

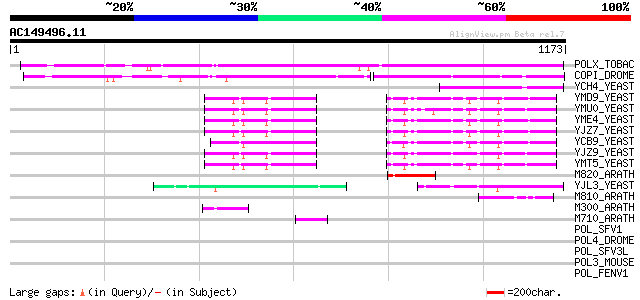
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC149496.11 - phase: 0 /pseudo
(1173 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
POLX_TOBAC (P10978) Retrovirus-related Pol polyprotein from tran... 645 0.0
COPI_DROME (P04146) Copia protein (Gag-int-pol protein) [Contain... 253 2e-66
YCH4_YEAST (P25600) Transposon Ty5-1 34.5 kDa hypothetical protein 132 4e-30
YMD9_YEAST (Q03434) Transposon Ty1 protein B 101 1e-20
YMU0_YEAST (Q04670) Transposon Ty1 protein B 100 4e-20
YME4_YEAST (Q04711) Transposon Ty1 protein B 100 4e-20
YJZ7_YEAST (P47098) Transposon Ty1 protein B 99 7e-20
YCB9_YEAST (P25384) Transposon Ty2 protein B (Ty1-17 protein B) 98 1e-19
YJZ9_YEAST (P47100) Transposon Ty1 protein B 96 7e-19
YMT5_YEAST (Q04214) Transposon Ty1 protein B 92 1e-17
M820_ARATH (P92520) Hypothetical mitochondrial protein AtMg00820... 89 9e-17
YJL3_YEAST (P47024) Transposon Ty4 207.7 kDa hypothetical protein 87 3e-16
M810_ARATH (P92519) Hypothetical mitochondrial protein AtMg00810... 80 2e-14
M300_ARATH (P93293) Hypothetical mitochondrial protein AtMg00300... 54 3e-06
M710_ARATH (P92512) Hypothetical mitochondrial protein AtMg00710... 49 8e-05
POL_SFV1 (P23074) Pol polyprotein [Contains: Protease (EC 3.4.23... 45 0.001
POL4_DROME (P10394) Retrovirus-related Pol polyprotein from tran... 44 0.003
POL_SFV3L (P27401) Pol polyprotein [Contains: Protease (EC 3.4.2... 43 0.004
POL3_MOUSE (P11367) Retrovirus-related Pol polyprotein (Endonucl... 43 0.006
POL_FENV1 (P31792) Pol polyprotein [Contains: Reverse transcript... 42 0.007
>POLX_TOBAC (P10978) Retrovirus-related Pol polyprotein from
transposon TNT 1-94 [Contains: Protease (EC 3.4.23.-);
Reverse transcriptase (EC 2.7.7.49); Endonuclease]
Length = 1328
Score = 645 bits (1665), Expect = 0.0
Identities = 418/1205 (34%), Positives = 631/1205 (51%), Gaps = 95/1205 (7%)
Query: 23 ETNFIDWSRNMRIVLTHE--KKLYVLDGPIPETPPAGASAALRNAHAK----HLNDSVEV 76
+ F W R MR +L + K+ +D P+T A A L A HL+D V
Sbjct: 14 DNGFSTWQRRMRDLLIQQGLHKVLDVDSKKPDTMKAEDWADLDERAASAIRLHLSDDVVN 73
Query: 77 SCIMLASMTPELQKQHEGMTAFDMIEHLKTLYEEQARHERFDVSKALFSTKLSEGGPVGP 136
+ I + TA + L++LY + + + K L++ +SEG
Sbjct: 74 NII-------------DEDTARGIWTRLESLYMSKTLTNKLYLKKQLYALHMSEGTNFLS 120
Query: 137 HVLKMIGYSENLARLGFVLEQELVVDLVLQSLPESLNGFVHNFLMNDMDKTLPQLAAMLR 196
H+ G LA LG +E+E L+L SLP S + L L + + L
Sbjct: 121 HLNVFNGLITQLANLGVKIEEEDKAILLLNSLPSSYDNLATTILHGKTTIELKDVTSALL 180
Query: 197 TAEKNMKGKGKVAAILMVNNGKFKKHHKKPN---KSKGNGKGKIVAKPSTKALKPTGGVA 253
EK M+ K + ++ G+ + + + N +S GK K +K +
Sbjct: 181 LNEK-MRKKPENQGQALITEGRGRSYQRSSNNYGRSGARGKSKNRSKSRVR--------- 230
Query: 254 KDDKCFYCNNAGHWK*NYPKYLEDKKNG----NVPTTSAGI-----FVIEIN-------- 296
C+ CN GH+K + P + K N T+A + V+ IN
Sbjct: 231 ---NCYNCNQPGHFKRDCPNPRKGKGETSGQKNDDNTAAMVQNNDNVVLFINEEEECMHL 287
Query: 297 MSTSTSWVLDTGCGSHICTDVQGLRKSRALAKGEVDLRVGNGAKVAALAVGTYVLTLPSG 356
+ WV+DT SH T V+ L R +A +++GN + +G + G
Sbjct: 288 SGPESEWVVDTAA-SHHATPVRDLF-CRYVAGDFGTVKMGNTSYSKIAGIGDICIKTNVG 345
Query: 357 LLLNLENCYYVPAISRNIISISCLDKAGFEFTIKNKCCSVYLDNILYANANLSNGLYVLD 416
L L++ +VP + N+IS LD+ G+E N+ + +++ A LY +
Sbjct: 346 CTLVLKDVRHVPDLRMNLISGIALDRDGYESYFANQKWRLTKGSLVIAKGVARGTLYRTN 405
Query: 417 LDMPIYNINAKRLKPNELNPTYLWHCRLGHINENRISKLHKDGFLDSFDFESYETCRSCL 476
++ +NA + +E++ LWH R+GH++E + L K + + + C CL
Sbjct: 406 AEICQGELNAAQ---DEISVD-LWHKRMGHMSEKGLQILAKKSLISYAKGTTVKPCDYCL 461
Query: 477 LGKMTKAPFTGQGERASDLLGLIHTDVCGPLNITARGGFHYFITLTDDFSRFGYVYLMKH 536
GK + F ER ++L L+++DVCGP+ I + GG YF+T DD SR +VY++K
Sbjct: 462 FGKQHRVSFQTSSERKLNILDLVYSDVCGPMEIESMGGNKYFVTFIDDASRKLWVYILKT 521
Query: 537 KSESFTFFKEFQNEVENQLGKKIKMLRSDRGGEYLSLEFDNHLKECGILSQLTPPGTPQW 596
K + F F++F VE + G+K+K LRSD GGEY S EF+ + GI + T PGTPQ
Sbjct: 522 KDQVFQVFQKFHALVERETGRKLKRLRSDNGGEYTSREFEEYCSSHGIRHEKTVPGTPQH 581
Query: 597 NGVSERRNRTLLDMVRSMMSHAELPNFLWGYALLTAAYTLNRVPSKAVE-KTPYEIWNGR 655
NGV+ER NRT+++ VRSM+ A+LP WG A+ TA Y +NR PS + + P +W +
Sbjct: 582 NGVAERMNRTIVEKVRSMLRMAKLPKSFWGEAVQTACYLINRSPSVPLAFEIPERVWTNK 641
Query: 656 KPNVGHFKIWGCEAY--IKRLMSTKLEQKSEKCFFVGYPKETRGYYFYKPPEGTIVVAKT 713
+ + H K++GC A+ + + TKL+ KS C F+GY E GY + P + ++ ++
Sbjct: 642 EVSYSHLKVFGCRAFAHVPKEQRTKLDDKSIPCIFIGYGDEEFGYRLWDPVKKKVIRSRD 701
Query: 714 GVFLEKDFVSRRISGSKVDLKEIQD-------------PQSTEIPVEEQGQDTQTVM--- 757
VF E + + KV I + +ST V EQG+ V+
Sbjct: 702 VVFRESEVRTAADMSEKVKNGIIPNFVTIPSTSNNPTSAESTTDEVSEQGEQPGEVIEQG 761
Query: 758 ---------TENPAPVTQEP---RRSSRIRQEPERYGYLISQEGDVLLMDQDEPVTYTEA 805
E+P ++ RRS R R E RY VL+ D EP + E
Sbjct: 762 EQLDEGVEEVEHPTQGEEQHQPLRRSERPRVESRRY----PSTEYVLISDDREPESLKEV 817
Query: 806 ITGPEFEKWLEAMKSEMDSMYTNQVWNLIDAPEGINPIGCKWVFKKKIDMDGKVSTYKAR 865
++ PE + ++AM+ EM+S+ N + L++ P+G P+ CKWVFK K D D K+ YKAR
Sbjct: 818 LSHPEKNQLMKAMQEEMESLQKNGTYKLVELPKGKRPLKCKWVFKLKKDGDCKLVRYKAR 877
