

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC149547.4 + phase: 0
(407 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
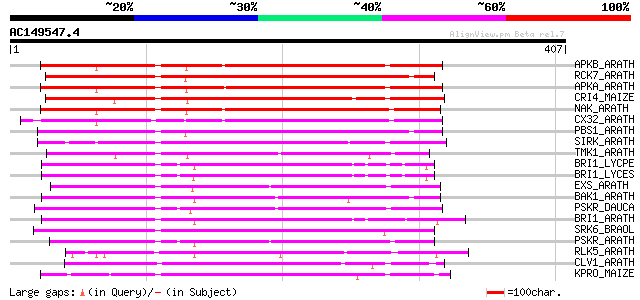
Score E
Sequences producing significant alignments: (bits) Value
APKB_ARATH (P46573) Protein kinase APK1B, chloroplast precursor ... 235 1e-61
RCK7_ARATH (Q9LQQ8) Putative serine/threonine-protein kinase RLC... 228 2e-59
APKA_ARATH (Q06548) Protein kinase APK1A, chloroplast precursor ... 228 2e-59
CRI4_MAIZE (O24585) Putative receptor protein kinase CRINKLY4 pr... 227 5e-59
NAK_ARATH (P43293) Probable serine/threonine-protein kinase NAK ... 223 5e-58
CX32_ARATH (P27450) Probable serine/threonine-protein kinase Cx3... 212 1e-54
PBS1_ARATH (Q9FE20) Serine/threonine-protein kinase PBS1 (EC 2.7... 211 3e-54
SIRK_ARATH (O64483) Senescence-induced receptor-like serine/thre... 204 3e-52
TMK1_ARATH (P43298) Putative receptor protein kinase TMK1 precur... 190 6e-48
BRI1_LYCPE (Q8L899) Systemin receptor SR160 precursor (EC 2.7.1.... 187 4e-47
BRI1_LYCES (Q8GUQ5) Brassinosteroid LRR receptor kinase precurso... 187 5e-47
EXS_ARATH (Q9LYN8) Leucine-rich repeat receptor protein kinase E... 186 9e-47
BAK1_ARATH (Q94F62) BRASSINOSTEROID INSENSITIVE 1-associated rec... 182 1e-45
PSKR_DAUCA (Q8LPB4) Phytosulfokine receptor precursor (EC 2.7.1.... 182 1e-45
BRI1_ARATH (O22476) BRASSINOSTEROID INSENSITIVE 1 precursor (EC ... 182 2e-45
SRK6_BRAOL (Q09092) Putative serine/threonine-protein kinase rec... 173 8e-43
PSKR_ARATH (Q9ZVR7) Putative phytosulfokine receptor precursor (... 170 7e-42
RLK5_ARATH (P47735) Receptor-like protein kinase 5 precursor (EC... 160 4e-39
CLV1_ARATH (Q9SYQ8) Receptor protein kinase CLAVATA1 precursor (... 158 2e-38
KPRO_MAIZE (P17801) Putative receptor protein kinase ZmPK1 precu... 156 1e-37
>APKB_ARATH (P46573) Protein kinase APK1B, chloroplast precursor (EC
2.7.1.-)
Length = 412
Score = 235 bits (600), Expect = 1e-61
Identities = 128/308 (41%), Positives = 190/308 (61%), Gaps = 20/308 (6%)
Query: 23 SLRCFTIEELERATKNFSQDCLLGSGAFCNVYKGIFELEG----------ILAIKRPHSE 72
+L+ FT EL+ AT+NF D +LG G F +V+KG + + ++A+K+ + +
Sbjct: 53 NLKSFTFAELKAATRNFRPDSVLGEGGFGSVFKGWIDEQTLTASKPGTGVVIAVKKLNQD 112
Query: 73 SFLSVEEFRNEVRLLSAVKHKNLIGLVGYCEEPERDGAKILVYEYVPNGSLLEYMM--GN 130
+ +E+ EV L H NL+ L+GYC E E ++LVYE++P GSL ++ G+
Sbjct: 113 GWQGHQEWLAEVNYLGQFSHPNLVKLIGYCLEDEH---RLLVYEFMPRGSLENHLFRRGS 169
Query: 131 RRRSLTWKQRINIAIGAAKGIAYLHEKVKPSIIHRDIKPSNILLGESFEAKVSDFGLVKS 190
+ L+W R+ +A+GAAKG+A+LH + S+I+RD K SNILL + AK+SDFGL K
Sbjct: 170 YFQPLSWTLRLKVALGAAKGLAFLHN-AETSVIYRDFKTSNILLDSEYNAKLSDFGLAKD 228
Query: 191 GPTGDQSHVSSQIKGTPGYLDPAYCSSCHLTKFSDVYSFGVILLQLISARPAVDNAENPS 250
GPTGD+SHVS++I GT GY P Y ++ HLT SDVYS+GV+LL+++S R AVD P
Sbjct: 229 GPTGDKSHVSTRIMGTYGYAAPEYLATGHLTTKSDVYSYGVVLLEVLSGRRAVDKNRPPG 288
Query: 251 NQHIIDWARPSI-EKGIIAEIMDANLFCQSEPCNMEVMLKVGQLGIRCVAQEPKHRPTMT 309
Q +++WARP + K + ++D L + +ME KV L +RC+ E K RP M
Sbjct: 289 EQKLVEWARPLLANKRKLFRVIDNRL---QDQYSMEEACKVATLALRCLTFEIKLRPNMN 345
Query: 310 QVCREIEH 317
+V +EH
Sbjct: 346 EVVSHLEH 353
>RCK7_ARATH (Q9LQQ8) Putative serine/threonine-protein kinase
RLCKVII (EC 2.7.1.37)
Length = 423
Score = 228 bits (582), Expect = 2e-59
Identities = 120/289 (41%), Positives = 183/289 (62%), Gaps = 10/289 (3%)
Query: 27 FTIEELERATKNFSQDCLLGSGAFCNVYKGIFE-LEGILAIKRPHSESFLSVEEFRNEVR 85
FT +EL AT NF DC LG G F V+KG E L+ ++AIK+ + EF EV
Sbjct: 91 FTFQELAEATGNFRSDCFLGEGGFGKVFKGTIEKLDQVVAIKQLDRNGVQGIREFVVEVL 150
