

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC149547.6 + phase: 0
(160 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
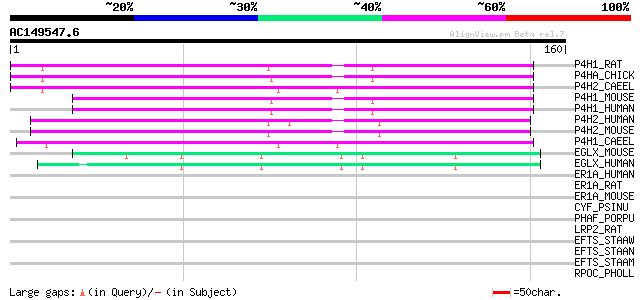
Score E
Sequences producing significant alignments: (bits) Value
P4H1_RAT (P54001) Prolyl 4-hydroxylase alpha-1 subunit precursor... 89 6e-18
P4HA_CHICK (P16924) Prolyl 4-hydroxylase alpha subunit (EC 1.14.... 88 7e-18
P4H2_CAEEL (Q20065) Prolyl 4-hydroxylase alpha-2 subunit precurs... 84 1e-16
P4H1_MOUSE (Q60715) Prolyl 4-hydroxylase alpha-1 subunit precurs... 80 2e-15
P4H1_HUMAN (P13674) Prolyl 4-hydroxylase alpha-1 subunit precurs... 79 6e-15
P4H2_HUMAN (O15460) Prolyl 4-hydroxylase alpha-2 subunit precurs... 77 2e-14
P4H2_MOUSE (Q60716) Prolyl 4-hydroxylase alpha-2 subunit precurs... 75 5e-14
P4H1_CAEEL (Q10576) Prolyl 4-hydroxylase alpha-1 subunit precurs... 75 6e-14
EGLX_MOUSE (Q8BG58) Putative HIF-prolyl hydroxylase PH-4 (EC 1.1... 50 2e-06
EGLX_HUMAN (Q9NXG6) Putative HIF-prolyl hydroxylase PH-4 (EC 1.1... 50 2e-06
ER1A_HUMAN (Q96HE7) ERO1-like protein alpha precursor (EC 1.8.4.... 32 0.61
ER1A_RAT (Q8R4A1) ERO1-like protein alpha precursor (EC 1.8.4.-)... 30 2.3
ER1A_MOUSE (Q8R180) ERO1-like protein alpha precursor (EC 1.8.4.... 30 2.3
CYF_PSINU (Q8WI07) Apocytochrome f precursor 30 2.3
PHAF_PORPU (P51215) Allophycocyanin beta 18 chain 30 3.0
LRP2_RAT (P98158) Low-density lipoprotein receptor-related prote... 28 6.8
EFTS_STAAW (Q8NWZ6) Elongation factor Ts (EF-Ts) 28 6.8
EFTS_STAAN (P99171) Elongation factor Ts (EF-Ts) 28 6.8
EFTS_STAAM (P64054) Elongation factor Ts (EF-Ts) 28 6.8
RPOC_PHOLL (Q7N9A3) DNA-directed RNA polymerase beta' chain (EC ... 28 8.9
>P4H1_RAT (P54001) Prolyl 4-hydroxylase alpha-1 subunit precursor
(EC 1.14.11.2) (4-PH alpha-1)
(Procollagen-proline,2-oxoglutarate-4-dioxygenase
alpha-1 subunit)
Length = 534
Score = 88.6 bits (218), Expect = 6e-18
Identities = 53/161 (32%), Positives = 82/161 (50%), Gaps = 13/161 (8%)
Query: 1 MHKSTVDD-ETGKSVDNSARTSSGTFINRGHDKILRNIEQRIADFTFIPVENGESVNILH 59
+ ++TV D ETGK R S +++ D ++ I RI D T + V E + + +
Sbjct: 360 LSRATVHDPETGKLTTAQYRVSKSAWLSGYEDPVVSRINMRIQDLTGLDVSTAEELQVAN 419
Query: 60 YEVGQKYEPHPDFF----TDEINTKNGGERVATMLMYLPWWNELSDCG-----KKGLSIK 110
Y VG +YEPH DF D G R+AT L Y+ +++S G + G S+
Sbjct: 420 YGVGGQYEPHFDFARKDEPDAFRELGTGNRIATWLFYM---SDVSAGGATVFPEVGASVW 476
Query: 111 PKMGDALLFWSMKPDGTLDPLSMHGACPVIKGDKWSCTKWM 151
