities = 115/368 (31%), Positives = 181/368 (48%), Gaps = 35/368 (9%)
Query: 119 CVVADMFFPWATDSAAKFGIPRIVFHGTSFFSLCASQCMKKYQPYKNVSSDTDLFE---- 174
C++ D F +A D + K G+P I + SLC + ++ ++ D+ E
Sbjct: 113 CLLTDAFLWFAADFSEKIGVPWIPVWTAASCSLCLHVYTDEI---RSRFAEFDIAEKAEK 169
Query: 175 -ITDLPGNIKMTRLQLPNTLTENDPISQS-FAKLFEEIKDSEVRSYGVIVNSFYELENVY 232
I +PG ++ LP L D SQS FA + ++ V VNSF E++ +
Sbjct: 170 TIDFIPGLSAISFSDLPEELIMED--SQSIFALTLHNMGLKLHKATAVAVNSFEEIDPII 227
Query: 233 ADYYREVLGIKEWHIGPFSIHNRNKEEEIPSYRGKEASIDKHECLKWLDTKNINSVVYMC 292
++ R + +IGP + IP + +ECLKWL T+ +SVVY+
Sbjct: 228 TNHLRSTNQLNILNIGPL----QTLSSSIPP--------EDNECLKWLQTQKESSVVYLS 275
Query: 293 FGSMTHFLNSQLKEIAMGLEASGHNFIWVVRTQTEDGDEWLPEGFEERTEGKGLIIRGWS 352
FG++ + +++ +A LE+ F+W +R ++ + LPE F +RT G I+ W+
Sbjct: 276 FGTVINPPPNEMAALASTLESRKIPFLWSLR---DEARKHLPENFIDRTSTFGKIV-SWA 331
Query: 353 PQVMILEHEAIGAFVTHCGWNSVLEGVVAGVPMITWPVAAEQFYNEKLVTEVLKTGVPVG 412
PQ+ +LE+ AIG FVTHCGWNS LE + VP+I P +Q N ++V +V K G VG
Sbjct: 332 PQLHVLENPAIGVFVTHCGWNSTLESIFCRVPVIGRPFFGDQKVNARMVEDVWKIG--VG 389
Query: 413 VKKWVMKVGDNVEWDAVEKAVKRVMEGEEAYEMRNKAKMLAEMAKKAVEEDGSSYSQLNA 472
VK G D + ++ V+ ++ EMR L E AK AV+ +GSS +
Sbjct: 390 VK------GGVFTEDETTRVLELVLFSDKGKEMRQNVGRLKEKAKDAVKANGSSTRNFES 443
Query: 473 LIEELRSL 480
L+ L
Sbjct: 444 LLAAFNKL 451
>UFOG_PETHY (Q43716) Flavonol 3-O-glucosyltransferase (EC 2.4.1.91)
(UDP-glucose flavonoid 3-O-glucosyltransferase)
(Anthocyanin rhamnosyl transferase)
Length = 473
Score = 141 bits (356), Expect = 3e-33
Identities = 123/468 (26%), Positives = 201/468 (42%), Gaps = 40/468 (8%)
Query: 1 MDSQSNPLHILVFPFMGHGHTIPTIDMAKLFASKGVRVTIVTTPLNKPPISKALEQSKIH 60
M ++ LH+++FPF GH P + +A +S GV+V+ T N + L +
Sbjct: 5 MKHSNDALHVVMFPFFAFGHISPFVQLANKLSSYGVKVSFFTASGNASRVKSMLNSAP-- 62
Query: 61 FNNIDIQTIKFPCVEAGLPEGCENVDSIPSVSFVPAFFAAIRLLQQPFEELLLQQKPHCV 120
I + P VE GLP G E+ + S A+ L+Q + LL KPH V
Sbjct: 63 --TTHIVPLTLPHVE-GLPPGAESTAELTPAS-AELLKVALDLMQPQIKTLLSHLKPHFV 118
Query: 121 VADMFFPWATDSAAKFGIPRIVFHGTSFFSLCASQCMKKYQPYKNVSSDTDLFEITDLPG 180