Query: 86 LLSAVKHKNLIGLVGYCEEPERDGAKILVYEYVPNGSLLEYM--MGNRRRSLTWKQRINI 143
LS H NL+ L+G+C E ++ ++LVYEY+P GSL +++ + + ++ L W R+ I
Sbjct: 151 TLSLADHPNLVKLIGFCAEGDQ---RLLVYEYMPQGSLEDHLHVLPSGKKPLDWNTRMKI 207
Query: 144 AIGAAKGIAYLHEKVKPSIIHRDIKPSNILLGESFEAKVSDFGLVKSGPTGDQSHVSSQI 203
A GAA+G+ YLH+++ P +I+RD+K SNILLGE ++ K+SDFGL K GP+GD++HVS+++
Sbjct: 208 AAGAARGLEYLHDRMTPPVIYRDLKCSNILLGEDYQPKLSDFGLAKVGPSGDKTHVSTRV 267
Query: 204 KGTPGYLDPAYCSSCHLTKFSDVYSFGVILLQLISARPAVDNAENPSNQHIIDWARPSI- 262
GT GY P Y + LT SD+YSFGV+LL+LI+ R A+DN + +Q+++ WARP
Sbjct: 268 MGTYGYCAPDYAMTGQLTFKSDIYSFGVVLLELITGRKAIDNTKTRKDQNLVGWARPLFK 327
Query: 263 EKGIIAEIMDANLFCQSEPCNMEVMLKVGQLGIRCVAQEPKHRPTMTQV 311
++ +++D L Q + L + + CV ++P RP ++ V
Sbjct: 328 DRRNFPKMVDPLLQGQYPVRGLYQALAISAM---CVQEQPTMRPVVSDV 373
>APKA_ARATH (Q06548) Protein kinase APK1A, chloroplast precursor (EC
2.7.1.-)
Length = 410
Score = 228 bits (581), Expect = 2e-59
Identities = 124/308 (40%), Positives = 189/308 (61%), Gaps = 20/308 (6%)
Query: 23 SLRCFTIEELERATKNFSQDCLLGSGAFCNVYKGIFELEG----------ILAIKRPHSE 72
+L+ F+ EL+ AT+NF D +LG G F V+KG + + ++A+K+ + +
Sbjct: 52 NLKSFSFAELKSATRNFRPDSVLGEGGFGCVFKGWIDEKSLTASRPGTGLVIAVKKLNQD 111
Query: 73 SFLSVEEFRNEVRLLSAVKHKNLIGLVGYCEEPERDGAKILVYEYVPNGSLLEYMM--GN 130
+ +E+ EV L H++L+ L+GYC E E ++LVYE++P GSL ++ G
Sbjct: 112 GWQGHQEWLAEVNYLGQFSHRHLVKLIGYCLEDEH---RLLVYEFMPRGSLENHLFRRGL 168
Query: 131 RRRSLTWKQRINIAIGAAKGIAYLHEKVKPSIIHRDIKPSNILLGESFEAKVSDFGLVKS 190
+ L+WK R+ +A+GAAKG+A+LH + +I+RD K SNILL + AK+SDFGL K
Sbjct: 169 YFQPLSWKLRLKVALGAAKGLAFLHSS-ETRVIYRDFKTSNILLDSEYNAKLSDFGLAKD 227
Query: 191 GPTGDQSHVSSQIKGTPGYLDPAYCSSCHLTKFSDVYSFGVILLQLISARPAVDNAENPS 250
GP GD+SHVS+++ GT GY P Y ++ HLT SDVYSFGV+LL+L+S R AVD
Sbjct: 228 GPIGDKSHVSTRVMGTHGYAAPEYLATGHLTTKSDVYSFGVVLLELLSGRRAVDKNRPSG 287
Query: 251 NQHIIDWARP-SIEKGIIAEIMDANLFCQSEPCNMEVMLKVGQLGIRCVAQEPKHRPTMT 309
+++++WA+P + K I ++D L + +ME KV L +RC+ E K RP M+
Sbjct: 288 ERNLVEWAKPYLVNKRKIFRVIDNRL---QDQYSMEEACKVATLSLRCLTTEIKLRPNMS 344
Query: 310 QVCREIEH 317
+V +EH
Sbjct: 345 EVVSHLEH 352
>CRI4_MAIZE (O24585) Putative receptor protein kinase CRINKLY4
precursor (EC 2.7.1.-)
Length = 901
Score = 227 bits (578), Expect = 5e-59
Identities = 128/298 (42%), Positives = 184/298 (60%), Gaps = 13/298 (4%)
Query: 27 FTIEELERATKNFSQDCLLGSGAFCNVYKGIFELEGILAIKRPHSESFL--SVEEFRNEV 84
F+ EELE+AT FS+D +G G+F V+KGI ++A+KR S + S +EF NE+
Sbjct: 493 FSYEELEQATGGFSEDSQVGKGSFSCVFKGILRDGTVVAVKRAIKASDVKKSSKEFHNEL 552
Query: 85 RLLSAVKHKNLIGLVGYCEEPERDGAKILVYEYVPNGSLLEYMMG---NRRRSLTWKQRI 141
LLS + H +L+ L+GYCE+ ++LVYE++ +GSL +++ G N ++ L W +R+
Sbjct: 553 DLLSRLNHAHLLNLLGYCEDGSE---RLLVYEFMAHGSLYQHLHGKDPNLKKRLNWARRV 609
Query: 142 NIAIGAAKGIAYLHEKVKPSIIHRDIKPSNILLGESFEAKVSDFGLVKSGPTGDQSHVSS 201
IA+ AA+GI YLH P +IHRDIK SNIL+ E A+V+DFGL GP + +S
Sbjct: 610 TIAVQAARGIEYLHGYACPPVIHRDIKSSNILIDEDHNARVADFGLSILGPADSGTPLSE 669
Query: 202 QIKGTPGYLDPAYCSSCHLTKFSDVYSFGVILLQLISARPAVDNAENPSNQHIIDWARPS 261
GT GYLDP Y +LT SDVYSFGV+LL+++S R A+D N I++WA P
Sbjct: 670 LPAGTLGYLDPEYYRLHYLTTKSDVYSFGVVLLEILSGRKAIDMQFEEGN--IVEWAVPL 727
Query: 262 IEKGIIAEIMDANLFCQSEPCNMEVMLKVGQLGIRCVAQEPKHRPTMTQVCREIEHAL 319
I+ G I I+D L S P ++E + K+ + +CV K RP+M +V +EHAL
Sbjct: 728 IKAGDIFAILDPVL---SPPSDLEALKKIASVACKCVRMRGKDRPSMDKVTTALEHAL 782
>NAK_ARATH (P43293) Probable serine/threonine-protein kinase NAK (EC
2.7.1.37)
Length = 389
Score = 223 bits (569), Expect = 5e-58
