

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC149490.2 - phase: 1
(317 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
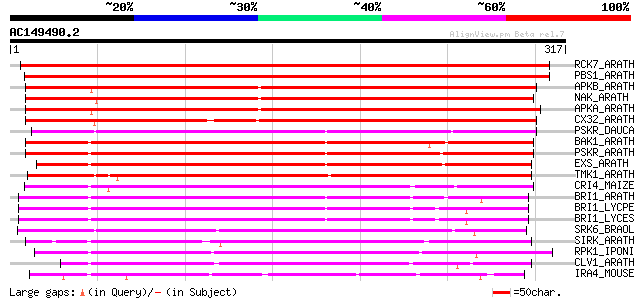
Score E
Sequences producing significant alignments: (bits) Value
RCK7_ARATH (Q9LQQ8) Putative serine/threonine-protein kinase RLC... 496 e-140
PBS1_ARATH (Q9FE20) Serine/threonine-protein kinase PBS1 (EC 2.7... 439 e-123
APKB_ARATH (P46573) Protein kinase APK1B, chloroplast precursor ... 291 1e-78
NAK_ARATH (P43293) Probable serine/threonine-protein kinase NAK ... 289 7e-78
APKA_ARATH (Q06548) Protein kinase APK1A, chloroplast precursor ... 284 2e-76
CX32_ARATH (P27450) Probable serine/threonine-protein kinase Cx3... 250 3e-66
PSKR_DAUCA (Q8LPB4) Phytosulfokine receptor precursor (EC 2.7.1.... 222 1e-57
BAK1_ARATH (Q94F62) BRASSINOSTEROID INSENSITIVE 1-associated rec... 222 1e-57
PSKR_ARATH (Q9ZVR7) Putative phytosulfokine receptor precursor (... 221 2e-57
EXS_ARATH (Q9LYN8) Leucine-rich repeat receptor protein kinase E... 219 7e-57
TMK1_ARATH (P43298) Putative receptor protein kinase TMK1 precur... 214 2e-55
CRI4_MAIZE (O24585) Putative receptor protein kinase CRINKLY4 pr... 209 6e-54
BRI1_ARATH (O22476) BRASSINOSTEROID INSENSITIVE 1 precursor (EC ... 209 6e-54
BRI1_LYCPE (Q8L899) Systemin receptor SR160 precursor (EC 2.7.1.... 201 3e-51
BRI1_LYCES (Q8GUQ5) Brassinosteroid LRR receptor kinase precurso... 200 4e-51
SRK6_BRAOL (Q09092) Putative serine/threonine-protein kinase rec... 190 3e-48
SIRK_ARATH (O64483) Senescence-induced receptor-like serine/thre... 182 7e-46
RPK1_IPONI (P93194) Receptor-like protein kinase precursor (EC 2... 182 1e-45
CLV1_ARATH (Q9SYQ8) Receptor protein kinase CLAVATA1 precursor (... 181 2e-45
IRA4_MOUSE (Q8R4K2) Interleukin-1 receptor-associated kinase-4 (... 173 4e-43
>RCK7_ARATH (Q9LQQ8) Putative serine/threonine-protein kinase
RLCKVII (EC 2.7.1.37)
Length = 423
Score = 496 bits (1276), Expect = e-140
Identities = 239/302 (79%), Positives = 267/302 (88%)
Query: 7 KVAKIFTFDELAAATKSFRVDCFVGEGGFGKVYKGYIKKINQFVAIKQLDPNGLQGTREF 66
K A+ FTF ELA AT +FR DCF+GEGGFGKV+KG I+K++Q VAIKQLD NG+QG REF