Query: 866 LVAKGFKQIHGVDYDETFSPVAMIKSIRILLAIAAYHDYEIWQMDVKTAFLNGNLLEDVY 925
LV KGF+Q G+D+DE FSPV + SIR +L++AA D E+ Q+DVKTAFL+G+L E++Y
Sbjct: 878 LVVKGFEQKKGIDFDEIFSPVVKMTSIRTILSLAASLDLEVEQLDVKTAFLHGDLEEEIY 937
Query: 926 MTQPEGFGDPKATKKVCKLQRSIYGLKQASRSWNLRFDETVQQYGFIKNEDEPCVY-KKV 984
M QPEGF VCKL +S+YGLKQA R W ++FD ++ ++K +PCVY K+
Sbjct: 938 MEQPEGFEVAGKKHMVCKLNKSLYGLKQAPRQWYMKFDSFMKSQTYLKTYSDPCVYFKRF 997
Query: 985 SGSIVSFLILYVDDILLIGNDIPTLQEIKTWLRKCFSMKDLGEASYILGIRIYRDRSQRL 1044
S + L+LYVDD+L++G D + ++K L K F MKDLG A ILG++I R+R+ R
Sbjct: 998 SENNFIILLLYVDDMLIVGKDKGLIAKLKGDLSKSFDMKDLGPAQQILGMKIVRERTSRK 1057
Query: 1045 LGLSQGRYIDKVLRRFNMHESKKGFIPMSSGLNLSKTQSPSTDDERDRMRDIPYASAIGS 1104
L LSQ +YI++VL RFNM +K P++ L LSK P+T +E+ M +PY+SA+GS
Sbjct: 1058 LWLSQEKYIERVLERFNMKNAKPVSTPLAGHLKLSKKMCPTTVEEKGNMAKVPYSSAVGS 1117
Query: 1105 IMYAMICTRPDVSYALSATSRYQSNPGNEYWIAVKNILKYLRRTKDTFLIYGG*EELSVI 1164
+MYAM+CTRPD+++A+ SR+ NPG E+W AVK IL+YLR T L +GG + + +
Sbjct: 1118 LMYAMVCTRPDIAHAVGVVSRFLENPGKEHWEAVKWILRYLRGTTGDCLCFGGSDPI-LK 1176
Query: 1165 GYIDA 1169
GY DA
Sbjct: 1177 GYTDA 1181
>COPI_DROME (P04146) Copia protein (Gag-int-pol protein) [Contains:
Copia VLP protein; Copia protease (EC 3.4.23.-)]
Length = 1409
Score = 253 bits (646), Expect = 2e-66
Identities = 146/412 (35%), Positives = 231/412 (55%), Gaps = 20/412 (4%)
Query: 769 RRSSRIRQEPE-RYGYLISQEGDVLL----MDQDEPVTYTEAITGPEFEKWLEAMKSEMD 823
RRS R++ +P+ Y + V+L + D P ++ E + W EA+ +E++
Sbjct: 856 RRSERLKTKPQISYNEEDNSLNKVVLNAHTIFNDVPNSFDEIQYRDDKSSWEEAINTELN 915
Query: 824 SMYTNQVWNLIDAPEGINPIGCKWVFKKKIDMDGKVSTYKARLVAKGFKQIHGVDYDETF 883
+ N W + PE N + +WVF K + G YKARLVA+GF Q + +DY+ETF
Sbjct: 916 AHKINNTWTITKRPENKNIVDSRWVFSVKYNELGNPIRYKARLVARGFTQKYQIDYEETF 975
Query: 884 SPVAMIKSIRILLAIAAYHDYEIWQMDVKTAFLNGNLLEDVYMTQPEGFGDPKATKKVCK 943
+PVA I S R +L++ ++ ++ QMDVKTAFLNG L E++YM P+G + VCK
Sbjct: 976 APVARISSFRFILSLVIQYNLKVHQMDVKTAFLNGTLKEEIYMRLPQGIS--CNSDNVCK 1033
Query: 944 LQRSIYGLKQASRSWNLRFDETVQQYGFIKNEDEPCVYKKVSGSIVS--FLILYVDDILL 1001
L ++IYGLKQA+R W F++ +++ F+ + + C+Y G+I +++LYVDD+++
Sbjct: 1034 LNKAIYGLKQAARCWFEVFEQALKECEFVNSSVDRCIYILDKGNINENIYVLLYVDDVVI 1093
Query: 1002 IGNDIPTLQEIKTWLRKCFSMKDLGEASYILGIRIYRDRSQRLLGLSQGRYIDKVLRRFN 1061
D+ + K +L + F M DL E + +GIRI + + + LSQ Y+ K+L +FN
Sbjct: 1094 ATGDMTRMNNFKRYLMEKFRMTDLNEIKHFIGIRI--EMQEDKIYLSQSAYVKKILSKFN 1151
Query: 1062 MHESKKGFIPMSSGLNLSKTQSPSTDDERDRMRDIPYASAIGSIMYAMICTRPDVSYALS 1121
M P+ S +N S D + P S IG +MY M+CTRPD++ A++
Sbjct: 1152 MENCNAVSTPLPSKINYELLNS-------DEDCNTPCRSLIGCLMYIMLCTRPDLTTAVN 1204
Query: 1122 ATSRYQSNPGNEYWIAVKNILKYLRRTKDTFLIY--GG*EELSVIGYIDASF 1171
SRY S +E W +K +L+YL+ T D LI+ E +IGY+D+ +
Sbjct: 1205 ILSRYSSKNNSELWQNLKRVLRYLKGTIDMKLIFKKNLAFENKIIGYVDSDW 1256
Score = 244 bits (622), Expect = 1e-63
Identities = 214/782 (27%), Positives = 350/782 (44%), Gaps = 104/782 (13%)
Query: 29 WSRNMRIVLTHEKKLYVLDGPIPETPPAGASAALRNAHA---KHLNDSVEVSCIMLASMT 85
W +R +L + L V+DG +P A R A + ++L+DS
Sbjct: 19 WKFRIRALLAEQDVLKVVDGLMPNEVDDSWKKAERCAKSTIIEYLSDSF----------- 67
Query: 86 PELQKQHEGMTAFDMIEHLKTLYEEQARHERFDVSKALFSTKLSEGGPVGPHVLKMIGYS 145
L +TA ++E+L +YE ++ + + K L S KLS + H
Sbjct: 68 --LNFATSDITARQILENLDAVYERKSLASQLALRKRLLSLKLSSEMSLLSHFHIFDELI 125
Query: 146 ENLARLGFVLEQELVVDLVLQSLPESLNGFVHNF-LMNDMDKTLPQLAAMLRTAEKNMKG 204
L G +E+ + +L +LP +G + +++ + TL + L E +K
Sbjct: 126 SELLAAGAKIEEMDKISHLLITLPSCYDGIITAIETLSEENLTLAFVKNRLLDQEIKIKN 185
Query: 205 ------KGKVAAILMVNNGK-----FKKHHKKPNKS-KGNGKGKIVAKPSTKALKPTGGV 252
K + AI+ NN FK KP K KGN K K+
Sbjct: 186 DHNDTSKKVMNAIVHNNNNTYKNNLFKNRVTKPKKIFKGNSKYKV--------------- 230
Query: 253 AKDDKCFYCNNAGHWK*N---YPKYLEDKKNGNVP----TTSAGI--FVIEINMST---S 300
KC +C GH K + Y + L +K N TS GI V E+N ++ +
Sbjct: 231 ----KCHHCGREGHIKKDCFHYKRILNNKNKENEKQVQTATSHGIAFMVKEVNNTSVMDN 286
Query: 301 TSWVLDTGCGSHICTDVQGLRKSRALAKGEVDLRVGNGAKVAALAVGTYVLTLPSGLL-- 358
+VLD+G H+ D S + V K+A G ++ G++
Sbjct: 287 CGFVLDSGASDHLINDESLYTDS---------VEVVPPLKIAVAKQGEFIYATKRGIVRL 337
Query: 359 -----LNLENCYYVPAISRNIISISCLDKAGFEFTIKNKCCSVYLDNILYA-NANLSNGL 412
+ LE+ + + N++S+ L +AG ++ + ++ N+ + N +
Sbjct: 338 RNDHEITLEDVLFCKEAAGNLMSVKRLQEAGMSIEFDKSGVTISKNGLMVVKNSGMLNNV 397
Query: 413 YVLDLDMPIYNINAKRLKPNELNPTYLWHCRLGHINENRISKLHK------DGFLDSFDF 466
V++ Y+INAK N LWH R GHI++ ++ ++ + L++ +
Sbjct: 398 PVINFQA--YSINAKHK-----NNFRLWHERFGHISDGKLLEIKRKNMFSDQSLLNNLEL 450
Query: 467 ESYETCRSCLLGKMTKAPFTGQGERA--SDLLGLIHTDVCGPLNITARGGFHYFITLTDD 524
S E C CL GK + PF ++ L ++H+DVCGP+ +YF+ D
Sbjct: 451 -SCEICEPCLNGKQARLPFKQLKDKTHIKRPLFVVHSDVCGPITPVTLDDKNYFVIFVDQ 509
Query: 525 FSRFGYVYLMKHKSESFTFFKEFQNEVENQLGKKIKMLRSDRGGEYLSLEFDNHLKECGI 584
F+ + YL+K+KS+ F+ F++F + E K+ L D G EYLS E + GI
Sbjct: 510 FTHYCVTYLIKYKSDVFSMFQDFVAKSEAHFNLKVVYLYIDNGREYLSNEMRQFCVKKGI 569
Query: 585 LSQLTPPGTPQWNGVSERRNRTLLDMVRSMMSHAELPNFLWGYALLTAAYTLNRVPSKAV 644