Identities = 119/307 (38%), Positives = 187/307 (60%), Gaps = 20/307 (6%)
Query: 23 SLRCFTIEELERATKNFSQDCLLGSGAFCNVYKGIFELEG----------ILAIKRPHSE 72
+L+ F++ EL+ AT+NF D ++G G F V+KG + ++A+KR + E
Sbjct: 52 NLKNFSLSELKSATRNFRPDSVVGEGGFGCVFKGWIDESSLAPSKPGTGIVIAVKRLNQE 111
Query: 73 SFLSVEEFRNEVRLLSAVKHKNLIGLVGYCEEPERDGAKILVYEYVPNGSLLEYMM--GN 130
F E+ E+ L + H NL+ L+GYC E E ++LVYE++ GSL ++ G
Sbjct: 112 GFQGHREWLAEINYLGQLDHPNLVKLIGYCLEEEH---RLLVYEFMTRGSLENHLFRRGT 168
Query: 131 RRRSLTWKQRINIAIGAAKGIAYLHEKVKPSIIHRDIKPSNILLGESFEAKVSDFGLVKS 190
+ L+W R+ +A+GAA+G+A+LH +P +I+RD K SNILL ++ AK+SDFGL +
Sbjct: 169 FYQPLSWNTRVRMALGAARGLAFLHN-AQPQVIYRDFKASNILLDSNYNAKLSDFGLARD 227
Query: 191 GPTGDQSHVSSQIKGTPGYLDPAYCSSCHLTKFSDVYSFGVILLQLISARPAVDNAENPS 250
GP GD SHVS+++ GT GY P Y ++ HL+ SDVYSFGV+LL+L+S R A+D +
Sbjct: 228 GPMGDNSHVSTRVMGTQGYAAPEYLATGHLSVKSDVYSFGVVLLELLSGRRAIDKNQPVG 287
Query: 251 NQHIIDWARPSI-EKGIIAEIMDANLFCQSEPCNMEVMLKVGQLGIRCVAQEPKHRPTMT 309
+++DWARP + K + +MD L Q ++ LK+ L + C++ + K RPTM
Sbjct: 288 EHNLVDWARPYLTNKRRLLRVMDPRLQGQ---YSLTRALKIAVLALDCISIDAKSRPTMN 344
Query: 310 QVCREIE 316
++ + +E
Sbjct: 345 EIVKTME 351
>CX32_ARATH (P27450) Probable serine/threonine-protein kinase Cx32,
chloroplast precursor (EC 2.7.1.37)
Length = 419
Score = 212 bits (540), Expect = 1e-54
Identities = 121/320 (37%), Positives = 188/320 (57%), Gaps = 24/320 (7%)
Query: 9 DAGKEYKSSKYDKISLRCFTIEELERATKNFSQDCLLGSGAFCNVYKGIFELEG------ 62
D+GK +S +L+ + +L+ ATKNF D +LG G F VY+G +
Sbjct: 61 DSGKLLESP-----NLKVYNFLDLKTATKNFKPDSMLGQGGFGKVYRGWVDATTLAPSRV 115
Query: 63 ----ILAIKRPHSESFLSVEEFRNEVRLLSAVKHKNLIGLVGYCEEPERDGAKILVYEYV 118
I+AIKR +SES E+R+EV L + H+NL+ L+GYC E D +LVYE++
Sbjct: 116 GSGMIVAIKRLNSESVQGFAEWRSEVNFLGMLSHRNLVKLLGYCRE---DKELLLVYEFM 172
Query: 119 PNGSLLEYMMGNRRRSLTWKQRINIAIGAAKGIAYLHEKVKPSIIHRDIKPSNILLGESF 178
P GSL ++ R W RI I IGAA+G+A+LH ++ +I+RD K SNILL ++
Sbjct: 173 PKGSLESHLF-RRNDPFPWDLRIKIVIGAARGLAFLHS-LQREVIYRDFKASNILLDSNY 230
Query: 179 EAKVSDFGLVKSGPTGDQSHVSSQIKGTPGYLDPAYCSSCHLTKFSDVYSFGVILLQLIS 238
+AK+SDFGL K GP ++SHV+++I GT GY P Y ++ HL SDV++FGV+LL++++
Sbjct: 231 DAKLSDFGLAKLGPADEKSHVTTRIMGTYGYAAPEYMATGHLYVKSDVFAFGVVLLEIMT 290
Query: 239 ARPAVDNAENPSNQHIIDWARPSI-EKGIIAEIMDANLFCQSEPCNMEVMLKVGQLGIRC 297
A + + ++DW RP + K + +IMD + Q +V ++ ++ + C
Sbjct: 291 GLTAHNTKRPRGQESLVDWLRPELSNKHRVKQIMDKGIKGQ---YTTKVATEMARITLSC 347
Query: 298 VAQEPKHRPTMTQVCREIEH 317
+ +PK+RP M +V +EH
Sbjct: 348 IEPDPKNRPHMKEVVEVLEH 367
>PBS1_ARATH (Q9FE20) Serine/threonine-protein kinase PBS1 (EC
2.7.1.37) (AvrPphB susceptible protein 1)
Length = 456
Score = 211 bits (537), Expect = 3e-54
Identities = 117/301 (38%), Positives = 174/301 (56%), Gaps = 10/301 (3%)
Query: 21 KISLRCFTIEELERATKNFSQDCLLGSGAFCNVYKGIFELEG-ILAIKRPHSESFLSVEE 79
+I+ F EL AT NF D LG G F VYKG + G ++A+K+ E
Sbjct: 68 QIAAHTFAFRELAAATMNFHPDTFLGEGGFGRVYKGRLDSTGQVVAVKQLDRNGLQGNRE 127
Query: 80 FRNEVRLLSAVKHKNLIGLVGYCEEPERDGAKILVYEYVPNGSLLEYM--MGNRRRSLTW 137
F EV +LS + H NL+ L+GYC + ++ ++LVYE++P GSL +++ + + +L W
Sbjct: 128 FLVEVLMLSLLHHPNLVNLIGYCADGDQ---RLLVYEFMPLGSLEDHLHDLPPDKEALDW 184
Query: 138 KQRINIAIGAAKGIAYLHEKVKPSIIHRDIKPSNILLGESFEAKVSDFGLVKSGPTGDQS 197
R+ IA GAAKG+ +LH+K P +I+RD K SNILL E F K+SDFGL K GPTGD+S
Sbjct: 185 NMRMKIAAGAAKGLEFLHDKANPPVIYRDFKSSNILLDEGFHPKLSDFGLAKLGPTGDKS 244
Query: 198 HVSSQIKGTPGYLDPAYCSSCHLTKFSDVYSFGVILLQLISARPAVDNAENPSNQHIIDW 257
HVS+++ GT GY P Y + LT SDVYSFGV+ L+LI+ R A+D+ Q+++ W
Sbjct: 245 HVSTRVMGTYGYCAPEYAMTGQLTVKSDVYSFGVVFLELITGRKAIDSEMPHGEQNLVAW 304