+ D W A GI + + S C + K S L ++ P
Sbjct: 119 LFDFAQEWLPKMANGLGIKTVYYSVVVALSTAFLTCPARVLEPKKYPS---LEDMKKPPL 175
Query: 181 NIKMTRLQLPNTLTEND--PISQSFAK---LFEEIKDSEVRSYGVIVNSFYELENVYADY 235
T + T D + +SF L++ I+ ++ + ++E Y Y
Sbjct: 176 GFPQTSVTSVRTFEARDFLYVFKSFHNGPTLYDRIQSGLRGCSAILAKTCSQMEGPYIKY 235
Query: 236 YREVLGIKEWHIGPFSIHNRNKEEEIPSYRGKEASIDKHECLKWLDTKNINSVVYMCFGS 295
+ IGP + PS + +E + WL+ +V+Y FGS
Sbjct: 236 VEAQFNKPVFLIGPVV-------PDPPSGKLEE------KWATWLNKFEGGTVIYCSFGS 282
Query: 296 MTHFLNSQLKEIAMGLEASGHNFIWVVRTQTE-----DGDEWLPEGFEERTEGKGLIIRG 350
T + Q+KE+A+GLE +G F V+ + + LPEGF ER + KG+I G
Sbjct: 283 ETFLTDDQVKELALGLEQTGLPFFLVLNFPANVDVSAELNRALPEGFLERVKDKGIIHSG 342
Query: 351 WSPQVMILEHEAIGAFVTHCGWNSVLEGVVAGVPMITWPVAAEQFYNEKLVTEVLKTGVP 410
W Q IL H ++G +V H G++SV+E +V ++ P +Q N KLV+ ++ GV
Sbjct: 343 WVQQQNILAHSSVGCYVCHAGFSSVIEALVNDCQVVMLPQKGDQILNAKLVSGDMEAGVE 402
Query: 411 VGVKKWVMKVGDNVEWDAVEKAVKRVMEGEEAYEMRNKAKMLAEMAKK 458
+ + G + +++AV++VM E ++ K++ E KK
Sbjct: 403 INRRDEDGYFGK----EDIKEAVEKVMVDVE----KDPGKLIRENQKK 442
>IAAG_MAIZE (Q41819) Indole-3-acetate beta-glucosyltransferase (EC
2.4.1.121) (IAA-Glu synthetase) ((Uridine
5'-diphosphate-glucose:indol-3-ylacetyl)-beta-D-glucosyl
transferase)
Length = 471
Score = 141 bits (356), Expect = 3e-33
Identities = 143/495 (28%), Positives = 205/495 (40%), Gaps = 56/495 (11%)
Query: 9 HILVFPFMGHGHTIPTIDMAKLFASKGVRVTIVTTPLNKPPISKALEQSKIHFNNIDIQT 68
H+LV PF G GH P + AK ASKGV T+VTT + ++++ ++D
Sbjct: 4 HVLVVPFPGQGHMNPMVQFAKRLASKGVATTLVTT--------RFIQRTA----DVDAHP 51
Query: 69 IKFPCVEAGLPEG-CENVDSIPSVSFVPAFFAAIRLLQQPFEELLLQQKPHCVVADMFFP 127
+ G EG + + A A+ L CVV D +
Sbjct: 52 AMVEAISDGHDEGGFASAAGVAEYLEKQAAAASASLASLVEARASSADAFTCVVYDSYED 111
Query: 128 WATDSAAKFGIPRIVFHGTSFFSLCA--------SQCMKKYQPYKNVSSDTDLFEITDLP 179
W A + G+P + F S CA SQ P L
Sbjct: 112 WVLPVARRMGLPAVPFSTQS----CAVSAVYYHFSQGRLAVPPGAAADGSDGGAGAAALS 167
Query: 180 ----GNIKMTRLQLPNTLTENDPISQSFAKLFEEIKDSEVRSYGVIVNSFYELENVYADY 235