LT P TPQ NGVSER RT+ + R+M+S A+L WG A+LTA Y +NR+PS+A+
Sbjct: 570 SYHLTVPHTPQLNGVSERMIRTITEKARTMVSGAKLDKSFWGEAVLTATYLINRIPSRAL 629
Query: 645 ---EKTPYEIWNGRKPNVGHFKIWGCEAYIK-RLMSTKLEQKSEKCFFVGYPKETRGYYF 700
KTPYE+W+ +KP + H +++G Y+ + K + KS K FVGY E G+
Sbjct: 630 VDSSKTPYEMWHNKKPYLKHLRVFGATVYVHIKNKQGKFDDKSFKSIFVGY--EPNGFKL 687
Query: 701 YKPPEGTIVVAKTGVFLEKDFV-SRRISGSKVDLKEIQDPQSTEIPVEEQGQDTQTVMTE 759
+ +VA+ V E + V SR + V LK+ ++ ++ P + + + + TE
Sbjct: 688 WDAVNEKFIVARDVVVDETNMVNSRAVKFETVFLKDSKESENKNFPNDSR----KIIQTE 743
Query: 760 NP 761
P
Sbjct: 744 FP 745
>YCH4_YEAST (P25600) Transposon Ty5-1 34.5 kDa hypothetical protein
Length = 308
Score = 132 bits (333), Expect = 4e-30
Identities = 83/263 (31%), Positives = 136/263 (51%), Gaps = 9/263 (3%)
Query: 909 MDVKTAFLNGNLLEDVYMTQPEGFGDPKATKKVCKLQRSIYGLKQASRSWNLRFDETVQQ 968
MDV TAFLN + E +Y+ QP GF + + V +L +YGLKQA WN + T+++
Sbjct: 1 MDVDTAFLNSTMDEPIYVKQPPGFVNERNPDYVWELYGGMYGLKQAPLLWNEHINNTLKK 60
Query: 969 YGFIKNEDEPCVYKKVSGSIVSFLILYVDDILLIGNDIPTLQEIKTWLRKCFSMKDLGEA 1028
GF ++E E +Y + + ++ +YVDD+L+ +K L K +SMKDLG+
Sbjct: 61 IGFCRHEGEHGLYFRSTSDGPIYIGVYVDDLLVAAPSPKIYDRVKQELTKLYSMKDLGKV 120
Query: 1029 SYILGIRIYRDRSQRLLGLSQGRYIDKVLRRFNMHESKKGFIPMSSGLNLSKTQSPSTDD 1088
LG+ I++ + + LS YI K ++ K P+ + L +T SP
Sbjct: 121 DKFLGLNIHQSTNGDIT-LSLQDYIAKAASESEINTFKLTQTPLCNSKPLFETTSP---- 175
Query: 1089 ERDRMRDI-PYASAIGSIMYAMICTRPDVSYALSATSRYQSNPGNEYWIAVKNILKYLRR 1147
++DI PY S +G +++ RPD+SY +S SR+ P + + + +L+YL
Sbjct: 176 ---HLKDITPYQSIVGQLLFCANTGRPDISYPVSLLSRFLREPRAIHLESARRVLRYLYT 232
Query: 1148 TKDTFLIYGG*EELSVIGYIDAS 1170
T+ L Y ++++ Y DAS
Sbjct: 233 TRSMCLKYRSGSQVALTVYCDAS 255
>YMD9_YEAST (Q03434) Transposon Ty1 protein B
Length = 1328
Score = 101 bits (251), Expect = 1e-20
Identities = 100/384 (26%), Positives = 184/384 (47%), Gaps = 50/384 (13%)
Query: 797 DEPVTYTEAITGPEFEKWLEAMKSEMDSMYTNQVWNL--------IDAPEGINPIGCKWV 848
DE +TY + I E EK++EA E++ + + W+ ID IN + ++
Sbjct: 808 DEAITYNKDIK--EKEKYIEAYHKEVNQLLKMKTWDTDEYYDRKEIDPKRVINSM---FI 862
Query: 849 FKKKIDMDGKVSTYKARLVAKGFKQIHGVDYDETF-SPVAMIKSIRILLAIAAYHDYEIW 907
F KK D T+KAR VA+G Q H YD S ++ L++A ++Y I
Sbjct: 863 FNKKRD-----GTHKARFVARGDIQ-HPDTYDSGMQSNTVHHYALMTSLSLALDNNYYIT 916
Query: 908 QMDVKTAFLNGNLLEDVYMTQPEGFGDPKATKKVCKLQRSIYGLKQASRSWNLRFDETVQ 967
Q+D+ +A+L ++ E++Y+ P G K+ +L++S+YGLKQ+ +W ET++
Sbjct: 917 QLDISSAYLYADIKEELYIRPPPHLG---MNDKLIRLKKSLYGLKQSGANWY----ETIK 969
Query: 968 QY-----GFIKNEDEPCVYKKVSGSIVSFLILYVDDILLIGNDIPTLQEIKTWLRKCFSM 1022
Y G + CV+K +I L+VDD++L D+ ++I T L+K +
Sbjct: 970 SYLIKQCGMEEVRGWSCVFKNSQVTI----CLFVDDMILFSKDLNANKKIITTLKKQYDT 1025
Query: 1023 K--DLGEASY-----ILGIRIYRDRSQRLLGLSQGRYIDKVLRRFNMHESKKG---FIPM 1072
K +LGE+ ILG+ I R + + L + + + + N+ + KG P
Sbjct: 1026 KIINLGESDNEIQYDILGLEIKYQRG-KYMKLGMENSLTEKIPKLNVPLNPKGRKLSAPG 1084
Query: 1073 SSGLNLSKTQSPSTDDE-RDRMRDIPYASAIGSIMYAMICTRPDVSYALSATSRYQSNPG 1131
GL + + + +DE ++++ ++ IG Y R D+ Y ++ +++ P
Sbjct: 1085 QPGLYIDQDELEIDEDEYKEKVHEM--QKLIGLASYVGYKFRFDLLYYINTLAQHILFPS 1142
Query: 1132 NEYWIAVKNILKYLRRTKDTFLIY 1155
+ +++++ T+D LI+
Sbjct: 1143 RQVLDMTYELIQFMWDTRDKQLIW 1166
Score = 87.8 bits (216), Expect = 2e-16
Identities = 69/253 (27%), Positives = 114/253 (44%), Gaps = 22/253 (8%)
Query: 413 YVLDLDMPIYNINAKRLKPNELNPTYLW-HCRLGHINENRISKLHKDGFLDSFDFESYET 471
Y+L ++ + IN + Y + H L H N I K+ + F+ +
Sbjct: 143 YLLPSNISVPTINNVHTSESTRKYPYPFIHRMLAHANAQTIRYSLKNNTITYFNESDVDR 202
Query: 472 -------CRSCLLGKMTKAPFTGQGERAS-----DLLGLIHTDVCGPLNITARGGFHYFI 519
C CL+GK TK +G R + +HTD+ GP++ + YFI
Sbjct: 203 SSAIDYQCPDCLIGKSTKHRHI-KGSRLKYQNSYEPFQYLHTDIFGPVHNLPKSAPSYFI 261
Query: 520 TLTDDFSRFGYVYLMKHKSES-----FTFFKEFQNEVENQLGKKIKMLRSDRGGEYLSLE 574
+ TD+ ++F +VY + + E FT F ++NQ + +++ DRG EY +
Sbjct: 262 SFTDETTKFRWVYPLHDRREDSILDVFTTILAF---IKNQFQASVLVIQMDRGSEYTNRT 318
Query: 575 FDNHLKECGILSQLTPPGTPQWNGVSERRNRTLLDMVRSMMSHAELPNFLWGYALLTAAY 634
L++ GI T + +GV+ER NRTLLD R+ + + LPN LW A+ +
Sbjct: 319 LHKFLEKNGITPCYTTTADSRAHGVAERLNRTLLDDCRTQLQCSGLPNHLWFSAIEFSTI 378
Query: 635 TLNRVPSKAVEKT 647
N + S +K+
Sbjct: 379 VRNSLASPKSKKS 391
>YMU0_YEAST (Q04670) Transposon Ty1 protein B
Length = 1328
Score = 99.8 bits (247), Expect = 4e-20
Identities = 95/387 (24%), Positives = 182/387 (46%), Gaps = 56/387 (14%)
Query: 797 DEPVTYTEAITGPEFEKWLEAMKSEMDSMYTNQVWNL--------IDAPEGINPIGCKWV 848
DE +TY + I E EK+++A E++ + + W+ ID IN + ++
Sbjct: 808 DEAITYNKDIK--EKEKYIQAYHKEVNQLLKMKTWDTDRYYDRKEIDPKRVINSM---FI 862
Query: 849 FKKKIDMDGKVSTYKARLVAKGFKQIHGVDYDETFSPVAMIKSIR-----ILLAIAAYHD 903
F +K D T+KAR VA+G + + +T+ P ++ L++A ++
Sbjct: 863 FNRKRD-----GTHKARFVARG-----DIQHPDTYDPGMQSNTVHHYALMTSLSLALDNN 912
Query: 904 YEIWQMDVKTAFLNGNLLEDVYMTQPEGFGDPKATKKVCKLQRSIYGLKQASRSWNLRFD 963
Y I Q+D+ +A+L ++ E++Y+ P G K+ +L++S+YGLKQ+ +W