Sbjct: 86 KKAQTFTFQELAEATGNFRSDCFLGEGGFGKVFKGTIEKLDQVVAIKQLDRNGVQGIREF 145
Query: 67 AVEVLTLSLAEHPNLVKLLGFCAEGEQRLLVYEYMPLGSLENHLHDLPPGKKPLDWNTRM 126
VEVLTLSLA+HPNLVKL+GFCAEG+QRLLVYEYMP GSLE+HLH LP GKKPLDWNTRM
Sbjct: 146 VVEVLTLSLADHPNLVKLIGFCAEGDQRLLVYEYMPQGSLEDHLHVLPSGKKPLDWNTRM 205
Query: 127 RIAAGVAKGLEYLHDEMKPPVIYRDLKCSNILLGDDYHPKLSDFGLAKVGPIGDMTHVST 186
+IAAG A+GLEYLHD M PPVIYRDLKCSNILLG+DY PKLSDFGLAKVGP GD THVST
Sbjct: 206 KIAAGAARGLEYLHDRMTPPVIYRDLKCSNILLGEDYQPKLSDFGLAKVGPSGDKTHVST 265
Query: 187 RVMGTYGYCAPDYAMTGQLTSKSDIYSLGVALLELITGRKAFDPSKPAKEQNLVAWAYPL 246
RVMGTYGYCAPDYAMTGQLT KSDIYS GV LLELITGRKA D +K K+QNLV WA PL
Sbjct: 266 RVMGTYGYCAPDYAMTGQLTFKSDIYSFGVVLLELITGRKAIDNTKTRKDQNLVGWARPL 325
Query: 247 FKEQRKFSKMVDPLLEGQYPARGLYQALAVAAMCVEEQSSMRPVIADVVAALDFIASHKY 306
FK++R F KMVDPLL+GQYP RGLYQALA++AMCV+EQ +MRPV++DVV AL+F+AS KY
Sbjct: 326 FKDRRNFPKMVDPLLQGQYPVRGLYQALAISAMCVQEQPTMRPVVSDVVLALNFLASSKY 385
Query: 307 DP 308
DP
Sbjct: 386 DP 387
>PBS1_ARATH (Q9FE20) Serine/threonine-protein kinase PBS1 (EC
2.7.1.37) (AvrPphB susceptible protein 1)
Length = 456
Score = 439 bits (1129), Expect = e-123
Identities = 209/300 (69%), Positives = 245/300 (81%)
Query: 9 AKIFTFDELAAATKSFRVDCFVGEGGFGKVYKGYIKKINQFVAIKQLDPNGLQGTREFAV 68
A F F ELAAAT +F D F+GEGGFG+VYKG + Q VA+KQLD NGLQG REF V
Sbjct: 71 AHTFAFRELAAATMNFHPDTFLGEGGFGRVYKGRLDSTGQVVAVKQLDRNGLQGNREFLV 130
Query: 69 EVLTLSLAEHPNLVKLLGFCAEGEQRLLVYEYMPLGSLENHLHDLPPGKKPLDWNTRMRI 128
EVL LSL HPNLV L+G+CA+G+QRLLVYE+MPLGSLE+HLHDLPP K+ LDWN RM+I
Sbjct: 131 EVLMLSLLHHPNLVNLIGYCADGDQRLLVYEFMPLGSLEDHLHDLPPDKEALDWNMRMKI 190
Query: 129 AAGVAKGLEYLHDEMKPPVIYRDLKCSNILLGDDYHPKLSDFGLAKVGPIGDMTHVSTRV 188
AAG AKGLE+LHD+ PPVIYRD K SNILL + +HPKLSDFGLAK+GP GD +HVSTRV
Sbjct: 191 AAGAAKGLEFLHDKANPPVIYRDFKSSNILLDEGFHPKLSDFGLAKLGPTGDKSHVSTRV 250
Query: 189 MGTYGYCAPDYAMTGQLTSKSDIYSLGVALLELITGRKAFDPSKPAKEQNLVAWAYPLFK 248
MGTYGYCAP+YAMTGQLT KSD+YS GV LELITGRKA D P EQNLVAWA PLF
Sbjct: 251 MGTYGYCAPEYAMTGQLTVKSDVYSFGVVFLELITGRKAIDSEMPHGEQNLVAWARPLFN 310
Query: 249 EQRKFSKMVDPLLEGQYPARGLYQALAVAAMCVEEQSSMRPVIADVVAALDFIASHKYDP 308
++RKF K+ DP L+G++P R LYQALAVA+MC++EQ++ RP+IADVV AL ++A+ YDP