G +M R +LP+ + ++ P + ++ + + V+ NSF ELE
Sbjct: 168 EAFLGLPEMERSELPSFVFDHGPYPTIAMQAIKQFAHAGKDDW-VLFNSFEELET----- 221
Query: 236 YREVLG-----IKEWHIGP---FSIHNRNKEEEIPSYRGKEASIDKHECLKWLDTKNINS 287
EVL +K IGP R G + C KWLDTK S
Sbjct: 222 --EVLAGLTKYLKARAIGPCVPLPTAGRTAGANGRITYGANLVKPEDACTKWLDTKPDRS 279
Query: 288 VVYMCFGSMTHFLNSQLKEIAMGLEASGHNFIWVVRTQTEDGDEWLPEGF-EERTEGKGL 346
V Y+ FGS+ N+Q +E+A GL A+G F+WVVR E +P E T
Sbjct: 280 VAYVSFGSLASLGNAQKEELARGLLAAGKPFLWVVRASDE---HQVPRYLLAEATATGAA 336
Query: 347 IIRGWSPQVMILEHEAIGAFVTHCGWNSVLEGVVAGVPMITWPVAAEQFYNEKLVTEVLK 406
++ W PQ+ +L H A+G FVTHCGWNS LE + GVPM+ + +Q N + V L
Sbjct: 337 MVVPWCPQLDVLAHPAVGCFVTHCGWNSTLEALSFGVPMVAMALWTDQPTNARNVE--LA 394
Query: 407 TGVPVGVKKWVMKVGDNVEW-DAVEKAVKRVMEGEEAYEMRNKAK-MLAEMAKKAVEEDG 464
G V ++ G V VE+ V+ VM+G EA KA + A+ AV G
Sbjct: 395 WGAGVRARR---DAGAGVFLRGEVERCVRAVMDGGEAASAARKAAGEWRDRARAAVAPGG 451
Query: 465 SSYSQLNALIEELRS 479
SS L+ ++ +R+
Sbjct: 452 SSDRNLDEFVQFVRA 466
>UFO7_MANES (Q40289) Flavonol 3-O-glucosyltransferase 7 (EC
2.4.1.91) (UDP-glucose flavonoid 3-O-glucosyltransferase
7) (Fragment)
Length = 287
Score = 138 bits (347), Expect = 4e-32
Identities = 98/309 (31%), Positives = 160/309 (51%), Gaps = 27/309 (8%)
Query: 178 LPGNIKMTRLQLPNTLTENDPISQSFAKLFEEIKDSEVRSYGVIVNSFYELE-NVYADYY 236
+PG K+ LP + + + F+++ + R+ V++NSF EL+ + +D
Sbjct: 5 IPGMSKIQIRDLPEGVLFGN-LESLFSQMLHNMGRMLPRAAAVLMNSFEELDPTIVSDLN 63
Query: 237 REVLGIKEWHIGPFSIHNRNKEEEIPSYRGKEASIDKHECLKWLDTKNINSVVYMCFGSM 296
+ I IGPF++ + +P D + C+ WLD + SV Y+ FGS+
Sbjct: 64 SKFNNIL--CIGPFNLVS--PPPPVP---------DTYGCMAWLDKQKPASVAYISFGSV 110
Query: 297 THFLNSQLKEIAMGLEASGHNFIWVVRTQTEDGDEWLPEGFEERTEGKGLIIRGWSPQVM 356
+L +A LEAS F+W ++ ++ LP GF +RT+ G+++ W+PQV
Sbjct: 111 ATPPPHELVALAEALEASKVPFLWSLKDHSK---VHLPNGFLDRTKSHGIVL-SWAPQVE 166
Query: 357 ILEHEAIGAFVTHCGWNSVLEGVVAGVPMITWPVAAEQFYNEKLVTEVLKTGVPVGVKKW 416
ILEH A+G FVTHCGWNS+LE +V GVPMI P +Q N ++V +V + G+
Sbjct: 167 ILEHAALGVFVTHCGWNSILESIVGGVPMICRPFFGDQRLNGRMVEDVWEIGL------- 219