Sbjct: 913 YYITQLDISSAYLYADIKEELYIRPPPHLG---MNDKLIRLKKSLYGLKQSGANWY---- 965
Query: 964 ETVQQY-----GFIKNEDEPCVYKKVSGSIVSFLILYVDDILLIGNDIPTLQEIKTWLRK 1018
ET++ Y G + CV+K +I L+VDD++L D+ ++I T L+K
Sbjct: 966 ETIKSYLIKQCGMEEVRGWSCVFKNSQVTI----CLFVDDMILFSKDLNANKKIITTLKK 1021
Query: 1019 CFSMK--DLGEASY-----ILGIRIYRDRSQRLLGLSQGRYIDKVLRRFNMHESKKG--- 1068
+ K +LGE+ ILG+ I R + + L + + + + N+ + KG
Sbjct: 1022 QYDTKIINLGESDNEIQYDILGLEIKYQRG-KYMKLGMENSLTEKIPKLNVPLNPKGRKL 1080
Query: 1069 FIPMSSGLNLSKTQSPSTDDERDRMRDIPYASAIGSIMYAMICTRPDVSYALSATSRYQS 1128
P GL + + + +D+ +M+ IG Y R D+ Y ++ +++
Sbjct: 1081 SAPGQPGLYIDQQELELEEDDY-KMKVHEMQKLIGLASYVGYKFRFDLLYYINTLAQHIL 1139
Query: 1129 NPGNEYWIAVKNILKYLRRTKDTFLIY 1155
P + +++++ T+D LI+
Sbjct: 1140 FPSKQVLDMTYELIQFIWNTRDKQLIW 1166
Score = 87.8 bits (216), Expect = 2e-16
Identities = 69/253 (27%), Positives = 114/253 (44%), Gaps = 22/253 (8%)
Query: 413 YVLDLDMPIYNINAKRLKPNELNPTYLW-HCRLGHINENRISKLHKDGFLDSFDFESYE- 470
Y+L ++ + IN + Y + H L H N I K+ + F+ +
Sbjct: 143 YLLPSNISVPTINNVHTSESTRKYPYPFIHRMLAHANAQTIRYSLKNNTITYFNESDVDW 202
Query: 471 ------TCRSCLLGKMTKAPFTGQGERAS-----DLLGLIHTDVCGPLNITARGGFHYFI 519
C CL+GK TK +G R + +HTD+ GP++ + YFI
Sbjct: 203 SSAIDYQCPDCLIGKSTKHRHI-KGSRLKYQNSYEPFQYLHTDIFGPVHNLPKSAPSYFI 261
Query: 520 TLTDDFSRFGYVYLMKHKSES-----FTFFKEFQNEVENQLGKKIKMLRSDRGGEYLSLE 574
+ TD+ ++F +VY + + E FT F ++NQ + +++ DRG EY +
Sbjct: 262 SFTDETTKFRWVYPLHDRREDSILDVFTTILAF---IKNQFQASVLVIQMDRGSEYTNRT 318
Query: 575 FDNHLKECGILSQLTPPGTPQWNGVSERRNRTLLDMVRSMMSHAELPNFLWGYALLTAAY 634
L++ GI T + +GV+ER NRTLLD R+ + + LPN LW A+ +
Sbjct: 319 LHKFLEKNGITPCYTTTADSRAHGVAERLNRTLLDDCRTQLQCSGLPNHLWFSAIEFSTI 378
Query: 635 TLNRVPSKAVEKT 647
N + S +K+
Sbjct: 379 VRNSLASPKSKKS 391
>YME4_YEAST (Q04711) Transposon Ty1 protein B
Length = 1328
Score = 99.8 bits (247), Expect = 4e-20
Identities = 99/384 (25%), Positives = 183/384 (46%), Gaps = 50/384 (13%)
Query: 797 DEPVTYTEAITGPEFEKWLEAMKSEMDSMYTNQVWNL--------IDAPEGINPIGCKWV 848
DE +TY + I E EK++EA E++ + W+ ID IN + ++
Sbjct: 808 DEAITYNKDIK--EKEKYIEAYHKEVNQLLKMNTWDTDKYYDRKEIDPKRVINSM---FI 862
Query: 849 FKKKIDMDGKVSTYKARLVAKGFKQIHGVDYDETF-SPVAMIKSIRILLAIAAYHDYEIW 907
F +K D T+KAR VA+G Q H YD S ++ L++A ++Y I
Sbjct: 863 FNRKRD-----GTHKARFVARGDIQ-HPDTYDSGMQSNTVHHYALMTSLSLALDNNYYIT 916
Query: 908 QMDVKTAFLNGNLLEDVYMTQPEGFGDPKATKKVCKLQRSIYGLKQASRSWNLRFDETVQ 967
Q+D+ +A+L ++ E++Y+ P G K+ +L++S+YGLKQ+ +W ET++
Sbjct: 917 QLDISSAYLYADIKEELYIRPPPHLG---MNDKLIRLKKSLYGLKQSGANWY----ETIK 969
Query: 968 QY-----GFIKNEDEPCVYKKVSGSIVSFLILYVDDILLIGNDIPTLQEIKTWLRKCFSM 1022
Y G + CV+K +I L+VDD++L D+ ++I T L+K +
Sbjct: 970 SYLIKQCGMEEVRGWSCVFKNSQVTI----CLFVDDMILFSKDLNANKKIITTLKKQYDT 1025
Query: 1023 K--DLGEASY-----ILGIRIYRDRSQRLLGLSQGRYIDKVLRRFNMHESKKG---FIPM 1072
K +LGE+ ILG+ I R + + L + + + + N+ + KG P
Sbjct: 1026 KIINLGESDNEIQYDILGLEIKYQRG-KYMKLGMENSLTEKIPKLNVPLNPKGRKLSAPG 1084
Query: 1073 SSGLNLSKTQSPSTDDE-RDRMRDIPYASAIGSIMYAMICTRPDVSYALSATSRYQSNPG 1131
GL + + + +DE ++++ ++ IG Y R D+ Y ++ +++ P
Sbjct: 1085 QPGLYIDQDELEIDEDEYKEKVHEM--QKLIGLASYVGYKFRFDLLYYINTLAQHILFPS 1142
Query: 1132 NEYWIAVKNILKYLRRTKDTFLIY 1155
+ +++++ T+D LI+
Sbjct: 1143 RQVLDMTYELIQFMWDTRDKQLIW 1166
Score = 87.8 bits (216), Expect = 2e-16
Identities = 69/253 (27%), Positives = 114/253 (44%), Gaps = 22/253 (8%)
Query: 413 YVLDLDMPIYNINAKRLKPNELNPTYLW-HCRLGHINENRISKLHKDGFLDSFDFESYE- 470
Y+L ++ + IN + Y + H L H N I K+ + F+ +
Sbjct: 143 YLLPSNISVPTINNVHTSESTRKYPYPFIHRMLAHANAQTIRYSLKNNTITYFNESDVDW 202
Query: 471 ------TCRSCLLGKMTKAPFTGQGERAS-----DLLGLIHTDVCGPLNITARGGFHYFI 519
C CL+GK TK +G R + +HTD+ GP++ + YFI
Sbjct: 203 SSAIDYQCPDCLIGKSTKHRHI-KGSRLKYQNSYEPFQYLHTDIFGPVHNLPKSAPSYFI 261
Query: 520 TLTDDFSRFGYVYLMKHKSES-----FTFFKEFQNEVENQLGKKIKMLRSDRGGEYLSLE 574
+ TD+ ++F +VY + + E FT F ++NQ + +++ DRG EY +
Sbjct: 262 SFTDETTKFRWVYPLHDRREDSILDVFTTILAF---IKNQFQASVLVIQMDRGSEYTNRT 318
Query: 575 FDNHLKECGILSQLTPPGTPQWNGVSERRNRTLLDMVRSMMSHAELPNFLWGYALLTAAY 634
L++ GI T + +GV+ER NRTLLD R+ + + LPN LW A+ +
Sbjct: 319 LHKFLEKNGITPCYTTTADSRAHGVAERLNRTLLDDCRTQLQCSGLPNHLWFSAIEFSTI 378
Query: 635 TLNRVPSKAVEKT 647
N + S +K+
Sbjct: 379 VRNSLASPKSKKS 391
>YJZ7_YEAST (P47098) Transposon Ty1 protein B
Length = 1755
Score = 99.0 bits (245), Expect = 7e-20
Identities = 100/384 (26%), Positives = 183/384 (47%), Gaps = 50/384 (13%)
Query: 797 DEPVTYTEAITGPEFEKWLEAMKSEMDSMYTNQVWNL--------IDAPEGINPIGCKWV 848
DE +TY + I E EK++EA E++ + + W+ ID IN + ++
Sbjct: 1235 DEAITYNKDIK--EKEKYIEAYHKEVNQLLKMKTWDTDEYYDRKEIDPKRVINSM---FI 1289
Query: 849 FKKKIDMDGKVSTYKARLVAKGFKQIHGVDYDETF-SPVAMIKSIRILLAIAAYHDYEIW 907
F KK D T+KAR VA+G Q H YD S ++ L++A ++Y I
Sbjct: 1290 FNKKRD-----GTHKARFVARGDIQ-HPDTYDTGMQSNTVHHYALMTSLSLALDNNYYIT 1343
Query: 908 QMDVKTAFLNGNLLEDVYMTQPEGFGDPKATKKVCKLQRSIYGLKQASRSWNLRFDETVQ 967
Q+D+ +A+L ++ E++Y+ P G K+ +L++S YGLKQ+ +W ET++