Query: 258 ARPSI-EKGIIAEIMDANLFCQSEPCNMEVMLKVGQLGIRCVAQEPKHRPTMTQVCREIE 316
ARP ++ ++ D L + + L V + C+ ++ RP + V +
Sbjct: 305 ARPLFNDRRKFIKLADPRLKGRFPTRALYQALAVASM---CIQEQAATRPLIADVVTALS 361
Query: 317 H 317
+
Sbjct: 362 Y 362
>SIRK_ARATH (O64483) Senescence-induced receptor-like
serine/threonine kinase precursor (FLG22-induced
receptor-like kinase 1)
Length = 876
Score = 204 bits (520), Expect = 3e-52
Identities = 115/300 (38%), Positives = 173/300 (57%), Gaps = 10/300 (3%)
Query: 21 KISLRCFTIEELERATKNFSQDCLLGSGAFCNVYKGIFELEGILAIKRPHSESFLSVEEF 80
K + R F E+ T NF + ++G G F VY G+ E + A+K ES +EF
Sbjct: 558 KTAKRYFKYSEVVNITNNFER--VIGKGGFGKVYHGVINGEQV-AVKVLSEESAQGYKEF 614
Query: 81 RNEVRLLSAVKHKNLIGLVGYCEEPERDGAKILVYEYVPNGSLLEYMMGNRRRSLTWKQR 140
R EV LL V H NL LVGYC E +L+YEY+ N +L +Y+ G R L+W++R
Sbjct: 615 RAEVDLLMRVHHTNLTSLVGYCNEINH---MVLIYEYMANENLGDYLAGKRSFILSWEER 671
Query: 141 INIAIGAAKGIAYLHEKVKPSIIHRDIKPSNILLGESFEAKVSDFGLVKSGPTGDQSHVS 200
+ I++ AA+G+ YLH KP I+HRD+KP+NILL E +AK++DFGL +S +S
Sbjct: 672 LKISLDAAQGLEYLHNGCKPPIVHRDVKPTNILLNEKLQAKMADFGLSRSFSVEGSGQIS 731
Query: 201 SQIKGTPGYLDPAYCSSCHLTKFSDVYSFGVILLQLISARPAVDNAENPSNQHIIDWARP 260
+ + G+ GYLDP Y S+ + + SDVYS GV+LL++I+ +PA+ +++ HI D R
Sbjct: 732 TVVAGSIGYLDPEYYSTRQMNEKSDVYSLGVVLLEVITGQPAIASSKT-EKVHISDHVRS 790
Query: 261 SIEKGIIAEIMDANLFCQSEPCNMEVMLKVGQLGIRCVAQEPKHRPTMTQVCREIEHALY 320
+ G I I+D L E ++ K+ ++ + C RPTM+QV E++ +Y
Sbjct: 791 ILANGDIRGIVDQRL---RERYDVGSAWKMSEIALACTEHTSAQRPTMSQVVMELKQIVY 847
>TMK1_ARATH (P43298) Putative receptor protein kinase TMK1 precursor
(EC 2.7.1.-)
Length = 942
Score = 190 bits (482), Expect = 6e-48
Identities = 109/288 (37%), Positives = 166/288 (56%), Gaps = 13/288 (4%)
Query: 28 TIEELERATKNFSQDCLLGSGAFCNVYKGIFELEGILAIKRPHSESFLS--VEEFRNEVR 85
+I+ L T NFS D +LGSG F VYKG +A+KR + EF++E+
Sbjct: 577 SIQVLRSVTNNFSSDNILGSGGFGVVYKGELHDGTKIAVKRMENGVIAGKGFAEFKSEIA 636
Query: 86 LLSAVKHKNLIGLVGYCEEPERDGAKILVYEYVPNGSLLEYMMG---NRRRSLTWKQRIN 142
+L+ V+H++L+ L+GYC + K+LVYEY+P G+L ++ + L WKQR+
Sbjct: 637 VLTKVRHRHLVTLLGYCLDGNE---KLLVYEYMPQGTLSRHLFEWSEEGLKPLLWKQRLT 693
Query: 143 IAIGAAKGIAYLHEKVKPSIIHRDIKPSNILLGESFEAKVSDFGLVKSGPTGDQSHVSSQ 202
+A+ A+G+ YLH S IHRD+KPSNILLG+ AKV+DFGLV+ P G S + ++
Sbjct: 694 LALDVARGVEYLHGLAHQSFIHRDLKPSNILLGDDMRAKVADFGLVRLAPEGKGS-IETR 752
Query: 203 IKGTPGYLDPAYCSSCHLTKFSDVYSFGVILLQLISARPAVDNAENPSNQHIIDWARPSI 262
I GT GYL P Y + +T DVYSFGVIL++LI+ R ++D ++ + H++ W +
Sbjct: 753 IAGTFGYLAPEYAVTGRVTTKVDVYSFGVILMELITGRKSLDESQPEESIHLVSWFKRMY 812
Query: 263 --EKGIIAEIMDANLFCQSEPCNMEVMLKVGQLGIRCVAQEPKHRPTM 308
++ + +D + E + + V +L C A+EP RP M
Sbjct: 813 INKEASFKKAIDTTIDLDEE--TLASVHTVAELAGHCCAREPYQRPDM 858
>BRI1_LYCPE (Q8L899) Systemin receptor SR160 precursor (EC 2.7.1.37)
(Brassinosteroid LRR receptor kinase)
Length = 1207
Score = 187 bits (475), Expect = 4e-47
Identities = 114/293 (38%), Positives = 170/293 (57%), Gaps = 14/293 (4%)
Query: 24 LRCFTIEELERATKNFSQDCLLGSGAFCNVYKGIFELEGILAIKRPHSESFLSVEEFRNE 83
LR T +L AT F D L+GSG F +VYK + ++AIK+ S EF E
Sbjct: 873 LRKLTFADLLEATNGFHNDSLVGSGGFGDVYKAQLKDGSVVAIKKLIHVSGQGDREFTAE 932
Query: 84 VRLLSAVKHKNLIGLVGYCEEPERDGAKILVYEYVPNGSLLEYMMGNRRRS---LTWKQR 140
+ + +KH+NL+ L+GYC+ E ++LVYEY+ GSL E ++ +R+++ L W R
Sbjct: 933 METIGKIKHRNLVPLLGYCKVGEE---RLLVYEYMKYGSL-EDVLHDRKKTGIKLNWPAR 988
Query: 141 INIAIGAAKGIAYLHEKVKPSIIHRDIKPSNILLGESFEAKVSDFGLVKSGPTGDQSHVS 200
IAIGAA+G+A+LH P IIHRD+K SN+LL E+ EA+VSDFG+ + D