Query: 417 VMKVGDNVEWDAVEKAVKRVMEGEEAYEMRNKAKMLAEMAKKAVEEDGSSYSQLNALIEE 476
+M G + A++ + +++G + +MR K L E+AK A E GSS L
Sbjct: 220 LMDGGVLTKNGAIDGLNQILLQG-KGKKMRENIKRLKELAKGATEPKGSSSKSFTELANL 278
Query: 477 LRSLSHHQH 485
+RS +++
Sbjct: 279 VRSRGSYEN 287
>UFOG_HORVU (P14726) Flavonol 3-O-glucosyltransferase (EC 2.4.1.91)
(UDP-glucose flavonoid 3-O-glucosyltransferase)
(Bronze-1)
Length = 455
Score = 132 bits (332), Expect = 2e-30
Identities = 81/214 (37%), Positives = 106/214 (48%), Gaps = 16/214 (7%)
Query: 262 PSYRGKEASIDKHECLKWLDTKNINSVVYMCFGSMTHFLNSQLKEIAMGLEASGHNFIWV 321
P+ EA D H CL WLD + SV Y+ FG+ +L+E+A GLEASG F+W
Sbjct: 253 PTADTNEAPADPHGCLAWLDRRPARSVAYVSFGTNATARPDELQELAAGLEASGAPFLWS 312
Query: 322 VRTQTEDGDEWLPEGFEERTEGKGLIIRGWSPQVMILEHEAIGAFVTHCGWNSVLEGVVA 381
+R P GF ER G ++ W+PQV +L H A+GAFVTH GW SV+EGV +
Sbjct: 313 LRGVVAAA----PRGFLERAPG---LVVPWAPQVGVLRHAAVGAFVTHAGWASVMEGVSS 365
Query: 382 GVPMITWPVAAEQFYNEKLVTEVLKTGVPVGVKKWVMKVGDNVEWDAVEKAVKRVMEGEE 441
GVPM P +Q N + V V G + AV AV ++ GE+
Sbjct: 366 GVPMACRPFFGDQTMNARSVASVWGFGT---------AFDGPMTRGAVANAVATLLRGED 416
Query: 442 AYEMRNKAKMLAEMAKKAVEEDGSSYSQLNALIE 475
MR KA+ L M KA E DG + +E
Sbjct: 417 GERMRAKAQELQAMVGKAFEPDGGCRKNFDEFVE 450
>UFOG_SOLME (Q43641) Flavonol 3-O-glucosyltransferase (EC 2.4.1.91)
(UDP-glucose flavonoid 3-O-glucosyltransferase)
Length = 433
Score = 131 bits (329), Expect = 4e-30
Identities = 128/479 (26%), Positives = 209/479 (42%), Gaps = 80/479 (16%)
Query: 5 SNPLHI--LVFPFMGHGHTIPTI--DMAKLFASKGVRVTIVTTPLNKPPISKALEQSKIH 60
++ LHI L FPF H + T+ ++ S + T+ N SK Q I
Sbjct: 3 TSQLHIAFLAFPFGTHATPLLTLVQKISPFLPSSTIFSFFNTSSSNSSIFSKVPNQENIK 62
Query: 61 FNNIDIQTIKFPCVEAGLPEGCENVDSIPSVS-FVPAFFAAIRLLQQPFEELLLQQKPHC 119
N V G+ EG + + ++ F+ + ++ ++ EE + K C
Sbjct: 63 IYN----------VWDGVKEGNDTPFGLEAIKLFIQSTLLISKITEEAEEETGV--KFSC 110
Query: 120 VVADMFFPWA--TDSAAKFGIPRIVFHGTSFFSLCASQCMKKYQPYKNVSSDTDLFEITD 177
+ +D F W K P + + SL TD
Sbjct: 111 IFSDAFL-WCFLVKLPKKMNAPGVAYWTGGSCSLAVHL-------------------YTD 150