Sbjct: 1344 QLDISSAYLYADIKEELYIRPPPHLG---MNDKLIRLKKSHYGLKQSGANWY----ETIK 1396
Query: 968 QY-----GFIKNEDEPCVYKKVSGSIVSFLILYVDDILLIGNDIPTLQEIKTWLRKCFSM 1022
Y G + CV+K +I L+VDD++L D+ ++I T L+K +
Sbjct: 1397 SYLIKQCGMEEVRGWSCVFKNSQVTI----CLFVDDMILFSKDLNANKKIITTLKKQYDT 1452
Query: 1023 K--DLGEASY-----ILGIRIYRDRSQRLLGLSQGRYIDKVLRRFNMHESKKG---FIPM 1072
K +LGE+ ILG+ I R + + L + + + + N+ + KG P
Sbjct: 1453 KIINLGESDNEIQYDILGLEIKYQRG-KYMKLGMENSLTEKIPKLNVPLNPKGRKLSAPG 1511
Query: 1073 SSGLNLSKTQSPSTDDE-RDRMRDIPYASAIGSIMYAMICTRPDVSYALSATSRYQSNPG 1131
GL + + + +DE ++++ ++ IG Y R D+ Y ++ +++ P
Sbjct: 1512 QPGLYIDQDELEIDEDEYKEKVHEM--QKLIGLASYVGYKFRFDLLYYINTLAQHILFPS 1569
Query: 1132 NEYWIAVKNILKYLRRTKDTFLIY 1155
+ +++++ T+D LI+
Sbjct: 1570 RQVLDMTYELIQFMWDTRDKQLIW 1593
Score = 87.8 bits (216), Expect = 2e-16
Identities = 69/253 (27%), Positives = 114/253 (44%), Gaps = 22/253 (8%)
Query: 413 YVLDLDMPIYNINAKRLKPNELNPTYLW-HCRLGHINENRISKLHKDGFLDSFDFESYE- 470
Y+L ++ + IN + Y + H L H N I K+ + F+ +
Sbjct: 570 YLLPSNISVPTINNVHTSESTRKYPYPFIHRMLAHANAQTIRYSLKNNTITYFNESDVDW 629
Query: 471 ------TCRSCLLGKMTKAPFTGQGERAS-----DLLGLIHTDVCGPLNITARGGFHYFI 519
C CL+GK TK +G R + +HTD+ GP++ + YFI
Sbjct: 630 SSAIDYQCPDCLIGKSTKHRHI-KGSRLKYQNSYEPFQYLHTDIFGPVHNLPKSAPSYFI 688
Query: 520 TLTDDFSRFGYVYLMKHKSES-----FTFFKEFQNEVENQLGKKIKMLRSDRGGEYLSLE 574
+ TD+ ++F +VY + + E FT F ++NQ + +++ DRG EY +
Sbjct: 689 SFTDETTKFRWVYPLHDRREDSILDVFTTILAF---IKNQFQASVLVIQMDRGSEYTNRT 745
Query: 575 FDNHLKECGILSQLTPPGTPQWNGVSERRNRTLLDMVRSMMSHAELPNFLWGYALLTAAY 634
L++ GI T + +GV+ER NRTLLD R+ + + LPN LW A+ +
Sbjct: 746 LHKFLEKNGITPCYTTTADSRAHGVAERLNRTLLDDCRTQLQCSGLPNHLWFSAIEFSTI 805
Query: 635 TLNRVPSKAVEKT 647
N + S +K+
Sbjct: 806 VRNSLASPKSKKS 818
>YCB9_YEAST (P25384) Transposon Ty2 protein B (Ty1-17 protein B)
Length = 1770
Score = 98.2 bits (243), Expect = 1e-19
Identities = 100/384 (26%), Positives = 182/384 (47%), Gaps = 50/384 (13%)
Query: 797 DEPVTYTEAITGPEFEKWLEAMKSEMDSMYTNQVW--------NLIDAPEGINPIGCKWV 848
DE +TY + E ++++EA E+ + W N ID + IN + ++
Sbjct: 1250 DEAITYNK--DNKEKDRYVEAYHKEISQLLKMNTWDTNKYYDRNDIDPKKVINSM---FI 1304
Query: 849 FKKKIDMDGKVSTYKARLVAKGFKQIHGVDYDETF-SPVAMIKSIRILLAIAAYHDYEIW 907
F KK D T+KAR VA+G Q H YD S ++ L+IA +DY I
Sbjct: 1305 FNKKRD-----GTHKARFVARGDIQ-HPDTYDSDMQSNTVHHYALMTSLSIALDNDYYIT 1358
Query: 908 QMDVKTAFLNGNLLEDVYMTQPEGFGDPKATKKVCKLQRSIYGLKQASRSWNLRFDETVQ 967
Q+D+ +A+L ++ E++Y+ P G K+ +L++S+YGLKQ+ +W ET++
Sbjct: 1359 QLDISSAYLYADIKEELYIRPPPHLG---LNDKLLRLRKSLYGLKQSGANWY----ETIK 1411
Query: 968 QY-----GFIKNEDEPCVYKKVSGSIVSFLILYVDDILLIGNDIPTLQEIKTWLRKCFSM 1022
Y + CV+K +I L+VDD++L D+ ++I T L+K +
Sbjct: 1412 SYLINCCDMQEVRGWSCVFKNSQVTI----CLFVDDMILFSKDLNANKKIITTLKKQYDT 1467
Query: 1023 K--DLGEASY-----ILGIRIYRDRSQRLLGLSQGRYIDKVLRRFNMHESKKG---FIPM 1072
K +LGE+ ILG+ I RS + + L + + + L + N+ + KG P
Sbjct: 1468 KIINLGESDNEIQYDILGLEIKYQRS-KYMKLGMEKSLTEKLPKLNVPLNPKGKKLRAPG 1526
Query: 1073 SSGLNLSKTQSPSTDDE-RDRMRDIPYASAIGSIMYAMICTRPDVSYALSATSRYQSNPG 1131
G + + + +DE ++++ ++ IG Y R D+ Y ++ +++ P
Sbjct: 1527 QPGHYIDQDELEIDEDEYKEKVHEM--QKLIGLASYVGYKFRFDLLYYINTLAQHILFPS 1584
Query: 1132 NEYWIAVKNILKYLRRTKDTFLIY 1155
+ +++++ T+D LI+
Sbjct: 1585 RQVLDMTYELIQFMWDTRDKQLIW 1608
Score = 93.2 bits (230), Expect = 4e-18
Identities = 70/237 (29%), Positives = 108/237 (45%), Gaps = 15/237 (6%)
Query: 425 NAKRLKPNELNPTYLWHCRLGHINENRISKLHKDG---FLDSFDFE----SYETCRSCLL 477
N + K P L H LGH N I K K +L D E S C CL+
Sbjct: 579 NVNKSKSVNKYPYPLIHRMLGHANFRSIQKSLKKNAVTYLKESDIEWSNASTYQCPDCLI 638
Query: 478 GKMTKAPFTGQGERAS-----DLLGLIHTDVCGPLNITARGGFHYFITLTDDFSRFGYVY 532
GK TK +G R + +HTD+ GP++ + YFI+ TD+ +RF +VY
Sbjct: 639 GKSTKHRHV-KGSRLKYQESYEPFQYLHTDIFGPVHHLPKSAPSYFISFTDEKTRFQWVY 697
Query: 533 LMKHKSES--FTFFKEFQNEVENQLGKKIKMLRSDRGGEYLSLEFDNHLKECGILSQLTP 590
+ + E F ++NQ ++ +++ DRG EY + GI + T
Sbjct: 698 PLHDRREESILNVFTSILAFIKNQFNARVLVIQMDRGSEYTNKTLHKFFTNRGITACYTT 757
Query: 591 PGTPQWNGVSERRNRTLLDMVRSMMSHAELPNFLWGYALLTAAYTLNRVPSKAVEKT 647
+ +GV+ER NRTLL+ R+++ + LPN LW A+ + N + S +K+
Sbjct: 758 TADSRAHGVAERLNRTLLNDCRTLLHCSGLPNHLWFSAVEFSTIIRNSLVSPKNDKS 814
>YJZ9_YEAST (P47100) Transposon Ty1 protein B
Length = 1755
Score = 95.5 bits (236), Expect = 7e-19
Identities = 96/379 (25%), Positives = 177/379 (46%), Gaps = 40/379 (10%)
Query: 797 DEPVTYTEAITGPEFEKWLEAMKSEMDSMYTNQVWNL--------IDAPEGINPIGCKWV 848
DE +TY + I E EK++EA E++ + + W+ ID IN + ++
Sbjct: 1235 DEAITYNKDIK--EKEKYIEAYHKEVNQLLKMKTWDTDEYYDRKEIDPKRVINSM---FI 1289
Query: 849 FKKKIDMDGKVSTYKARLVAKGFKQIHGVDYDETF-SPVAMIKSIRILLAIAAYHDYEIW 907
F KK D T+KAR VA+G Q H YD S ++ L++A ++Y I
Sbjct: 1290 FNKKRD-----GTHKARFVARGDIQ-HPDTYDSGMQSNTVHHYALMTSLSLALDNNYYIT 1343
Query: 908 QMDVKTAFLNGNLLEDVYMTQPEGFGDPKATKKVCKLQRSIYGLKQASRSWNLRFDE-TV 966
Q+D+ +A+L ++ E++Y+ P G K+ +L++S+YGLKQ+ +W +
Sbjct: 1344 QLDISSAYLYADIKEELYIRPPPHLG---MNDKLIRLKKSLYGLKQSGANWYETIKSYLI 1400