Sbjct: 989 RKIAIGAARGLAFLHHNCIPHIIHRDMKSSNVLLDENLEARVSDFGMARLMSAMDTHLSV 1048
Query: 201 SQIKGTPGYLDPAYCSSCHLTKFSDVYSFGVILLQLISARPAVDNAENPSNQHIIDWARP 260
S + GTPGY+ P Y S + DVYS+GV+LL+L++ + D+A+ N +++ W +
Sbjct: 1049 STLAGTPGYVPPEYYQSFRCSTKGDVYSYGVVLLELLTGKQPTDSADFGDN-NLVGWVKL 1107
Query: 261 SIEKGIIAEIMDANLFCQSEPCNMEVMLKVGQLGIRCVAQEPKH--RPTMTQV 311
KG I ++ D L E ++E+ L + L + C + +H RPTM QV
Sbjct: 1108 H-AKGKITDVFDRELL--KEDASIEIEL-LQHLKVACACLDDRHWKRPTMIQV 1156
>BRI1_LYCES (Q8GUQ5) Brassinosteroid LRR receptor kinase precursor (EC
2.7.1.37) (tBRI1) (Altered brassinolide sensitivity 1)
(Systemin receptor SR160)
Length = 1207
Score = 187 bits (474), Expect = 5e-47
Identities = 114/293 (38%), Positives = 169/293 (56%), Gaps = 14/293 (4%)
Query: 24 LRCFTIEELERATKNFSQDCLLGSGAFCNVYKGIFELEGILAIKRPHSESFLSVEEFRNE 83
LR T +L AT F D L+GSG F +VYK + ++AIK+ S EF E
Sbjct: 873 LRKLTFADLLEATNGFHNDSLVGSGGFGDVYKAQLKDGSVVAIKKLIHVSGQGDREFTAE 932
Query: 84 VRLLSAVKHKNLIGLVGYCEEPERDGAKILVYEYVPNGSLLEYMMGNRRR---SLTWKQR 140
+ + +KH+NL+ L+GYC+ E ++LVYEY+ GSL E ++ +R++ L W R
Sbjct: 933 METIGKIKHRNLVPLLGYCKVGEE---RLLVYEYMKYGSL-EDVLHDRKKIGIKLNWPAR 988
Query: 141 INIAIGAAKGIAYLHEKVKPSIIHRDIKPSNILLGESFEAKVSDFGLVKSGPTGDQSHVS 200
IAIGAA+G+A+LH P IIHRD+K SN+LL E+ EA+VSDFG+ + D
Sbjct: 989 RKIAIGAARGLAFLHHNCIPHIIHRDMKSSNVLLDENLEARVSDFGMARLMSAMDTHLSV 1048
Query: 201 SQIKGTPGYLDPAYCSSCHLTKFSDVYSFGVILLQLISARPAVDNAENPSNQHIIDWARP 260
S + GTPGY+ P Y S + DVYS+GV+LL+L++ + D+A+ N +++ W +
Sbjct: 1049 STLAGTPGYVPPEYYQSFRCSTKGDVYSYGVVLLELLTGKQPTDSADFGDN-NLVGWVKL 1107
Query: 261 SIEKGIIAEIMDANLFCQSEPCNMEVMLKVGQLGIRCVAQEPKH--RPTMTQV 311
KG I ++ D L E ++E+ L + L + C + +H RPTM QV
Sbjct: 1108 H-AKGKITDVFDRELL--KEDASIEIEL-LQHLKVACACLDDRHWKRPTMIQV 1156
>EXS_ARATH (Q9LYN8) Leucine-rich repeat receptor protein kinase EXS
precursor (EC 2.7.1.37) (Extra sporogenous cells protein)
(EXCESS MICROSPOROCYTES1 protein)
Length = 1192
Score = 186 bits (472), Expect = 9e-47
Identities = 110/289 (38%), Positives = 160/289 (55%), Gaps = 10/289 (3%)
Query: 31 ELERATKNFSQDCLLGSGAFCNVYKGIFELEGILAIKRPHSESFLSVEEFRNEVRLLSAV 90
++ AT +FS+ ++G G F VYK E +A+K+ EF E+ L V
Sbjct: 909 DIVEATDHFSKKNIIGDGGFGTVYKACLPGEKTVAVKKLSEAKTQGNREFMAEMETLGKV 968
Query: 91 KHKNLIGLVGYCEEPERDGAKILVYEYVPNGSLLEYMMGNRR--RSLTWKQRINIAIGAA 148
KH NL+ L+GYC E K+LVYEY+ NGSL ++ L W +R+ IA+GAA
Sbjct: 969 KHPNLVSLLGYCSFSEE---KLLVYEYMVNGSLDHWLRNQTGMLEVLDWSKRLKIAVGAA 1025
Query: 149 KGIAYLHEKVKPSIIHRDIKPSNILLGESFEAKVSDFGLVKSGPTGDQSHVSSQIKGTPG 208
+G+A+LH P IIHRDIK SNILL FE KV+DFGL + + +SHVS+ I GT G
Sbjct: 1026 RGLAFLHHGFIPHIIHRDIKASNILLDGDFEPKVADFGLARL-ISACESHVSTVIAGTFG 1084
Query: 209 YLDPAYCSSCHLTKFSDVYSFGVILLQLISAR-PAVDNAENPSNQHIIDWARPSIEKGII 267
Y+ P Y S T DVYSFGVILL+L++ + P + + +++ WA I +G
Sbjct: 1085 YIPPEYGQSARATTKGDVYSFGVILLELVTGKEPTGPDFKESEGGNLVGWAIQKINQGKA 1144
Query: 268 AEIMDANLFCQSEPCNMEVMLKVGQLGIRCVAQEPKHRPTMTQVCREIE 316
+++D L L++ Q+ + C+A+ P RP M V + ++
Sbjct: 1145 VDVIDPLLV---SVALKNSQLRLLQIAMLCLAETPAKRPNMLDVLKALK 1190
>BAK1_ARATH (Q94F62) BRASSINOSTEROID INSENSITIVE 1-associated
receptor kinase 1 precursor (EC 2.7.1.37)
(BRI1-associated receptor kinase 1) (Somatic
embryogenesis receptor-like kinase 3)
Length = 615
Score = 182 bits (463), Expect = 1e-45
Identities = 108/298 (36%), Positives = 173/298 (57%), Gaps = 12/298 (4%)
Query: 24 LRCFTIEELERATKNFSQDCLLGSGAFCNVYKGIFELEGILAIKRPHSESFLSVE-EFRN 82
L+ F++ EL+ A+ NFS +LG G F VYKG ++A+KR E E +F+
Sbjct: 274 LKRFSLRELQVASDNFSNKNILGRGGFGKVYKGRLADGTLVAVKRLKEERTQGGELQFQT 333