Query: 178 LPGNIKMTRLQLP---NTLTENDPISQSFAKLFEEIKDSEV--------RSYGVIVNSFY 226
L + K T L++P +TL+ ND + A+ E S + ++ V++NSF
Sbjct: 151 LIRSNKETSLKIPGFSSTLSINDIPPEVTAEDLEGPMSSMLYNMALNLHKADAVVLNSFQ 210
Query: 227 ELEN---VYADYYREVLGIKEWHIGPFSIHNRNKEEEIPSYRGKEASIDKHECLKWLDTK 283
EL+ + D + + K ++IGP + + K +D+ C++WLD +
Sbjct: 211 ELDRDPLINKDLQKNLQ--KVFNIGPLVLQSSRK-------------LDESGCIQWLDKQ 255
Query: 284 NINSVVYMCFGSMTHFLNSQLKEIAMGLEASGHNFIWVVRTQTEDGDEWLPEGFEERTEG 343
SVVY+ FG++T +++ IA LE FIW +R +G + LP+GF ERT+
Sbjct: 256 KEKSVVYLSFGTVTTLPPNEIGSIAEALETKKTPFIWSLRN---NGVKNLPKGFLERTKE 312
Query: 344 KGLIIRGWSPQVMILEHEAIGAFVTHCGWNSVLEGVVAGVPMITWPVAAEQFYNEKLVTE 403
G I+ W+PQ+ IL H+++G FVTHCGWNS+LEG+ GVPMI P +Q N ++V
Sbjct: 313 FGKIV-SWAPQLEILAHKSVGVFVTHCGWNSILEGISFGVPMICRPFFGDQKLNSRMVES 371
Query: 404 VLKTGVPVGVKKWVMKVGDNVEWDAVEKAVKRVMEGEEAYEMRNKAKMLAEMAKKAVEE 462
V + G+ + G + A+ E+ +R + L E A +AV +
Sbjct: 372 VWEIGLQI--------EGGIFTKSGIISALDTFFNEEKGKILRENVEGLKEKALEAVNQ 422
>UFO3_MAIZE (P16167) Flavonol 3-O-glucosyltransferase (EC 2.4.1.91)
(UDP-glucose flavonoid 3-O-glucosyltransferase)
(Bronze-1) (Bz-W22 allele)
Length = 471
Score = 128 bits (321), Expect = 4e-29
Identities = 130/487 (26%), Positives = 208/487 (42%), Gaps = 38/487 (7%)
Query: 2 DSQSNP---LHILVFPFMGHGHTIPTID--MAKLFASKGVRVTIVTTPLNKPPISKALEQ 56
D +S+P + ++ FPF H + +I +A A G ++ ++T + + KA
Sbjct: 5 DGESSPPPHVAVVAFPFSSHAAVLLSIARALAAAAAPSGATLSFLSTASSLAQLRKASSA 64
Query: 57 SKIHFNNIDIQTIKFPCVEAGLPEGCENVDSIPSVSFVPAFFAA-----IRLLQQPFEEL 111
S H +++ ++ P G P E S+P + F A ++ +
Sbjct: 65 SAGHGLPGNLRFVEVP---DGAPAAEE---SVPVPRQMQLFMEAAEAGGVKAWLEAARAA 118
Query: 112 LLQQKPHCVVADMFFPWATDSAAKFGIPRI-VFHGTSFFSLCASQCMKKYQPYKNVSSDT 170
+ CVV D F A D+AA G P + V+ S L + + + +++
Sbjct: 119 AGGARVTCVVGDAFVWPAADAAASAGAPWVPVWTAASCALLAHIRTDALREDVGDQAANR 178
Query: 171 DLFEITDLPGNIKMTRLQLPNTLTENDPISQSFAKLFEEIKDSEVRSYGVI-VNSFYELE 229
+ PG LP+ + D + L + RS + +N+F L+
Sbjct: 179 VDEPLISHPGLASYRVRDLPDGVVSGD-FNYVINLLVHRMGQCLPRSAAAVALNTFPGLD 237
Query: 230 NVYADYYREVLGIKEWHIGPFSIHNRNKEEEIPSYRGKEASIDKHECLKWLDTKNINSVV 289