PK G A+ ++++ G D + H ACPV+ G+KW KW+
Sbjct: 477 PKKGTAVFWYNLFASGEGDYSTRHAACPVLVGNKWVSNKWL 517
>P4HA_CHICK (P16924) Prolyl 4-hydroxylase alpha subunit (EC
1.14.11.2) (4-PH alpha)
(Procollagen-proline,2-oxoglutarate-4-dioxygenase alpha
subunit)
Length = 516
Score = 88.2 bits (217), Expect = 7e-18
Identities = 53/161 (32%), Positives = 82/161 (50%), Gaps = 13/161 (8%)
Query: 1 MHKSTVDD-ETGKSVDNSARTSSGTFINRGHDKILRNIEQRIADFTFIPVENGESVNILH 59
+ ++TV D ETGK R S +++ ++ I RI D T + V E + + +
Sbjct: 342 LSRATVHDPETGKLTTAHYRVSKSAWLSGYESPVVSRINTRIQDLTGLDVSTAEELQVAN 401
Query: 60 YEVGQKYEPHPDFFT----DEINTKNGGERVATMLMYLPWWNELSDCG-----KKGLSIK 110
Y VG +YEPH DF D G R+AT L Y+ +++S G + G S+
Sbjct: 402 YGVGGQYEPHFDFGRKDEPDAFKELGTGNRIATWLFYM---SDVSAGGATVFPEVGASVW 458
Query: 111 PKMGDALLFWSMKPDGTLDPLSMHGACPVIKGDKWSCTKWM 151
PK G A+ ++++ P G D + H ACPV+ G+KW KW+
Sbjct: 459 PKKGTAVFWYNLFPSGEGDYSTRHAACPVLVGNKWVSNKWL 499
>P4H2_CAEEL (Q20065) Prolyl 4-hydroxylase alpha-2 subunit precursor
(EC 1.14.11.2) (4-PH alpha-2)
(Procollagen-proline,2-oxoglutarate-4-dioxygenase
alpha-2 subunit)
Length = 539
Score = 84.0 bits (206), Expect = 1e-16
Identities = 48/158 (30%), Positives = 81/158 (50%), Gaps = 7/158 (4%)
Query: 1 MHKSTVDD-ETGKSVDNSARTSSGTFINRGHDKILRNIEQRIADFTFIPVENGESVNILH 59
+ ++TV + +TG+ + R S ++ D ++ + +RI DFT + E + + +
Sbjct: 350 LKRATVQNSKTGELEHATYRISKSAWLKGDLDPVIDRVNRRIEDFTNLNQATSEELQVAN 409
Query: 60 YEVGQKYEPHPDFFTDE----INTKNGGERVATMLMYL--PWWNELSDCGKKGLSIKPKM 113
Y +G Y+PH DF E T N G R+AT+L Y+ P + G ++ P
Sbjct: 410 YGLGGHYDPHFDFARKEEKNAFKTLNTGNRIATVLFYMSQPERGGATVFNHLGTAVFPSK 469
Query: 114 GDALLFWSMKPDGTLDPLSMHGACPVIKGDKWSCTKWM 151
DAL +++++ DG D + H ACPV+ G KW KW+
Sbjct: 470 NDALFWYNLRRDGEGDLRTRHAACPVLLGVKWVSNKWI 507
>P4H1_MOUSE (Q60715) Prolyl 4-hydroxylase alpha-1 subunit precursor
(EC 1.14.11.2) (4-PH alpha-1)
(Procollagen-proline,2-oxoglutarate-4-dioxygenase
alpha-1 subunit)
Length = 534
Score = 80.5 bits (197), Expect = 2e-15
Identities = 46/142 (32%), Positives = 72/142 (50%), Gaps = 12/142 (8%)
Query: 19 RTSSGTFINRGHDKILRNIEQRIADFTFIPVENGESVNILHYEVGQKYEPHPDFFT---- 74
R S +++ D ++ I RI D T + V E + + +Y VG +YEPH DF
Sbjct: 379 RISKSAWLSGYEDPVVSRINMRIQDLTGLDVSTAEELQVANYGVGGQYEPHFDFARKDEP 438
Query: 75 DEINTKNGGERVATMLMYLPWWNELSDCG-----KKGLSIKPKMGDALLFWSMKPDGTLD 129
D G R+AT L Y+ +++S G + G S+ PK G A+ ++++ G D
Sbjct: 439 DAFRELGTGNRIATWLFYM---SDVSAGGATVFPEVGASVWPKKGTAVFWYNLFASGEGD 495