Sbjct: 311 DRRKFIKLADPRLKGRFPTRALYQALAVASMCIQEQAATRPLIADVVTALSYLANQAYDP 370
>APKB_ARATH (P46573) Protein kinase APK1B, chloroplast precursor (EC
2.7.1.-)
Length = 412
Score = 291 bits (745), Expect = 1e-78
Identities = 150/301 (49%), Positives = 200/301 (65%), Gaps = 10/301 (3%)
Query: 10 KIFTFDELAAATKSFRVDCFVGEGGFGKVYKGYIKK---------INQFVAIKQLDPNGL 60
K FTF EL AAT++FR D +GEGGFG V+KG+I + +A+K+L+ +G
Sbjct: 55 KSFTFAELKAATRNFRPDSVLGEGGFGSVFKGWIDEQTLTASKPGTGVVIAVKKLNQDGW 114
Query: 61 QGTREFAVEVLTLSLAEHPNLVKLLGFCAEGEQRLLVYEYMPLGSLENHLHDLPPGKKPL 120
QG +E+ EV L HPNLVKL+G+C E E RLLVYE+MP GSLENHL +PL
Sbjct: 115 QGHQEWLAEVNYLGQFSHPNLVKLIGYCLEDEHRLLVYEFMPRGSLENHLFRRGSYFQPL 174
Query: 121 DWNTRMRIAAGVAKGLEYLHDEMKPPVIYRDLKCSNILLGDDYHPKLSDFGLAKVGPIGD 180
W R+++A G AKGL +LH+ + VIYRD K SNILL +Y+ KLSDFGLAK GP GD
Sbjct: 175 SWTLRLKVALGAAKGLAFLHNA-ETSVIYRDFKTSNILLDSEYNAKLSDFGLAKDGPTGD 233
Query: 181 MTHVSTRVMGTYGYCAPDYAMTGQLTSKSDIYSLGVALLELITGRKAFDPSKPAKEQNLV 240
+HVSTR+MGTYGY AP+Y TG LT+KSD+YS GV LLE+++GR+A D ++P EQ LV
Sbjct: 234 KSHVSTRIMGTYGYAAPEYLATGHLTTKSDVYSYGVVLLEVLSGRRAVDKNRPPGEQKLV 293
Query: 241 AWAYPLFKEQRKFSKMVDPLLEGQYPARGLYQALAVAAMCVEEQSSMRPVIADVVAALDF 300
WA PL +RK +++D L+ QY + +A C+ + +RP + +VV+ L+
Sbjct: 294 EWARPLLANKRKLFRVIDNRLQDQYSMEEACKVATLALRCLTFEIKLRPNMNEVVSHLEH 353
Query: 301 I 301
I
Sbjct: 354 I 354
>NAK_ARATH (P43293) Probable serine/threonine-protein kinase NAK (EC
2.7.1.37)
Length = 389
Score = 289 bits (739), Expect = 7e-78
Identities = 146/299 (48%), Positives = 199/299 (65%), Gaps = 10/299 (3%)
Query: 10 KIFTFDELAAATKSFRVDCFVGEGGFGKVYKGYIKKINQ---------FVAIKQLDPNGL 60
K F+ EL +AT++FR D VGEGGFG V+KG+I + + +A+K+L+ G
Sbjct: 54 KNFSLSELKSATRNFRPDSVVGEGGFGCVFKGWIDESSLAPSKPGTGIVIAVKRLNQEGF 113
Query: 61 QGTREFAVEVLTLSLAEHPNLVKLLGFCAEGEQRLLVYEYMPLGSLENHLHDLPPGKKPL 120
QG RE+ E+ L +HPNLVKL+G+C E E RLLVYE+M GSLENHL +PL
Sbjct: 114 QGHREWLAEINYLGQLDHPNLVKLIGYCLEEEHRLLVYEFMTRGSLENHLFRRGTFYQPL 173
Query: 121 DWNTRMRIAAGVAKGLEYLHDEMKPPVIYRDLKCSNILLGDDYHPKLSDFGLAKVGPIGD 180
WNTR+R+A G A+GL +LH+ +P VIYRD K SNILL +Y+ KLSDFGLA+ GP+GD
Sbjct: 174 SWNTRVRMALGAARGLAFLHNA-QPQVIYRDFKASNILLDSNYNAKLSDFGLARDGPMGD 232