+ GP+ H E++ + A D H CL WL + V
Sbjct: 238 PPDVTAALAEILPNCVPFGPY--HLLLAEDDADT----AAPADPHGCLAWLGRQPARGVA 291
Query: 290 YMCFGSMTHFLNSQLKEIAMGLEASGHNFIWVVRTQTEDGDEWLPEGFEERTEGKGL-II 348
Y+ FG++ +L+E+A GLEASG F+W +R ED LP GF +R G G ++
Sbjct: 292 YVSFGTVACPRPDELRELAAGLEASGAPFLWSLR---EDSWTLLPPGFLDRAAGTGSGLV 348
Query: 349 RGWSPQVMILEHEAIGAFVTHCGWNSVLEGVVAGVPMITWPVAAEQFYNEKLVTEVLKTG 408
W+PQV +L H ++GAFVTH GW SVLEGV +GVPM P +Q N + V V G
Sbjct: 349 VPWAPQVAVLRHPSVGAFVTHAGWASVLEGVSSGVPMACRPFFGDQRMNARSVAHV--WG 406
Query: 409 VPVGVKKWVMKVGDNVEWDAVEKAVKRVMEGEEAYEMRNKAKMLAEMAKKAVEEDGSSYS 468
+ + G V AV+ ++ GEE MR +AK+L + +A G
Sbjct: 407 FGAAFEGAMTSAG-------VAAAVEELLRGEEGARMRARAKVLQALVAEAFGPGGECRK 459
Query: 469 QLNALIE 475
+ +E
Sbjct: 460 NFDRFVE 466
>UFO2_MAIZE (P16165) Flavonol 3-O-glucosyltransferase (EC 2.4.1.91)
(UDP-glucose flavonoid 3-O-glucosyltransferase)
(Bronze-1) (Bz-Mc2 allele)
Length = 471
Score = 124 bits (312), Expect = 4e-28
Identities = 129/484 (26%), Positives = 207/484 (42%), Gaps = 32/484 (6%)
Query: 2 DSQSNP---LHILVFPFMGHGHTIPTID--MAKLFASKGVRVTIVTTPLNKPPISKALEQ 56
D +S+P + ++ FPF H + +I +A A G ++ ++T + + KA
Sbjct: 5 DGESSPPPHVAVVAFPFSSHAAVLLSIARALAAAAAPSGATLSFLSTASSLAQLRKASSA 64
Query: 57 SKIHFNNIDIQTIKFPCVEAGLPEGCENVDSIPSVS-FVPAFFAA-IRLLQQPFEELLLQ 114
S H +++ ++ P G P E V + F+ A A ++ +
Sbjct: 65 SAGHGLPGNLRFVEVP---DGAPAAEETVPVPRQMQLFMEAAEAGGVKAWLEAARAAAGG 121
Query: 115 QKPHCVVADMFFPWATDSAAKFGIPRI-VFHGTSFFSLCASQCMKKYQPYKNVSSDTDLF 173
+ CVV D F A D+AA G P + V+ S L + + + +++
Sbjct: 122 ARVTCVVGDAFVWPAADAAASAGAPWVPVWTAASCALLAHIRTDSLREDVGDQAANRVDE 181
Query: 174 EITDLPGNIKMTRLQLPNTLTENDPISQSFAKLFEEIKDSEVRSYGVI-VNSFYELENVY 232
+ PG LP+ + D + + L + RS + +N+F L+
Sbjct: 182 PLISHPGLASYRVRDLPDGVVSGD-FNYVISLLVHRMGQCLPRSAAAVALNTFPGLDPPD 240
Query: 233 ADYYREVLGIKEWHIGPFSIHNRNKEEEIPSYRGKEASIDKHECLKWLDTKNINSVVYMC 292
+ GP+ H E++ + A D H CL WL + V Y+
Sbjct: 241 VTAALAEILPNCVPFGPY--HLLLAEDDADT----AAPADPHGCLAWLGRQPARGVAYVS 294