Query: 130 PLSMHGACPVIKGDKWSCTKWM 151
+ H ACPV+ G+KW KW+
Sbjct: 496 YSTRHAACPVLVGNKWVSNKWL 517
>P4H1_HUMAN (P13674) Prolyl 4-hydroxylase alpha-1 subunit precursor
(EC 1.14.11.2) (4-PH alpha-1)
(Procollagen-proline,2-oxoglutarate-4-dioxygenase
alpha-1 subunit)
Length = 534
Score = 78.6 bits (192), Expect = 6e-15
Identities = 45/142 (31%), Positives = 72/142 (50%), Gaps = 12/142 (8%)
Query: 19 RTSSGTFINRGHDKILRNIEQRIADFTFIPVENGESVNILHYEVGQKYEPHPDFFT---- 74
R S +++ + ++ I RI D T + V E + + +Y VG +YEPH DF
Sbjct: 379 RISKSAWLSGYENPVVSRINMRIQDLTGLDVSTAEELQVANYGVGGQYEPHFDFARKDEP 438
Query: 75 DEINTKNGGERVATMLMYLPWWNELSDCG-----KKGLSIKPKMGDALLFWSMKPDGTLD 129
D G R+AT L Y+ +++S G + G S+ PK G A+ ++++ G D
Sbjct: 439 DAFKELGTGNRIATWLFYM---SDVSAGGATVFPEVGASVWPKKGTAVFWYNLFASGEGD 495
Query: 130 PLSMHGACPVIKGDKWSCTKWM 151
+ H ACPV+ G+KW KW+
Sbjct: 496 YSTRHAACPVLVGNKWVSNKWL 517
>P4H2_HUMAN (O15460) Prolyl 4-hydroxylase alpha-2 subunit precursor
(EC 1.14.11.2) (4-PH alpha-2)
(Procollagen-proline,2-oxoglutarate-4-dioxygenase
alpha-2 subunit) (UNQ290/PRO330)
Length = 535
Score = 76.6 bits (187), Expect = 2e-14
Identities = 49/153 (32%), Positives = 77/153 (50%), Gaps = 12/153 (7%)
Query: 7 DDETGKSVDNSARTSSGTFINRGHDKILRNIEQRIADFTFIPVENGESVNILHYEVGQKY 66
D +TG S R S +++ D ++ + +R+ T + V+ E + + +Y VG +Y
Sbjct: 368 DPKTGVLTVASYRVSKSSWLEEDDDPVVARVNRRMQHITGLTVKTAELLQVANYGVGGQY 427
Query: 67 EPHPDFF-TDEINT---KNGGERVATMLMYLPWWNELSDCGKK-----GLSIKPKMGDAL 117
EPH DF DE +T G RVAT L Y+ +++ G G +I PK G A+
Sbjct: 428 EPHFDFSRNDERDTFKHLGTGNRVATFLNYM---SDVEAGGATVFPDLGAAIWPKKGTAV 484
Query: 118 LFWSMKPDGTLDPLSMHGACPVIKGDKWSCTKW 150
++++ G D + H ACPV+ G KW KW
Sbjct: 485 FWYNLLRSGEGDYRTRHAACPVLVGCKWVSNKW 517
>P4H2_MOUSE (Q60716) Prolyl 4-hydroxylase alpha-2 subunit precursor
(EC 1.14.11.2) (4-PH alpha-2)
(Procollagen-proline,2-oxoglutarate-4-dioxygenase
alpha-2 subunit)
Length = 537
Score = 75.5 bits (184), Expect = 5e-14
Identities = 46/153 (30%), Positives = 74/153 (48%), Gaps = 12/153 (7%)
Query: 7 DDETGKSVDNSARTSSGTFINRGHDKILRNIEQRIADFTFIPVENGESVNILHYEVGQKY 66
D +TG S R S +++ D ++ + +R+ T + V+ E + + +Y +G +Y
Sbjct: 370 DPKTGVLTVASYRVSKSSWLEEDDDPVVARVNRRMQHITGLTVKTAELLQVANYGMGGQY 429
Query: 67 EPHPDFF----TDEINTKNGGERVATMLMYLPWWNELSDCGKK-----GLSIKPKMGDAL 117
EPH DF D G RVAT L Y+ +++ G G +I PK G A+
Sbjct: 430 EPHFDFSRSDDEDAFKRLGTGNRVATFLNYM---SDVEAGGATVFPDLGAAIWPKKGTAV 486