Query: 181 MTHVSTRVMGTYGYCAPDYAMTGQLTSKSDIYSLGVALLELITGRKAFDPSKPAKEQNLV 240
+HVSTRVMGT GY AP+Y TG L+ KSD+YS GV LLEL++GR+A D ++P E NLV
Sbjct: 233 NSHVSTRVMGTQGYAAPEYLATGHLSVKSDVYSFGVVLLELLSGRRAIDKNQPVGEHNLV 292
Query: 241 AWAYPLFKEQRKFSKMVDPLLEGQYPARGLYQALAVAAMCVEEQSSMRPVIADVVAALD 299
WA P +R+ +++DP L+GQY + +A C+ + RP + ++V ++
Sbjct: 293 DWARPYLTNKRRLLRVMDPRLQGQYSLTRALKIAVLALDCISIDAKSRPTMNEIVKTME 351
>APKA_ARATH (Q06548) Protein kinase APK1A, chloroplast precursor (EC
2.7.1.-)
Length = 410
Score = 284 bits (726), Expect = 2e-76
Identities = 147/303 (48%), Positives = 202/303 (66%), Gaps = 10/303 (3%)
Query: 10 KIFTFDELAAATKSFRVDCFVGEGGFGKVYKGYIKK---------INQFVAIKQLDPNGL 60
K F+F EL +AT++FR D +GEGGFG V+KG+I + +A+K+L+ +G
Sbjct: 54 KSFSFAELKSATRNFRPDSVLGEGGFGCVFKGWIDEKSLTASRPGTGLVIAVKKLNQDGW 113
Query: 61 QGTREFAVEVLTLSLAEHPNLVKLLGFCAEGEQRLLVYEYMPLGSLENHLHDLPPGKKPL 120
QG +E+ EV L H +LVKL+G+C E E RLLVYE+MP GSLENHL +PL
Sbjct: 114 QGHQEWLAEVNYLGQFSHRHLVKLIGYCLEDEHRLLVYEFMPRGSLENHLFRRGLYFQPL 173
Query: 121 DWNTRMRIAAGVAKGLEYLHDEMKPPVIYRDLKCSNILLGDDYHPKLSDFGLAKVGPIGD 180
W R+++A G AKGL +LH + VIYRD K SNILL +Y+ KLSDFGLAK GPIGD
Sbjct: 174 SWKLRLKVALGAAKGLAFLHSS-ETRVIYRDFKTSNILLDSEYNAKLSDFGLAKDGPIGD 232
Query: 181 MTHVSTRVMGTYGYCAPDYAMTGQLTSKSDIYSLGVALLELITGRKAFDPSKPAKEQNLV 240
+HVSTRVMGT+GY AP+Y TG LT+KSD+YS GV LLEL++GR+A D ++P+ E+NLV
Sbjct: 233 KSHVSTRVMGTHGYAAPEYLATGHLTTKSDVYSFGVVLLELLSGRRAVDKNRPSGERNLV 292
Query: 241 AWAYPLFKEQRKFSKMVDPLLEGQYPARGLYQALAVAAMCVEEQSSMRPVIADVVAALDF 300
WA P +RK +++D L+ QY + ++ C+ + +RP +++VV+ L+
Sbjct: 293 EWAKPYLVNKRKIFRVIDNRLQDQYSMEEACKVATLSLRCLTTEIKLRPNMSEVVSHLEH 352
Query: 301 IAS 303
I S
Sbjct: 353 IQS 355
>CX32_ARATH (P27450) Probable serine/threonine-protein kinase Cx32,
chloroplast precursor (EC 2.7.1.37)
Length = 419
Score = 250 bits (639), Expect = 3e-66
Identities = 128/301 (42%), Positives = 186/301 (61%), Gaps = 13/301 (4%)
Query: 10 KIFTFDELAAATKSFRVDCFVGEGGFGKVYKGYIKKIN---------QFVAIKQLDPNGL 60
K++ F +L ATK+F+ D +G+GGFGKVY+G++ VAIK+L+ +
Sbjct: 72 KVYNFLDLKTATKNFKPDSMLGQGGFGKVYRGWVDATTLAPSRVGSGMIVAIKRLNSESV 131