Query: 293 FGSMTHFLNSQLKEIAMGLEASGHNFIWVVRTQTEDGDEWLPEGFEERTEGKGL-IIRGW 351
FG++ +L+E+A GLEAS F+W +R ED LP GF +R G G ++ W
Sbjct: 295 FGTVACPRPDELRELAAGLEASAAPFLWSLR---EDSWTLLPPGFLDRAAGTGSGLVVPW 351
Query: 352 SPQVMILEHEAIGAFVTHCGWNSVLEGVVAGVPMITWPVAAEQFYNEKLVTEVLKTGVPV 411
+PQV +L H ++GAFVTH GW SVLEGV +GVPM P +Q N + V V G
Sbjct: 352 APQVAVLRHPSVGAFVTHAGWASVLEGVSSGVPMACRPFFGDQRMNARSVAHV--WGFGA 409
Query: 412 GVKKWVMKVGDNVEWDAVEKAVKRVMEGEEAYEMRNKAKMLAEMAKKAVEEDGSSYSQLN 471
+ + G V AV+ ++ GEE MR +AK L + +A G +
Sbjct: 410 AFEGAMTSAG-------VAAAVEELLRGEEGAGMRARAKELQALVAEAFGPGGECRKNFD 462
Query: 472 ALIE 475
+E
Sbjct: 463 RFVE 466
>UFO1_MAIZE (P16166) Flavonol 3-O-glucosyltransferase (EC 2.4.1.91)
(UDP-glucose flavonoid 3-O-glucosyltransferase)
(Bronze-1) (Bz-McC allele)
Length = 471
Score = 123 bits (308), Expect = 1e-27
Identities = 131/488 (26%), Positives = 205/488 (41%), Gaps = 40/488 (8%)
Query: 2 DSQSNP---LHILVFPFMGHGHTIPTID--MAKLFASKGVRVTIVTTPLNKPPISKALEQ 56
D +S+P + ++ FPF H + +I +A A G ++ ++T + + KA
Sbjct: 5 DGESSPPPHVAVVAFPFSSHAAVLLSIARALAAAAAPSGATLSFLSTASSLAQLRKASSA 64
Query: 57 SKIHFNNIDIQTIKFPCVEAGLPEGCENVDSIPSVS-FVPAFFAA-IRLLQQPFEELLLQ 114
S H +++ ++ P G P E V + F+ A A ++ +
Sbjct: 65 SAGHGLPGNLRFVEVP---DGAPAAEETVPVPRQMQLFMEAAEAGGVKAWLEAARAAAGG 121
Query: 115 QKPHCVVADMFFPWATDSAAKFGIPRI-VFHGTSFFSLCASQCMKKYQPYKNVSSDTDLF 173
+ CVV D F A D+AA G P + V+ S CA + + D
Sbjct: 122 ARVTCVVGDAFVWPAADAAASAGAPWVPVWTAAS----CALLAHIRTDALREDVGDQAAN 177
Query: 174 EITDL----PGNIKMTRLQLPNTLTENDPISQSFAKLFEEIKDSEVRSYGVI-VNSFYEL 228
+ L PG LP+ + D + L + RS + +N+F L
Sbjct: 178 RVDGLLISHPGLASYRVRDLPDGVVSGD-FNYVINLLVHRMGQCLPRSAAAVALNTFPGL 236
Query: 229 ENVYADYYREVLGIKEWHIGPFSIHNRNKEEEIPSYRGKEASIDKHECLKWLDTKNINSV 288
+ + GP+ H E++ + A D H CL WL + V
Sbjct: 237 DPPDVTAALAEILPNCVPFGPY--HLLLAEDDADT----AAPADPHGCLAWLGRQPARGV 290
Query: 289 VYMCFGSMTHFLNSQLKEIAMGLEASGHNFIWVVRTQTEDGDEWLPEGFEERTEGKGL-I 347
Y+ FG++ +L+E+A GLE SG F+W +R ED LP GF +R G G +
Sbjct: 291 AYVSFGTVACPRPDELRELAAGLEDSGAPFLWSLR---EDSWPHLPPGFLDRAAGTGSGL 347