Query: 118 LFWSMKPDGTLDPLSMHGACPVIKGDKWSCTKW 150
++++ G D + H ACPV+ G KW KW
Sbjct: 487 FWYNLLRSGEGDYRTRHAACPVLVGCKWVSNKW 519
>P4H1_CAEEL (Q10576) Prolyl 4-hydroxylase alpha-1 subunit precursor
(EC 1.14.11.2) (4-PH alpha-1)
(Procollagen-proline,2-oxoglutarate-4-dioxygenase
alpha-1 subunit)
Length = 559
Score = 75.1 bits (183), Expect = 6e-14
Identities = 46/156 (29%), Positives = 76/156 (48%), Gaps = 7/156 (4%)
Query: 3 KSTVDDE-TGKSVDNSARTSSGTFINRGHDKILRNIEQRIADFTFIPVENGESVNILHYE 61
++TV D TGK V + R S ++ ++ + +RI T + +E E + I +Y
Sbjct: 355 RATVHDSVTGKLVTATYRISKSAWLKEWEGDVVETVNKRIGYMTNLEMETAEELQIANYG 414
Query: 62 VGQKYEPHPDFFTDE----INTKNGGERVATMLMYL--PWWNELSDCGKKGLSIKPKMGD 115
+G Y+PH D E + G R+AT+L Y+ P + + +I P D
Sbjct: 415 IGGHYDPHFDHAKKEESKSFESLGTGNRIATVLFYMSQPSHGGGTVFTEAKSTILPTKND 474
Query: 116 ALLFWSMKPDGTLDPLSMHGACPVIKGDKWSCTKWM 151
AL ++++ G +P + H ACPV+ G KW KW+
Sbjct: 475 ALFWYNLYKQGDGNPDTRHAACPVLVGIKWVSNKWI 510
>EGLX_MOUSE (Q8BG58) Putative HIF-prolyl hydroxylase PH-4 (EC
1.14.11.-) (Hypoxia-inducible factor prolyl
4-hydroxylase)
Length = 503
Score = 50.1 bits (118), Expect = 2e-06
Identities = 46/188 (24%), Positives = 72/188 (37%), Gaps = 53/188 (28%)
Query: 19 RTSSGTFINRGHDK--ILRNIEQRIADFTFIP---VENGESVNILHYEVGQKYEPHPD-- 71
R S T++++G ++R I QR+ T + VE E + ++ Y G Y H D
Sbjct: 274 RNSHHTWLHQGEGAHHVMRAIRQRVLRLTRLSPEIVEFSEPLQVVRYGEGGHYHAHVDSG 333
Query: 72 ------------FFTDEINTKNGGERVATMLMYLP----------------WWNELS--- 100
+E R T+L YL ++E+S
Sbjct: 334 PVYPETICSHTKLVANESVPFETSCRYMTVLFYLNNVTGGGETVFPVADNRTYDEMSLIQ 393
Query: 101 ----------DCGKKGLSIKPKMGDALLFWSMKPDGT-----LDPLSMHGACPVIKGDKW 145
C K L +KP+ G A+ +++ PDG +D S+HG C V +G KW
Sbjct: 394 DDVDLRDTRRHCDKGNLRVKPQQGTAVFWYNYLPDGQGWVGEVDDYSLHGGCLVTRGTKW 453
Query: 146 SCTKWMRV 153
W+ V
Sbjct: 454 IANNWINV 461
>EGLX_HUMAN (Q9NXG6) Putative HIF-prolyl hydroxylase PH-4 (EC
1.14.11.-) (Hypoxia-inducible factor prolyl
4-hydroxylase)
Length = 502
Score = 50.1 bits (118), Expect = 2e-06
Identities = 49/196 (25%), Positives = 73/196 (37%), Gaps = 53/196 (27%)
Query: 9 ETGKSVDNSARTSSGTFINRGHDKILRNIEQRIADFTFIP---VENGESVNILHYEVGQK 65
E+ + V NS T + G I+R I QR+ T + VE E + ++ Y G
Sbjct: 267 ESSELVRNSHHT--WLYQGEGAHHIMRAIRQRVLRLTRLSPEIVELSEPLQVVRYGEGGH 324
Query: 66 YEPHPD--------------FFTDEINTKNGGERVATMLMYLP----------------W 95
Y H D +E R T+L YL
Sbjct: 325 YHAHVDSGPVYPETICSHTKLVANESVPFETSCRYMTVLFYLNNVTGGGETVFPVADNRT 384