Query: 61 QGTREFAVEVLTLSLAEHPNLVKLLGFCAEGEQRLLVYEYMPLGSLENHLHDLPPGKKPL 120
QG E+ EV L + H NLVKLLG+C E ++ LLVYE+MP GSLE+HL P
Sbjct: 132 QGFAEWRSEVNFLGMLSHRNLVKLLGYCREDKELLLVYEFMPKGSLESHLFRR---NDPF 188
Query: 121 DWNTRMRIAAGVAKGLEYLHDEMKPPVIYRDLKCSNILLGDDYHPKLSDFGLAKVGPIGD 180
W+ R++I G A+GL +LH ++ VIYRD K SNILL +Y KLSDFGLAK+GP +
Sbjct: 189 PWDLRIKIVIGAARGLAFLHS-LQREVIYRDFKASNILLDSNYDAKLSDFGLAKLGPADE 247
Query: 181 MTHVSTRVMGTYGYCAPDYAMTGQLTSKSDIYSLGVALLELITGRKAFDPSKPAKEQNLV 240
+HV+TR+MGTYGY AP+Y TG L KSD+++ GV LLE++TG A + +P +++LV
Sbjct: 248 KSHVTTRIMGTYGYAAPEYMATGHLYVKSDVFAFGVVLLEIMTGLTAHNTKRPRGQESLV 307
Query: 241 AWAYPLFKEQRKFSKMVDPLLEGQYPARGLYQALAVAAMCVEEQSSMRPVIADVVAALDF 300
W P + + +++D ++GQY + + + C+E RP + +VV L+
Sbjct: 308 DWLRPELSNKHRVKQIMDKGIKGQYTTKVATEMARITLSCIEPDPKNRPHMKEVVEVLEH 367
Query: 301 I 301
I
Sbjct: 368 I 368
>PSKR_DAUCA (Q8LPB4) Phytosulfokine receptor precursor (EC 2.7.1.37)
(Phytosulfokine LRR receptor kinase)
Length = 1021
Score = 222 bits (565), Expect = 1e-57
Identities = 123/289 (42%), Positives = 173/289 (59%), Gaps = 3/289 (1%)
Query: 13 TFDELAAATKSFRVDCFVGEGGFGKVYKGYIKKINQFVAIKQLDPNGLQGTREFAVEVLT 72
+ D++ +T SF +G GGFG VYK + + VAIK+L + Q REF EV T
Sbjct: 732 SLDDILKSTSSFNQANIIGCGGFGLVYKATLPDGTK-VAIKRLSGDTGQMDREFQAEVET 790
Query: 73 LSLAEHPNLVKLLGFCAEGEQRLLVYEYMPLGSLENHLHDLPPGKKPLDWNTRMRIAAGV 132
LS A+HPNLV LLG+C +LL+Y YM GSL+ LH+ G LDW TR+RIA G
Sbjct: 791 LSRAQHPNLVHLLGYCNYKNDKLLIYSYMDNGSLDYWLHEKVDGPPSLDWKTRLRIARGA 850
Query: 133 AKGLEYLHDEMKPPVIYRDLKCSNILLGDDYHPKLSDFGLAKVGPIGDMTHVSTRVMGTY 192
A+GL YLH +P +++RD+K SNILL D + L+DFGLA++ D THV+T ++GT
Sbjct: 851 AEGLAYLHQSCEPHILHRDIKSSNILLSDTFVAHLADFGLARLILPYD-THVTTDLVGTL 909
Query: 193 GYCAPDYAMTGQLTSKSDIYSLGVALLELITGRKAFDPSKPAKEQNLVAWAYPLFKEQRK 252
GY P+Y T K D+YS GV LLEL+TGR+ D KP ++L++W + E+R+
Sbjct: 910 GYIPPEYGQASVATYKGDVYSFGVVLLELLTGRRPMDVCKPRGSRDLISWVLQMKTEKRE 969
Query: 253 FSKMVDPLLEGQYPARGLYQALAVAAMCVEEQSSMRPVIADVVAALDFI 301
S++ DP + + A + L +A C+ E RP +V+ L+ I
Sbjct: 970 -SEIFDPFIYDKDHAEEMLLVLEIACRCLGENPKTRPTTQQLVSWLENI 1017
>BAK1_ARATH (Q94F62) BRASSINOSTEROID INSENSITIVE 1-associated
receptor kinase 1 precursor (EC 2.7.1.37)