Query: 348 IRGWSPQVMILEHEAIGAFVTHCGWNSVLEGVVAGVPMITWPVAAEQFYNEKLVTEVLKT 407
+ W+PQV +L H ++GAFVTH GW SVLEG+ +GVPM P +Q N + V V
Sbjct: 348 VVPWAPQVAVLRHPSVGAFVTHAGWASVLEGLSSGVPMACRPFFGDQRMNARSVAHV--W 405
Query: 408 GVPVGVKKWVMKVGDNVEWDAVEKAVKRVMEGEEAYEMRNKAKMLAEMAKKAVEEDGSSY 467
G + + G V AV+ ++ GEE MR +AK L + +A G
Sbjct: 406 GFGAAFEGAMTSAG-------VATAVEELLRGEEGARMRARAKELQALVAEAFGPGGECR 458
Query: 468 SQLNALIE 475
+ +E
Sbjct: 459 KNFDRFVE 466
>UFOG_VITVI (P51094) Flavonol 3-O-glucosyltransferase (EC 2.4.1.91)
(UDP-glucose flavonoid 3-O-glucosyltransferase)
(Fragment)
Length = 154
Score = 91.7 bits (226), Expect = 4e-18
Identities = 50/143 (34%), Positives = 75/143 (51%), Gaps = 9/143 (6%)
Query: 333 LPEGFEERTEGKGLIIRGWSPQVMILEHEAIGAFVTHCGWNSVLEGVVAGVPMITWPVAA 392
LPEGF E+T G G+++ W+PQ +L GAFVTHCGWNS+ E V GVP+I P
Sbjct: 13 LPEGFLEKTRGYGMVVP-WAPQAEVLALRQFGAFVTHCGWNSLWESVAGGVPLICRPFFG 71
Query: 393 EQFYNEKLVTEVLKTGVPVGVKKWVMKVGDNVEWDAVEKAVKRVMEGEEAYEMRNKAKML 452
+Q N ++V +VL+ GV + G + +++ E+ ++R + L
Sbjct: 72 DQRLNGRMVEDVLEIGVRI--------EGGVFTKSGLMSCFDQILSQEKGKKLRENLRAL 123
Query: 453 AEMAKKAVEEDGSSYSQLNALIE 475
E A +AV GSS L++
Sbjct: 124 RETADRAVGPKGSSTENFKTLVD 146
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.319 0.135 0.408
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 58,376,371
Number of Sequences: 164201
Number of extensions: 2508904
Number of successful extensions: 6981
Number of sequences better than 10.0: 101
Number of HSP's better than 10.0 without gapping: 89
Number of HSP's successfully gapped in prelim test: 12
Number of HSP's that attempted gapping in prelim test: 6819
Number of HSP's gapped (non-prelim): 116
length of query: 489
length of database: 59,974,054
effective HSP length: 114
effective length of query: 375
effective length of database: 41,255,140
effective search space: 15470677500
effective search space used: 15470677500
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 68 (30.8 bits)
Medicago: description of AC149574.3