Query: 96 WNELS-------------DCGKKGLSIKPKMGDALLFWSMKPDGT-----LDPLSMHGAC 137
++E+S C K L +KP+ G A+ +++ PDG +D S+HG C
Sbjct: 385 YDEMSLIQDDVDLRDTRRHCDKGNLRVKPQQGTAVFWYNYLPDGQGWVGDVDDYSLHGGC 444
Query: 138 PVIKGDKWSCTKWMRV 153
V +G KW W+ V
Sbjct: 445 LVTRGTKWIANNWINV 460
>ER1A_HUMAN (Q96HE7) ERO1-like protein alpha precursor (EC 1.8.4.-)
(ERO1-Lalpha) (Oxidoreductin 1-lalpha) (Endoplasmic
oxidoreductin 1-like protein) (ERO1-L) (UNQ434/PRO865)
Length = 468
Score = 32.0 bits (71), Expect = 0.61
Identities = 12/33 (36%), Positives = 20/33 (60%), Gaps = 6/33 (18%)
Query: 94 PWWNELSDCGKKGLSIKPKMGDALLFWSMKPDG 126
P+WN++S CG++ ++KP D + PDG
Sbjct: 86 PFWNDISQCGRRDCAVKPCQSDEV------PDG 112
>ER1A_RAT (Q8R4A1) ERO1-like protein alpha precursor (EC 1.8.4.-)
(ERO1-Lalpha) (Oxidoreductin 1-lalpha) (Endoplasmic
oxidoreductin 1-like protein) (ERO1-L) (Global
ischemia-induced protein 11)
Length = 464
Score = 30.0 bits (66), Expect = 2.3
Identities = 11/33 (33%), Positives = 20/33 (60%), Gaps = 6/33 (18%)
Query: 94 PWWNELSDCGKKGLSIKPKMGDALLFWSMKPDG 126
P+WN+++ CG++ ++KP D + PDG
Sbjct: 86 PFWNDINQCGRRDCAVKPCHSDEV------PDG 112
>ER1A_MOUSE (Q8R180) ERO1-like protein alpha precursor (EC 1.8.4.-)
(ERO1-Lalpha) (Oxidoreductin 1-lalpha) (Endoplasmic
oxidoreductin 1-like protein) (ERO1-L)
Length = 464
Score = 30.0 bits (66), Expect = 2.3
Identities = 11/33 (33%), Positives = 20/33 (60%), Gaps = 6/33 (18%)
Query: 94 PWWNELSDCGKKGLSIKPKMGDALLFWSMKPDG 126
P+WN+++ CG++ ++KP D + PDG
Sbjct: 86 PFWNDINQCGRRDCAVKPCHSDEV------PDG 112
>CYF_PSINU (Q8WI07) Apocytochrome f precursor
Length = 321
Score = 30.0 bits (66), Expect = 2.3
Identities = 18/48 (37%), Positives = 26/48 (53%), Gaps = 1/48 (2%)
Query: 78 NTKNGGERVATMLMYLPWWNELSDCGKKGLSIKPKMGDALLFWSMKPD 125
N K GG V +L+ LP EL+ + +K K+GD LF + +PD
Sbjct: 98 NGKKGGINVGAVLI-LPEGFELAPYNRIPAEMKDKIGDLALFQNYRPD 144
>PHAF_PORPU (P51215) Allophycocyanin beta 18 chain
Length = 169
Score = 29.6 bits (65), Expect = 3.0
Identities = 20/64 (31%), Positives = 32/64 (49%), Gaps = 11/64 (17%)
Query: 8 DETGKSVDNSARTSSGTFINRGHDKILRNIEQRIADFTFIPVENGESVNILHYEVGQKYE 67
D TG+ +D +A T +F + G D+I +IA+ + N ++ NIL Q YE
Sbjct: 13 DLTGRYLDKAAVTQLESFFSSGLDRI------KIAE-----IINNQATNILKEASAQLYE 61
Query: 68 PHPD 71
P+
Sbjct: 62 EQPE 65
>LRP2_RAT (P98158) Low-density lipoprotein receptor-related protein 2
precursor (Megalin) (Glycoprotein 330) (gp330)
Length = 4660
Score = 28.5 bits (62), Expect = 6.8