(BRI1-associated receptor kinase 1) (Somatic
embryogenesis receptor-like kinase 3)
Length = 615
Score = 222 bits (565), Expect = 1e-57
Identities = 125/293 (42%), Positives = 178/293 (60%), Gaps = 6/293 (2%)
Query: 10 KIFTFDELAAATKSFRVDCFVGEGGFGKVYKGYIKKINQFVAIKQLDPNGLQGTR-EFAV 68
K F+ EL A+ +F +G GGFGKVYKG + VA+K+L QG +F
Sbjct: 275 KRFSLRELQVASDNFSNKNILGRGGFGKVYKGRLAD-GTLVAVKRLKEERTQGGELQFQT 333
Query: 69 EVLTLSLAEHPNLVKLLGFCAEGEQRLLVYEYMPLGSLENHLHDLPPGKKPLDWNTRMRI 128
EV +S+A H NL++L GFC +RLLVY YM GS+ + L + P + PLDW R RI
Sbjct: 334 EVEMISMAVHRNLLRLRGFCMTPTERLLVYPYMANGSVASCLRERPESQPPLDWPKRQRI 393
Query: 129 AAGVAKGLEYLHDEMKPPVIYRDLKCSNILLGDDYHPKLSDFGLAKVGPIGDMTHVSTRV 188
A G A+GL YLHD P +I+RD+K +NILL +++ + DFGLAK+ D THV+T V
Sbjct: 394 ALGSARGLAYLHDHCDPKIIHRDVKAANILLDEEFEAVVGDFGLAKLMDYKD-THVTTAV 452
Query: 189 MGTYGYCAPDYAMTGQLTSKSDIYSLGVALLELITGRKAFDPSKPAKEQN--LVAWAYPL 246
GT G+ AP+Y TG+ + K+D++ GV LLELITG++AFD ++ A + + L+ W L
Sbjct: 453 RGTIGHIAPEYLSTGKSSEKTDVFGYGVMLLELITGQRAFDLARLANDDDVMLLDWVKGL 512
Query: 247 FKEQRKFSKMVDPLLEGQYPARGLYQALAVAAMCVEEQSSMRPVIADVVAALD 299
KE +K +VD L+G Y + Q + VA +C + RP +++VV L+
Sbjct: 513 LKE-KKLEALVDVDLQGNYKDEEVEQLIQVALLCTQSSPMERPKMSEVVRMLE 564
>PSKR_ARATH (Q9ZVR7) Putative phytosulfokine receptor precursor (EC
2.7.1.37) (Phytosulfokine LRR receptor kinase)
Length = 1008
Score = 221 bits (563), Expect = 2e-57
Identities = 122/290 (42%), Positives = 176/290 (60%), Gaps = 3/290 (1%)
Query: 10 KIFTFDELAAATKSFRVDCFVGEGGFGKVYKGYIKKINQFVAIKQLDPNGLQGTREFAVE 69
K ++D+L +T SF +G GGFG VYK + + VAIK+L + Q REF E
Sbjct: 720 KELSYDDLLDSTNSFDQANIIGCGGFGMVYKATLPD-GKKVAIKKLSGDCGQIEREFEAE 778
Query: 70 VLTLSLAEHPNLVKLLGFCAEGEQRLLVYEYMPLGSLENHLHDLPPGKKPLDWNTRMRIA 129
V TLS A+HPNLV L GFC RLL+Y YM GSL+ LH+ G L W TR+RIA
Sbjct: 779 VETLSRAQHPNLVLLRGFCFYKNDRLLIYSYMENGSLDYWLHERNDGPALLKWKTRLRIA 838
Query: 130 AGVAKGLEYLHDEMKPPVIYRDLKCSNILLGDDYHPKLSDFGLAKVGPIGDMTHVSTRVM 189
G AKGL YLH+ P +++RD+K SNILL ++++ L+DFGLA++ + THVST ++
Sbjct: 839 QGAAKGLLYLHEGCDPHILHRDIKSSNILLDENFNSHLADFGLARLMSPYE-THVSTDLV 897
Query: 190 GTYGYCAPDYAMTGQLTSKSDIYSLGVALLELITGRKAFDPSKPAKEQNLVAWAYPLFKE 249