Query: 83 EVRLLSAVKHKNLIGLVGYCEEPERDGAKILVYEYVPNGSLLEYMMGNRRRS--LTWKQR 140
EV ++S H+NL+ L G+C P ++LVY Y+ NGS+ + L W +R
Sbjct: 334 EVEMISMAVHRNLLRLRGFCMTPTE---RLLVYPYMANGSVASCLRERPESQPPLDWPKR 390
Query: 141 INIAIGAAKGIAYLHEKVKPSIIHRDIKPSNILLGESFEAKVSDFGLVKSGPTGDQSHVS 200
IA+G+A+G+AYLH+ P IIHRD+K +NILL E FEA V DFGL K D +HV+
Sbjct: 391 QRIALGSARGLAYLHDHCDPKIIHRDVKAANILLDEEFEAVVGDFGLAKLMDYKD-THVT 449
Query: 201 SQIKGTPGYLDPAYCSSCHLTKFSDVYSFGVILLQLISARPAVDNAE--NPSNQHIIDWA 258
+ ++GT G++ P Y S+ ++ +DV+ +GV+LL+LI+ + A D A N + ++DW
Sbjct: 450 TAVRGTIGHIAPEYLSTGKSSEKTDVFGYGVMLLELITGQRAFDLARLANDDDVMLLDWV 509
Query: 259 RPSIEKGIIAEIMDANLFCQSEPCNMEVMLKVGQLGIRCVAQEPKHRPTMTQVCREIE 316
+ +++ + ++D +L + +E +++V L C P RP M++V R +E
Sbjct: 510 KGLLKEKKLEALVDVDLQGNYKDEEVEQLIQVALL---CTQSSPMERPKMSEVVRMLE 564
>PSKR_DAUCA (Q8LPB4) Phytosulfokine receptor precursor (EC 2.7.1.37)
(Phytosulfokine LRR receptor kinase)
Length = 1021
Score = 182 bits (462), Expect = 1e-45
Identities = 110/302 (36%), Positives = 175/302 (57%), Gaps = 11/302 (3%)
Query: 19 YDKISLRCFTIEELERATKNFSQDCLLGSGAFCNVYKGIFELEGILAIKRPHSESFLSVE 78
++K S +++++ ++T +F+Q ++G G F VYK +AIKR ++
Sbjct: 723 HNKDSNNELSLDDILKSTSSFNQANIIGCGGFGLVYKATLPDGTKVAIKRLSGDTGQMDR 782
Query: 79 EFRNEVRLLSAVKHKNLIGLVGYCEEPERDGAKILVYEYVPNGSLLEYMMGNR---RRSL 135
EF+ EV LS +H NL+ L+GYC K+L+Y Y+ NGSL +Y + + SL
Sbjct: 783 EFQAEVETLSRAQHPNLVHLLGYCNYKND---KLLIYSYMDNGSL-DYWLHEKVDGPPSL 838
Query: 136 TWKQRINIAIGAAKGIAYLHEKVKPSIIHRDIKPSNILLGESFEAKVSDFGLVKSGPTGD 195
WK R+ IA GAA+G+AYLH+ +P I+HRDIK SNILL ++F A ++DFGL + D
Sbjct: 839 DWKTRLRIARGAAEGLAYLHQSCEPHILHRDIKSSNILLSDTFVAHLADFGLARLILPYD 898
Query: 196 QSHVSSQIKGTPGYLDPAYCSSCHLTKFSDVYSFGVILLQLISARPAVDNAENPSNQHII 255
+HV++ + GT GY+ P Y + T DVYSFGV+LL+L++ R +D + ++ +I
Sbjct: 899 -THVTTDLVGTLGYIPPEYGQASVATYKGDVYSFGVVLLELLTGRRPMDVCKPRGSRDLI 957
Query: 256 DWARPSIEKGIIAEIMDANLFCQSEPCNMEVMLKVGQLGIRCVAQEPKHRPTMTQVCREI 315
W + +EI D ++ + + E ML V ++ RC+ + PK RPT Q+ +
Sbjct: 958 SWVLQMKTEKRESEIFDPFIY---DKDHAEEMLLVLEIACRCLGENPKTRPTTQQLVSWL 1014
Query: 316 EH 317
E+
Sbjct: 1015 EN 1016
>BRI1_ARATH (O22476) BRASSINOSTEROID INSENSITIVE 1 precursor (EC
2.7.1.37) (AtBRI1) (Brassinosteroid LRR receptor kinase)
Length = 1196
Score = 182 bits (461), Expect = 2e-45
Identities = 110/316 (34%), Positives = 173/316 (53%), Gaps = 11/316 (3%)
Query: 24 LRCFTIEELERATKNFSQDCLLGSGAFCNVYKGIFELEGILAIKRPHSESFLSVEEFRNE 83
LR T +L +AT F D L+GSG F +VYK I + +AIK+ S EF E
Sbjct: 868 LRKLTFADLLQATNGFHNDSLIGSGGFGDVYKAILKDGSAVAIKKLIHVSGQGDREFMAE 927
Query: 84 VRLLSAVKHKNLIGLVGYCEEPERDGAKILVYEYVPNGSLLEYMMGNRRRS--LTWKQRI 141
+ + +KH+NL+ L+GYC+ + ++LVYE++ GSL + + ++ L W R
Sbjct: 928 METIGKIKHRNLVPLLGYCKVGDE---RLLVYEFMKYGSLEDVLHDPKKAGVKLNWSTRR 984
Query: 142 NIAIGAAKGIAYLHEKVKPSIIHRDIKPSNILLGESFEAKVSDFGLVKSGPTGDQSHVSS 201
IAIG+A+G+A+LH P IIHRD+K SN+LL E+ EA+VSDFG+ + D S
Sbjct: 985 KIAIGSARGLAFLHHNCSPHIIHRDMKSSNVLLDENLEARVSDFGMARLMSAMDTHLSVS 1044
Query: 202 QIKGTPGYLDPAYCSSCHLTKFSDVYSFGVILLQLISARPAVDNAENPSNQHIIDWARPS 261
+ GTPGY+ P Y S + DVYS+GV+LL+L++ + D+ + N +++ W +
Sbjct: 1045 TLAGTPGYVPPEYYQSFRCSTKGDVYSYGVVLLELLTGKRPTDSPDFGDN-NLVGWVKQH 1103
Query: 262 IEKGIIAEIMDANLFCQSEPCNMEVMLKVGQLGIRCVAQEPKHRPTMTQVC---REIEHA 318
K I+++ D L + +E++ + ++ + C+ RPTM QV +EI+
Sbjct: 1104 -AKLRISDVFDPELMKEDPALEIELLQHL-KVAVACLDDRAWRRPTMVQVMAMFKEIQAG 1161
Query: 319 LYSDDSFTSKDSETLG 334
D T + E G
Sbjct: 1162 SGIDSQSTIRSIEDGG 1177
>SRK6_BRAOL (Q09092) Putative serine/threonine-protein kinase