Query: 967 QQYGFIKNEDEPCVYKKVSGSIVSFLILYVDDILLIGNDIPTLQEIKTWLRKCFSMK--D 1024
QQ G + CV+K +I L+VDD++L ++ + + I L+ + K +
Sbjct: 1401 QQCGMEEVRGWSCVFKNSQVTI----CLFVDDMVLFSKNLNSNKRIIEKLKMQYDTKIIN 1456
Query: 1025 LGEASY-----ILGIRIYRDRSQRLLGLSQGRYIDKVLRRFNMHESKKG---FIPMSSGL 1076
LGE+ ILG+ I R + + L + + + + N+ + KG P GL
Sbjct: 1457 LGESDEEIQYDILGLEIKYQRG-KYMKLGMENSLTEKIPKLNVPLNPKGRKLSAPGQPGL 1515
Query: 1077 NLSKTQSPSTDDERDRMRDIPYASAIGSIMYAMICTRPDVSYALSATSRYQSNPGNEYWI 1136
+ + + +D+ +M+ IG Y R D+ Y ++ +++ P +
Sbjct: 1516 YIDQQELELEEDDY-KMKVHEMQKLIGLASYVGYKFRFDLLYYINTLAQHILFPSKQVLD 1574
Query: 1137 AVKNILKYLRRTKDTFLIY 1155
+++++ T+D LI+
Sbjct: 1575 MTYELIQFIWNTRDKQLIW 1593
Score = 87.8 bits (216), Expect = 2e-16
Identities = 69/253 (27%), Positives = 114/253 (44%), Gaps = 22/253 (8%)
Query: 413 YVLDLDMPIYNINAKRLKPNELNPTYLW-HCRLGHINENRISKLHKDGFLDSFDFESYE- 470
Y+L ++ + IN + Y + H L H N I K+ + F+ +
Sbjct: 570 YLLPSNISVPTINNVHTSESTRKYPYPFIHRMLAHANAQTIRYSLKNNTITYFNESDVDW 629
Query: 471 ------TCRSCLLGKMTKAPFTGQGERAS-----DLLGLIHTDVCGPLNITARGGFHYFI 519
C CL+GK TK +G R + +HTD+ GP++ + YFI
Sbjct: 630 SSAIDYQCPDCLIGKSTKHRHI-KGSRLKYQNSYEPFQYLHTDIFGPVHNLPKSAPSYFI 688
Query: 520 TLTDDFSRFGYVYLMKHKSES-----FTFFKEFQNEVENQLGKKIKMLRSDRGGEYLSLE 574
+ TD+ ++F +VY + + E FT F ++NQ + +++ DRG EY +
Sbjct: 689 SFTDETTKFRWVYPLHDRREDSILDVFTTILAF---IKNQFQASVLVIQMDRGSEYTNRT 745
Query: 575 FDNHLKECGILSQLTPPGTPQWNGVSERRNRTLLDMVRSMMSHAELPNFLWGYALLTAAY 634
L++ GI T + +GV+ER NRTLLD R+ + + LPN LW A+ +
Sbjct: 746 LHKFLEKNGITPCYTTTADSRAHGVAERLNRTLLDDCRTQLQCSGLPNHLWFSAIEFSTI 805
Query: 635 TLNRVPSKAVEKT 647
N + S +K+
Sbjct: 806 VRNSLASPKSKKS 818
>YMT5_YEAST (Q04214) Transposon Ty1 protein B
Length = 1328
Score = 91.7 bits (226), Expect = 1e-17
Identities = 95/383 (24%), Positives = 179/383 (45%), Gaps = 48/383 (12%)
Query: 797 DEPVTYTEAITGPEFEKWLEAMKSEMDSMYTNQVWNL--------IDAPEGINPIGCKWV 848
DE +TY + I E EK++EA E++ + + W+ ID IN + ++
Sbjct: 808 DEAITYNKDIK--EKEKYIEAYHKEVNQLLKMKTWDTDKYYDRKEIDPKRVINSM---FI 862
Query: 849 FKKKIDMDGKVSTYKARLVAKGFKQIHGVDYDETF-SPVAMIKSIRILLAIAAYHDYEIW 907
F +K D T+KAR VA+G Q H YD S ++ L++A ++Y I
Sbjct: 863 FNRKRD-----GTHKARFVARGDIQ-HPDTYDSGMQSNTVHHYALMTSLSLALDNNYYIT 916
Query: 908 QMDVKTAFLNGNLLEDVYMTQPEGFGDPKATKKVCKLQRSIYGLKQASRSWNLRFDETVQ 967
Q+D+ +A+L ++ E++Y+ P G K+ +L++S+YGLKQ+ +W ET++
Sbjct: 917 QLDISSAYLYADIKEELYIRPPPHLG---MNDKLIRLKKSLYGLKQSGANWY----ETIK 969
Query: 968 QY-----GFIKNEDEPCVYKKVSGSIVSFLILYVDDILLIGNDIPTLQEIKTWLRKCFSM 1022
Y G + CV++ +I L+VDD++L ++ + + I L+ +
Sbjct: 970 SYLIKQCGMEEVRGWSCVFENSQVTI----CLFVDDMVLFSKNLNSNKRIIDKLKMQYDT 1025
Query: 1023 K--DLGEASY-----ILGIRIYRDRSQRLLGLSQGRYIDKVLRRFNMHESKKG---FIPM 1072
K +LGE+ ILG+ I R + + L + + + + N+ + KG P
Sbjct: 1026 KIINLGESDEEIQYDILGLEIKYQRG-KYMKLGMENSLTEKIPKLNVPLNPKGRKLSAPG 1084
Query: 1073 SSGLNLSKTQSPSTDDERDRMRDIPYASAIGSIMYAMICTRPDVSYALSATSRYQSNPGN 1132
GL + + + +D+ +M+ IG Y R D+ Y ++ +++ P
Sbjct: 1085 QPGLYIDQQELELEEDDY-KMKVHEMQKLIGLASYVGYKFRFDLLYYINTLAQHILFPSK 1143
Query: 1133 EYWIAVKNILKYLRRTKDTFLIY 1155
+ +++++ T+D LI+
Sbjct: 1144 QVLDMTYELIQFIWNTRDKQLIW 1166
Score = 87.8 bits (216), Expect = 2e-16
Identities = 69/253 (27%), Positives = 114/253 (44%), Gaps = 22/253 (8%)
Query: 413 YVLDLDMPIYNINAKRLKPNELNPTYLW-HCRLGHINENRISKLHKDGFLDSFDFESYE- 470
Y+L ++ + IN + Y + H L H N I K+ + F+ +
Sbjct: 143 YLLPSNISVPTINNVHTSESTRKYPYPFIHRMLAHANAQTIRYSLKNNTITYFNESDVDW 202
Query: 471 ------TCRSCLLGKMTKAPFTGQGERAS-----DLLGLIHTDVCGPLNITARGGFHYFI 519
C CL+GK TK +G R + +HTD+ GP++ + YFI
Sbjct: 203 SSAIDYQCPDCLIGKSTKHRHI-KGSRLKYQNSYEPFQYLHTDIFGPVHNLPKSAPSYFI 261
Query: 520 TLTDDFSRFGYVYLMKHKSES-----FTFFKEFQNEVENQLGKKIKMLRSDRGGEYLSLE 574
+ TD+ ++F +VY + + E FT F ++NQ + +++ DRG EY +
Sbjct: 262 SFTDETTKFRWVYPLHDRREDSILDVFTTILAF---IKNQFQASVLVIQMDRGSEYTNRT 318
Query: 575 FDNHLKECGILSQLTPPGTPQWNGVSERRNRTLLDMVRSMMSHAELPNFLWGYALLTAAY 634
L++ GI T + +GV+ER NRTLLD R+ + + LPN LW A+ +
Sbjct: 319 LHKFLEKNGITPCYTTTADSRAHGVAERLNRTLLDDCRTQLQCSGLPNHLWFSAIEFSTI 378
Query: 635 TLNRVPSKAVEKT 647
N + S +K+
Sbjct: 379 VRNSLASPKSKKS 391
>M820_ARATH (P92520) Hypothetical mitochondrial protein AtMg00820
(ORF170)
Length = 170
Score = 88.6 bits (218), Expect = 9e-17
Identities = 44/102 (43%), Positives = 62/102 (60%), Gaps = 3/102 (2%)
Query: 798 EPVTYTEAITGPEFEKWLEAMKSEMDSMYTNQVWNLIDAPEGINPIGCKWVFKKKIDMDG 857
EP + A+ P W +AM+ E+D++ N+ W L+ P N +GCKWVFK K+ DG
Sbjct: 27 EPKSVIFALKDPG---WCQAMQEELDALSRNKTWILVPPPVNQNILGCKWVFKTKLHSDG 83
Query: 858 KVSTYKARLVAKGFKQIHGVDYDETFSPVAMIKSIRILLAIA 899
+ KARLVAKGF Q G+ + ET+SPV +IR +L +A
Sbjct: 84 TLDRLKARLVAKGFHQEEGIYFVETYSPVVRTATIRTILNVA 125
>YJL3_YEAST (P47024) Transposon Ty4 207.7 kDa hypothetical protein
Length = 1803
Score = 87.0 bits (214), Expect = 3e-16
Identities = 77/320 (24%), Positives = 143/320 (44%), Gaps = 20/320 (6%)