Identities = 9/17 (52%), Positives = 15/17 (87%)
Query: 105 KGLSIKPKMGDALLFWS 121
+GL++ P+MGD ++FWS
Sbjct: 1556 RGLALDPRMGDNVMFWS 1572
>EFTS_STAAW (Q8NWZ6) Elongation factor Ts (EF-Ts)
Length = 293
Score = 28.5 bits (62), Expect = 6.8
Identities = 11/32 (34%), Positives = 19/32 (59%)
Query: 46 FIPVENGESVNILHYEVGQKYEPHPDFFTDEI 77
F+ + G+ V+ + YEVG+ E + F DE+
Sbjct: 257 FLKTKGGKLVDFVRYEVGEGMEKREENFADEV 288
>EFTS_STAAN (P99171) Elongation factor Ts (EF-Ts)
Length = 293
Score = 28.5 bits (62), Expect = 6.8
Identities = 11/32 (34%), Positives = 19/32 (59%)
Query: 46 FIPVENGESVNILHYEVGQKYEPHPDFFTDEI 77
F+ + G+ V+ + YEVG+ E + F DE+
Sbjct: 257 FLKTKGGKLVDFVRYEVGEGMEKREENFADEV 288
>EFTS_STAAM (P64054) Elongation factor Ts (EF-Ts)
Length = 293
Score = 28.5 bits (62), Expect = 6.8
Identities = 11/32 (34%), Positives = 19/32 (59%)
Query: 46 FIPVENGESVNILHYEVGQKYEPHPDFFTDEI 77
F+ + G+ V+ + YEVG+ E + F DE+
Sbjct: 257 FLKTKGGKLVDFVRYEVGEGMEKREENFADEV 288
>RPOC_PHOLL (Q7N9A3) DNA-directed RNA polymerase beta' chain (EC
2.7.7.6) (RNAP beta' subunit) (Transcriptase beta'
chain) (RNA polymerase beta' subunit)
Length = 1406
Score = 28.1 bits (61), Expect = 8.9
Identities = 27/107 (25%), Positives = 49/107 (45%), Gaps = 12/107 (11%)
Query: 31 DKILRNIEQRIADFTFIPVENG-ESVNILHYEVGQKYEPHPDFFTDEINTKNGGERVATM 89
D LR+IE+ + +++ VE G S+ ++Y + F DE + K G E + ++
Sbjct: 129 DMPLRDIERVLYFESYVVVEGGMTSLERSQILTEEQYLDALEEFGDEFDAKMGAEAIQSL 188
Query: 90 LMYLPWWNEL-----------SDCGKKGLSIKPKMGDALLFWSMKPD 125
L L NE S+ +K L+ + K+ +A + KP+
Sbjct: 189 LKNLDLENECETLREELNETNSETKRKKLTKRIKLLEAFIQSGNKPE 235
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.317 0.136 0.438
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 21,440,039
Number of Sequences: 164201
Number of extensions: 903178
Number of successful extensions: 1862
Number of sequences better than 10.0: 20
Number of HSP's better than 10.0 without gapping: 17
Number of HSP's successfully gapped in prelim test: 3
Number of HSP's that attempted gapping in prelim test: 1834
Number of HSP's gapped (non-prelim): 20
length of query: 160
length of database: 59,974,054
effective HSP length: 101
effective length of query: 59
effective length of database: 43,389,753
effective search space: 2559995427
effective search space used: 2559995427
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 61 (28.1 bits)
Medicago: description of AC149547.6