receptor precursor (EC 2.7.1.37) (S-receptor kinase)
(SRK)
Length = 849
Score = 173 bits (438), Expect = 8e-43
Identities = 104/299 (34%), Positives = 166/299 (54%), Gaps = 8/299 (2%)
Query: 18 KYDKISLRCFTIEELERATKNFSQDCLLGSGAFCNVYKGIFELEGILAIKRPHSESFLSV 77
K++++ L +E + +AT+NFS LG G F VYKG +A+KR S
Sbjct: 507 KFEELELPLIEMETVVKATENFSSCNKLGQGGFGIVYKGRLLDGKEIAVKRLSKTSVQGT 566
Query: 78 EEFRNEVRLLSAVKHKNLIGLVGYCEEPERDGAKILVYEYVPNGSLLEYMMGNRRRS-LT 136
+EF NEV L++ ++H NL+ ++G C E + K+L+YEY+ N SL Y+ G RRS L
Sbjct: 567 DEFMNEVTLIARLQHINLVQVLGCCIEGDE---KMLIYEYLENLSLDSYLFGKTRRSKLN 623
Query: 137 WKQRINIAIGAAKGIAYLHEKVKPSIIHRDIKPSNILLGESFEAKVSDFGLVKSGPTGDQ 196
W +R +I G A+G+ YLH+ + IIHRD+K SNILL ++ K+SDFG+ + +
Sbjct: 624 WNERFDITNGVARGLLYLHQDSRFRIIHRDLKVSNILLDKNMIPKISDFGMARIFERDET 683
Query: 197 SHVSSQIKGTPGYLDPAYCSSCHLTKFSDVYSFGVILLQLISARPAVDNAENPSNQHIID 256
+ ++ GT GY+ P Y ++ SDV+SFGVI+L+++S + ++
Sbjct: 684 EANTMKVVGTYGYMSPEYAMYGIFSEKSDVFSFGVIVLEIVSGKKNRGFYNLDYENDLLS 743
Query: 257 WARPSIEKGIIAEIMDA----NLFCQSEPCNMEVMLKVGQLGIRCVAQEPKHRPTMTQV 311
+ ++G EI+D +L Q + +LK Q+G+ CV + +HRP M+ V
Sbjct: 744 YVWSRWKEGRALEIVDPVIVDSLSSQPSIFQPQEVLKCIQIGLLCVQELAEHRPAMSSV 802
>PSKR_ARATH (Q9ZVR7) Putative phytosulfokine receptor precursor (EC
2.7.1.37) (Phytosulfokine LRR receptor kinase)
Length = 1008
Score = 170 bits (430), Expect = 7e-42
Identities = 103/285 (36%), Positives = 158/285 (55%), Gaps = 11/285 (3%)
Query: 30 EELERATKNFSQDCLLGSGAFCNVYKGIFELEGILAIKRPHSESFLSVEEFRNEVRLLSA 89
++L +T +F Q ++G G F VYK +AIK+ + EF EV LS
Sbjct: 725 DDLLDSTNSFDQANIIGCGGFGMVYKATLPDGKKVAIKKLSGDCGQIEREFEAEVETLSR 784
Query: 90 VKHKNLIGLVGYCEEPERDGAKILVYEYVPNGSLLEYMMGNRRRS---LTWKQRINIAIG 146
+H NL+ L G+C ++L+Y Y+ NGSL +Y + R L WK R+ IA G
Sbjct: 785 AQHPNLVLLRGFCFYKND---RLLIYSYMENGSL-DYWLHERNDGPALLKWKTRLRIAQG 840
Query: 147 AAKGIAYLHEKVKPSIIHRDIKPSNILLGESFEAKVSDFGLVKSGPTGDQSHVSSQIKGT 206
AAKG+ YLHE P I+HRDIK SNILL E+F + ++DFGL + + ++HVS+ + GT
Sbjct: 841 AAKGLLYLHEGCDPHILHRDIKSSNILLDENFNSHLADFGLARL-MSPYETHVSTDLVGT 899
Query: 207 PGYLDPAYCSSCHLTKFSDVYSFGVILLQLISARPAVDNAENPSNQHIIDWARPSIEKGI 266
GY+ P Y + T DVYSFGV+LL+L++ + VD + + +I W +
Sbjct: 900 LGYIPPEYGQASVATYKGDVYSFGVVLLELLTDKRPVDMCKPKGCRDLISWVVKMKHESR 959
Query: 267 IAEIMDANLFCQSEPCNMEVMLKVGQLGIRCVAQEPKHRPTMTQV 311
+E+ D ++ + N + M +V ++ C+++ PK RPT Q+
Sbjct: 960 ASEVFDPLIYSKE---NDKEMFRVLEIACLCLSENPKQRPTTQQL 1001
>RLK5_ARATH (P47735) Receptor-like protein kinase 5 precursor (EC
2.7.1.37)
Length = 999
Score = 160 bits (406), Expect = 4e-39
Identities = 106/319 (33%), Positives = 173/319 (54%), Gaps = 35/319 (10%)
Query: 42 DCL-----LGSGAFCNVYKGIFELEG--ILAIKR----------PHSESFLSVEEFRNEV 84
DCL +G G+ VYK EL G ++A+K+ +S L+ + F EV
Sbjct: 681 DCLDEKNVIGFGSSGKVYK--VELRGGEVVAVKKLNKSVKGGDDEYSSDSLNRDVFAAEV 738
Query: 85 RLLSAVKHKNLIGLVGYCEEPERDGAKILVYEYVPNGSLLEYMMGNRRRS--LTWKQRIN 142
L ++HK+++ L C + K+LVYEY+PNGSL + + G+R+ L W +R+
Sbjct: 739 ETLGTIRHKSIVRLWCCCSSGD---CKLLVYEYMPNGSLADVLHGDRKGGVVLGWPERLR 795
Query: 143 IAIGAAKGIAYLHEKVKPSIIHRDIKPSNILLGESFEAKVSDFGLVKSGPTGDQS--HVS 200
IA+ AA+G++YLH P I+HRD+K SNILL + AKV+DFG+ K G
Sbjct: 796 IALDAAEGLSYLHHDCVPPIVHRDVKSSNILLDSDYGAKVADFGIAKVGQMSGSKTPEAM 855
Query: 201 SQIKGTPGYLDPAYCSSCHLTKFSDVYSFGVILLQLISARPAVDNAENPSNQHIIDWARP 260
S I G+ GY+ P Y + + + SD+YSFGV+LL+L++ + D+ ++ + W
Sbjct: 856 SGIAGSCGYIAPEYVYTLRVNEKSDIYSFGVVLLELVTGKQPTDS--ELGDKDMAKWVCT 913
Query: 261 SIEKGIIAEIMDANLFCQSEPCNMEVMLKVGQLGIRCVAQEPKHRPTMTQV---CREIEH 317