Query: 862 YKARLVAKGFKQIHGVDYDETFSPVAMIKSIRILLAIAAYHDYEIWQMDVKTAFLNGNLL 921
YKAR+V +G Q Y + I+I L IA + + +D+ AFL L
Sbjct: 1337 YKARIVCRGDTQSPDT-YSVITTESLNHNHIKIFLMIANNRNMFMKTLDINHAFLYAKLE 1395
Query: 922 EDVYMTQPEGFGDPKATKKVCKLQRSIYGLKQASRSWNLRFDETVQQYGFIKNEDEPCVY 981
E++Y+ P + V KL +++YGLKQ+ + WN + + G N P +Y
Sbjct: 1396 EEIYIPHPHD------RRCVVKLNKALYGLKQSPKEWNDHLRQYLNGIGLKDNSYTPGLY 1449
Query: 982 KKVSGSIVSFLILYVDDILLIGNDIPTLQEIKTWLRKCFSMKDLGEA------SYILGIR 1035
+ +++ + +YVDD ++ ++ L E L+ F +K G + ILG+
Sbjct: 1450 QTEDKNLM--IAVYVDDCVIAASNEQRLDEFINKLKSNFELKITGTLIDDVLDTDILGMD 1507
Query: 1036 IYRDRSQRLLGLSQGRYIDKVLRRFN--MHESKKGFIPMSSGLNLS-KTQSPSTDDERDR 1092
+ ++ + L+ +I+++ +++N + + +K IP S + K +E R
Sbjct: 1508 LVYNKRLGTIDLTLKSFINRMDKKYNEELKKIRKSSIPHMSTYKIDPKKDVLQMSEEEFR 1567
Query: 1093 MRDIPYASAIGSIMYAMICTRPDVSYALSATSRYQSNPGNEYWIAVKNILKYLRRTKDTF 1152
+ +G + Y R D+ +A+ +R + P + + I++YL R KD
Sbjct: 1568 QGVLKLQQLLGELNYVRHKCRYDIEFAVKKVARLVNYPHERVFYMIYKIIQYLVRYKDIG 1627
Query: 1153 LIYGG--*EELSVIGYIDAS 1170
+ Y ++ VI DAS
Sbjct: 1628 IHYDRDCNKDKKVIAITDAS 1647
Score = 72.4 bits (176), Expect = 7e-12
Identities = 97/437 (22%), Positives = 175/437 (39%), Gaps = 34/437 (7%)
Query: 304 VLDTGCGSHICTDVQGLRKSRALAKGEVDLRVGNGAKVAALAVGTYVLTLPSGLLLNLEN 363
++DTG G +I D L + +G + V+ G + + +G
Sbjct: 413 IIDTGSGVNITNDKTLLHNYEDSNRSTRFFGIGKNSSVSVKGYG--YIKIKNGHNNTDNK 470
Query: 364 C---YYVPAISRNIISISCLDKAGFEFTIKNKCCSVYLDNILYANANLSNGLYVLDL--- 417
C YYVP IIS C D A + ++ + + I+ + NG+ + +
Sbjct: 471 CLLTYYVPEEESTIIS--CYDLAKKTKMVLSRKYTRLGNKIIKIKTKIVNGVIHVKMNEL 528
Query: 418 -DMPIYNINAKRLKPN-----ELNPTYLW----HCRLGHIN----ENRISKLHKDGFLDS 463
+ P + +KP +LN + H R+GH EN I H + LD
Sbjct: 529 IERPSDDSKINAIKPTSSPGFKLNKRSITLEDAHKRMGHTGIQQIENSIKHNHYEESLDL 588
Query: 464 FDFESYETCRSCLLGKMTKAP-FTGQGERAS---DLLGLIHTDVCGPLNITARGGFHYFI 519
+ C++C + K TK +TG S + D+ GP++ + Y +
Sbjct: 589 IKEPNEFWCQTCKISKATKRNHYTGSMNNHSTDHEPGSSWCMDIFGPVSSSNADTKRYML 648
Query: 520 TLTDDFSRFGYVYLMKHKSESFTFFKEFQN--EVENQLGKKIKMLRSDRGGEYLSLEFDN 577
+ D+ +R+ +K+ + +N VE Q +K++ + SDRG E+ + + +
Sbjct: 649 IMVDNNTRYCMTSTHFNKNAETILAQVRKNIQYVETQFDRKVREINSDRGTEFTNDQIEE 708
Query: 578 HLKECGILSQLTPPGTPQWNGVSERRNRTLLDMVRSMMSHAELPNFLWGYALLTAAYTLN 637
+ GI LT NG +ER RT++ +++ + L W YA+ +A N
Sbjct: 709 YFISKGIHHILTSTQDHAANGRAERYIRTIITDATTLLRQSNLRVKFWEYAVTSATNIRN 768
Query: 638 RVPSKAVEKTPYEIWNGRKP---NVGHFKIWGCEAYIKRLMSTKLEQKSEKCFFVGYPKE 694
+ K+ K P + + R+P + F +G + I KL+ +
Sbjct: 769 YLEHKSTGKLPLKAIS-RQPVTVRLMSFLPFGEKGIIWNHNHKKLKPSGLPSIILCKDPN 827
Query: 695 TRGYYFYKPPEGTIVVA 711
+ GY F+ P + IV +
Sbjct: 828 SYGYKFFIPSKNKIVTS 844
>M810_ARATH (P92519) Hypothetical mitochondrial protein AtMg00810
(ORF240b)
Length = 240
Score = 80.5 bits (197), Expect = 2e-14
Identities = 57/158 (36%), Positives = 83/158 (52%), Gaps = 10/158 (6%)
Query: 991 FLILYVDDILLIGNDIPTLQEIKTWLRKCFSMKDLGEASYILGIRIYRDRSQRLLGLSQG 1050
+L+LYVDDILL G+ L + L FSMKDLG Y LGI+I S L LSQ
Sbjct: 2 YLLLYVDDILLTGSSNTLLNMLIFQLSSTFSMKDLGPVHYFLGIQIKTHPSG--LFLSQT 59
Query: 1051 RYIDKVLRRFNMHESKKGFIPMSSGLNLSKTQSPSTDDERDRMRDIPYASAIGSIMYAMI 1110
+Y +++L M + K PMS+ L L S ST D + S +G++ Y +
Sbjct: 60 KYAEQILNNAGMLDCK----PMSTPLPLKLNSSVSTAKYPD---PSDFRSIVGALQY-LT 111
Query: 1111 CTRPDVSYALSATSRYQSNPGNEYWIAVKNILKYLRRT 1148
TRPD+SYA++ + P + +K +L+Y++ T
Sbjct: 112 LTRPDISYAVNIVCQRMHEPTLADFDLLKRVLRYVKGT 149
>M300_ARATH (P93293) Hypothetical mitochondrial protein AtMg00300
(ORF145a) (ORF1451)
Length = 145
Score = 53.5 bits (127), Expect = 3e-06
Identities = 30/99 (30%), Positives = 46/99 (46%), Gaps = 3/99 (3%)
Query: 407 NLSNGLYVLDLDMPIYNINAKRLKPNELNPTYLWHCRLGHINENRISKLHKDGFLDSFDF 466
N + LY+L + N +E T LWH RL H+++ + L K GFLDS
Sbjct: 42 NRHDSLYILQGSVETGESNLAETAKDE---TRLWHSRLAHMSQRGMELLVKKGFLDSSKV 98
Query: 467 ESYETCRSCLLGKMTKAPFTGQGERASDLLGLIHTDVCG 505
S + C C+ GK + F+ + L +H+D+ G
Sbjct: 99 SSLKFCEDCIYGKTHRVNFSTGQHTTKNPLDYVHSDLWG 137
>M710_ARATH (P92512) Hypothetical mitochondrial protein AtMg00710
(ORF120)
Length = 120
Score = 48.9 bits (115), Expect = 8e-05
Identities = 25/69 (36%), Positives = 37/69 (53%), Gaps = 1/69 (1%)
Query: 604 NRTLLDMVRSMMSHAELPNFLWGYALLTAAYTLNRVPSKAVE-KTPYEIWNGRKPNVGHF 662
NRT+++ VRSM+ LP A TA + +N+ PS A+ P E+W P +
Sbjct: 2 NRTIIEKVRSMLCECGLPKTFRADAANTAVHIINKYPSTAINFHVPDEVWFQSVPTYSYL 61
Query: 663 KIWGCEAYI 671
+ +GC AYI
Sbjct: 62 RRFGCVAYI 70
>POL_SFV1 (P23074) Pol polyprotein [Contains: Protease (EC 3.4.23.-);
Reverse transcriptase/ribonuclease H (EC 2.7.7.49) (EC
3.1.26.4) (RT); Integrase (IN)]
Length = 1161
Score = 45.1 bits (105), Expect = 0.001
Identities = 47/196 (23%), Positives = 80/196 (39%), Gaps = 15/196 (7%)
Query: 467 ESYETCRSCLLGKMTK--APFTGQGERASDLLGLIHTDVCGPLNITARGGFHYFITLTDD 524
+S C+ CL+ T +P + + + D GPL G+ + + + D
Sbjct: 853 KSIRQCKQCLVTNATNLTSPPILRPVKPLKPFDKFYIDYIGPL--PPSNGYLHVLVVVDS 910