+++K + ++D L + + E + KV +G+ C + P +RP+M +V +E+
Sbjct: 914 ALDKCGLEPVIDPKLDLKFK----EEISKVIHIGLLCTSPLPLNRPSMRKVVIMLQEVSG 969
Query: 318 ALYSDDSFTSKDSETLGSV 336
A+ TSK S+T G +
Sbjct: 970 AVPCSSPNTSKRSKTGGKL 988
>CLV1_ARATH (Q9SYQ8) Receptor protein kinase CLAVATA1 precursor (EC
2.7.1.-)
Length = 980
Score = 158 bits (400), Expect = 2e-38
Identities = 101/287 (35%), Positives = 154/287 (53%), Gaps = 21/287 (7%)
Query: 41 QDCLLGSGAFCNVYKGIFELEGILAIKRPHSESF-LSVEEFRNEVRLLSAVKHKNLIGLV 99
++ ++G G VY+G +AIKR S F E++ L ++H++++ L+
Sbjct: 694 EENIIGKGGAGIVYRGSMPNNVDVAIKRLVGRGTGRSDHGFTAEIQTLGRIRHRHIVRLL 753
Query: 100 GYCEEPERDGAKILVYEYVPNGSLLEYMMGNRRRSLTWKQRINIAIGAAKGIAYLHEKVK 159
GY + + +L+YEY+PNGSL E + G++ L W+ R +A+ AAKG+ YLH
Sbjct: 754 GYVANKDTN---LLLYEYMPNGSLGELLHGSKGGHLQWETRHRVAVEAAKGLCYLHHDCS 810
Query: 160 PSIIHRDIKPSNILLGESFEAKVSDFGLVKSGPTGDQSHVSSQIKGTPGYLDPAYCSSCH 219
P I+HRD+K +NILL FEA V+DFGL K G S S I G+ GY+ P Y +
Sbjct: 811 PLILHRDVKSNNILLDSDFEAHVADFGLAKFLVDGAASECMSSIAGSYGYIAPEYAYTLK 870
Query: 220 LTKFSDVYSFGVILLQLISARPAVDNAENPSNQHIIDWARPSIEK-------GIIAEIMD 272
+ + SDVYSFGV+LL+LI+ + V E I+ W R + E+ I+ I+D
Sbjct: 871 VDEKSDVYSFGVVLLELIAGKKPV--GEFGEGVDIVRWVRNTEEEITQPSDAAIVVAIVD 928
Query: 273 ANLFCQSEPCNMEVMLKVGQLGIRCVAQEPKHRPTMTQVCREIEHAL 319
L + ++ V ++ + CV +E RPTM RE+ H L
Sbjct: 929 PRL----TGYPLTSVIHVFKIAMMCVEEEAAARPTM----REVVHML 967
>KPRO_MAIZE (P17801) Putative receptor protein kinase ZmPK1
precursor (EC 2.7.1.37)
Length = 817
Score = 156 bits (394), Expect = 1e-37
Identities = 101/310 (32%), Positives = 161/310 (51%), Gaps = 21/310 (6%)
Query: 23 SLRCFTIEELERATKNFSQDCLLGSGAFCNVYKGIFELEGILAIKRPHSESFLSVEEFRN 82
+ R ++ EL +AT+ F + LG G VYKG+ E + +A+K+ + E F+
Sbjct: 520 NFRRYSYRELVKATRKFKVE--LGRGESGTVYKGVLEDDRHVAVKKLENVR-QGKEVFQA 576
Query: 83 EVRLLSAVKHKNLIGLVGYCEEPERDGAKILVYEYVPNGSLLEYMMGNRRRSLT-WKQRI 141
E+ ++ + H NL+ + G+C E ++LV EYV NGSL + L W+ R
Sbjct: 577 ELSVIGRINHMNLVRIWGFCSEGSH---RLLVSEYVENGSLANILFSEGGNILLDWEGRF 633
Query: 142 NIAIGAAKGIAYLHEKVKPSIIHRDIKPSNILLGESFEAKVSDFGLVKSGPTGDQSHVSS 201
NIA+G AKG+AYLH + +IH D+KP NILL ++FE K++DFGLVK G + S
Sbjct: 634 NIALGVAKGLAYLHHECLEWVIHCDVKPENILLDQAFEPKITDFGLVKLLNRGGSTQNVS 693
Query: 202 QIKGTPGYLDPAYCSSCHLTKFSDVYSFGVILLQLISARPAVDNAENPSNQH-------- 253
++GT GY+ P + SS +T DVYS+GV+LL+L++ + H
Sbjct: 694 HVRGTLGYIAPEWVSSLPITAKVDVYSYGVVLLELLTGTRVSELVGGTDEVHSMLRKLVR 753
Query: 254 IIDWARPSIEKGIIAEIMDANLFCQSEPCNMEVMLKVGQLGIRCVAQEPKHRPTMTQVCR 313
++ E+ I +D+ L + P N + +L + C+ ++ RPTM
Sbjct: 754 MLSAKLEGEEQSWIDGYLDSKL---NRPVNYVQARTLIKLAVSCLEEDRSKRPTMEHA-- 808
Query: 314 EIEHALYSDD 323
++ L +DD
Sbjct: 809 -VQTLLSADD 817
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.321 0.138 0.409
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 44,339,606
Number of Sequences: 164201
Number of extensions: 1825998
Number of successful extensions: 7146
Number of sequences better than 10.0: 1644
Number of HSP's better than 10.0 without gapping: 1060
Number of HSP's successfully gapped in prelim test: 585
Number of HSP's that attempted gapping in prelim test: 4213
Number of HSP's gapped (non-prelim): 1808
length of query: 407
length of database: 59,974,054
effective HSP length: 113
effective length of query: 294
effective length of database: 41,419,341
effective search space: 12177286254
effective search space used: 12177286254
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 67 (30.4 bits)
Medicago: description of AC149547.4