Query: 525 FSRFGYVYLMKHKSESFTFFKEFQNEVENQLGKKI--KMLRSDRGGEYLSLEFDNHLKEC 582
+ F ++Y K S S T + N L K+L SD+G + S F + KE
Sbjct: 911 MTGFVWLYPTKAPSTSATV------KALNMLTSIAIPKVLHSDQGAAFTSSTFADWAKEK 964
Query: 583 GILSQLTPPGTPQWNGVSERRNRTLLDMVRSMMSHAELPNFLWGYALLTAAYTLNRVPSK 642
GI + + P PQ +G ER+N + ++ ++ W L LN S
Sbjct: 965 GIQLEFSTPYHPQSSGKVERKNSDIKRLLTKLLIGRPAK---WYDLLPVVQLALNNSYSP 1021
Query: 643 AVEKTPYEIWNGRKPN 658
+ + TP+++ G N
Sbjct: 1022 SSKYTPHQLLFGVDSN 1037
>POL4_DROME (P10394) Retrovirus-related Pol polyprotein from
transposon 412 [Contains: Protease (EC 3.4.23.-); Reverse
transcriptase (EC 2.7.7.49); Endonuclease]
Length = 1237
Score = 43.5 bits (101), Expect = 0.003
Identities = 48/190 (25%), Positives = 74/190 (38%), Gaps = 7/190 (3%)
Query: 470 ETCRSCLLGKMTKAPFTGQGERASDLLGLIHTDVCGPLNITARGGFHYFITLTDDFSRFG 529
+ C+ K TK P T E + D GPL + G Y +TL D +++
Sbjct: 940 QKCQKAKTTKHTKTPMTIT-ETPEHAFDRVVVDTIGPLPKSENGN-EYAVTLICDLTKYL 997
Query: 530 YVYLMKHKSESFTFFKEFQNEVENQLGKKIKMLRSDRGGEYLSLEFDNHLKECGILSQLT 589
+ +KS F++ + +K +D G EY + + K I + +
Sbjct: 998 VAIPIANKSAKTVAKAIFESFILKY--GPMKTFITDMGTEYKNSIITDLCKYLKIKNITS 1055
Query: 590 PPGTPQWNGVSERRNRTLLDMVRSMMSHAELPNFLWGYALLTAAYTLNRVPSKAVEKTPY 649
Q GV ER +RTL + +RS +S + W L Y N S PY
Sbjct: 1056 TAHHHQTVGVVERSHRTLNEYIRSYISTDKTD---WDVWLQYFVYCFNTTQSMVHNYCPY 1112
Query: 650 EIWNGRKPNV 659
E+ GR N+
Sbjct: 1113 ELVFGRTSNL 1122
>POL_SFV3L (P27401) Pol polyprotein [Contains: Protease (EC 3.4.23.-);
Reverse transcriptase/ribonuclease H (EC 2.7.7.49) (EC
3.1.26.4) (RT); Integrase (IN)]
Length = 1157
Score = 43.1 bits (100), Expect = 0.004
Identities = 46/191 (24%), Positives = 76/191 (39%), Gaps = 15/191 (7%)
Query: 472 CRSCLLGKMTK--APFTGQGERASDLLGLIHTDVCGPLNITARGGFHYFITLTDDFSRFG 529
C+ CL+ AP + ER D GPL G+ + + + D + F
Sbjct: 860 CKQCLVTNAATLAAPPILRPERPVKPFDKFFIDYIGPL--PPSNGYLHVLVVVDSMTGFV 917
Query: 530 YVYLMKHKSESFTFFKEFQNEVENQLGKKI--KMLRSDRGGEYLSLEFDNHLKECGILSQ 587
++Y K S S T + N L K++ SD+G + S F + K GI +
Sbjct: 918 WLYPTKAPSTSATV------KALNMLTSIAVPKVIHSDQGAAFTSATFADWAKNKGIQLE 971
Query: 588 LTPPGTPQWNGVSERRNRTLLDMVRSMMSHAELPNFLWGYALLTAAYTLNRVPSKAVEKT 647
+ P PQ +G ER+N + ++ ++ W L LN S + + T
Sbjct: 972 FSTPYHPQSSGKVERKNSDIKRLLTKLLVGRPAK---WYDLLPVVQLALNNSYSPSSKYT 1028
Query: 648 PYEIWNGRKPN 658
P+++ G N
Sbjct: 1029 PHQLLFGIDSN 1039
>POL3_MOUSE (P11367) Retrovirus-related Pol polyprotein
(Endonuclease) (Fragment)
Length = 390
Score = 42.7 bits (99), Expect = 0.006
Identities = 53/212 (25%), Positives = 86/212 (40%), Gaps = 18/212 (8%)
Query: 455 LHKDGFLDSFDFESYETCRSCLLGKMTKAPFTG----QGERASDLLGLIHTDVCGPLNIT 510
L+KD L E E+C++C+ +K +G R + T+V L
Sbjct: 81 LNKDKILH----EVAESCQACVQVNASKTKIRAGTRVRGHRLGTHWEIDFTEVKPGLY-- 134
Query: 511 ARGGFHYFITLTDDFSRFGYVYLMKHKSESFTFFKEFQNEVENQLGKKIKMLRSDRGGEY 570
G+ Y + D FS + + KH++ K+ E+ + G ++L +D G +
Sbjct: 135 ---GYKYLLVFVDTFSGWVEAFPTKHETAKIVT-KKLLEEIFPRFGMP-QVLGTDNGPAF 189
Query: 571 LSLEFDNHLKECGILSQLTPPGTPQWNGVSERRNRTLLDMVRSMMSHAELPNFLWGYALL 630
+S + K GI +L PQ +G ER NRT+ + + + + W L
Sbjct: 190 VSQVSQSVAKLLGIDWKLHCAYRPQSSGQVERMNRTIKETLTKLTLATGTRD--WVLLLP 247
Query: 631 TAAYTLNRVPSKAVEKTPYEIWNGRKPNVGHF 662
A Y P TPYEI G P + +F
Sbjct: 248 LALYRARNTPGPH-GLTPYEILYGAPPPLVNF 278
>POL_FENV1 (P31792) Pol polyprotein [Contains: Reverse
transcriptase/ribonuclease H (EC 2.7.7.49) (EC 3.1.26.4)
(RT); Integrase (IN)] (Fragment)
Length = 1046
Score = 42.4 bits (98), Expect = 0.007
Identities = 41/146 (28%), Positives = 63/146 (43%), Gaps = 9/146 (6%)
Query: 514 GFHYFITLTDDFSRFGYVYLMKHKSESFTFFKEFQNEVENQLGKKIKMLRSDRGGEYLSL 573
G+ Y + D FS + Y + ++ K+ E+ + G K++ SD G ++S
Sbjct: 778 GYKYLLVFVDTFSGWVEAYPTRQETAHMVA-KKILEEIFPRFGLP-KVIGSDNGPAFVSQ 835
Query: 574 EFDNHLKECGILSQLTPPGTPQWNGVSERRNRTLLDMVRSMMSHAELPNF--LWGYALLT 631
+ GI +L PQ +G ER NRT+ + + + L ++ L ALL
Sbjct: 836 VSQGLARTLGINWKLHCAYRPQSSGQVERMNRTIKETLTKLTLETGLKDWRRLLSLALLR 895
Query: 632 AAYTLNRVPSKAVEKTPYEIWNGRKP 657
A T NR TPYEI G P
Sbjct: 896 ARNTPNR-----FGLTPYEILYGGPP 916
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.320 0.137 0.410
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 142,253,451
Number of Sequences: 164201
Number of extensions: 6346689
Number of successful extensions: 14067
Number of sequences better than 10.0: 51
Number of HSP's better than 10.0 without gapping: 22
Number of HSP's successfully gapped in prelim test: 29
Number of HSP's that attempted gapping in prelim test: 13975
Number of HSP's gapped (non-prelim): 70
length of query: 1173
length of database: 59,974,054
effective HSP length: 121
effective length of query: 1052
effective length of database: 40,105,733
effective search space: 42191231116
effective search space used: 42191231116
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 72 (32.3 bits)
Medicago: description of AC149496.11