GT GY P+Y T K D+YS GV LLEL+T ++ D KP ++L++W + K
Sbjct: 898 GTLGYIPPEYGQASVATYKGDVYSFGVVLLELLTDKRPVDMCKPKGCRDLISWVVKM-KH 956
Query: 250 QRKFSKMVDPLLEGQYPARGLYQALAVAAMCVEEQSSMRPVIADVVAALD 299
+ + S++ DPL+ + + +++ L +A +C+ E RP +V+ LD
Sbjct: 957 ESRASEVFDPLIYSKENDKEMFRVLEIACLCLSENPKQRPTTQQLVSWLD 1006
>EXS_ARATH (Q9LYN8) Leucine-rich repeat receptor protein kinase EXS
precursor (EC 2.7.1.37) (Extra sporogenous cells protein)
(EXCESS MICROSPOROCYTES1 protein)
Length = 1192
Score = 219 bits (558), Expect = 7e-57
Identities = 120/284 (42%), Positives = 174/284 (61%), Gaps = 4/284 (1%)
Query: 16 ELAAATKSFRVDCFVGEGGFGKVYKGYIKKINQFVAIKQLDPNGLQGTREFAVEVLTLSL 75
++ AT F +G+GGFG VYK + + VA+K+L QG REF E+ TL
Sbjct: 909 DIVEATDHFSKKNIIGDGGFGTVYKACLPG-EKTVAVKKLSEAKTQGNREFMAEMETLGK 967
Query: 76 AEHPNLVKLLGFCAEGEQRLLVYEYMPLGSLENHLHDLPPGKKPLDWNTRMRIAAGVAKG 135
+HPNLV LLG+C+ E++LLVYEYM GSL++ L + + LDW+ R++IA G A+G
Sbjct: 968 VKHPNLVSLLGYCSFSEEKLLVYEYMVNGSLDHWLRNQTGMLEVLDWSKRLKIAVGAARG 1027
Query: 136 LEYLHDEMKPPVIYRDLKCSNILLGDDYHPKLSDFGLAKVGPIGDMTHVSTRVMGTYGYC 195
L +LH P +I+RD+K SNILL D+ PK++DFGLA++ + +HVST + GT+GY
Sbjct: 1028 LAFLHHGFIPHIIHRDIKASNILLDGDFEPKVADFGLARLISACE-SHVSTVIAGTFGYI 1086
Query: 196 APDYAMTGQLTSKSDIYSLGVALLELITGRKAFDPS-KPAKEQNLVAWAYPLFKEQRKFS 254
P+Y + + T+K D+YS GV LLEL+TG++ P K ++ NLV WA Q K
Sbjct: 1087 PPEYGQSARATTKGDVYSFGVILLELVTGKEPTGPDFKESEGGNLVGWAIQKI-NQGKAV 1145
Query: 255 KMVDPLLEGQYPARGLYQALAVAAMCVEEQSSMRPVIADVVAAL 298
++DPLL + L +A +C+ E + RP + DV+ AL
Sbjct: 1146 DVIDPLLVSVALKNSQLRLLQIAMLCLAETPAKRPNMLDVLKAL 1189
>TMK1_ARATH (P43298) Putative receptor protein kinase TMK1 precursor
(EC 2.7.1.-)
Length = 942
Score = 214 bits (546), Expect = 2e-55
Identities = 118/294 (40%), Positives = 179/294 (60%), Gaps = 9/294 (3%)
Query: 11 IFTFDELAAATKSFRVDCFVGEGGFGKVYKGYIKKINQFVAIKQLDPNGL---QGTREFA 67
+ + L + T +F D +G GGFG VYKG + + +A+K+++ NG+ +G EF
Sbjct: 575 LISIQVLRSVTNNFSSDNILGSGGFGVVYKGELHDGTK-IAVKRME-NGVIAGKGFAEFK 632
Query: 68 VEVLTLSLAEHPNLVKLLGFCAEGEQRLLVYEYMPLGSLENHLHDLPP-GKKPLDWNTRM 126
E+ L+ H +LV LLG+C +G ++LLVYEYMP G+L HL + G KPL W R+
Sbjct: 633 SEIAVLTKVRHRHLVTLLGYCLDGNEKLLVYEYMPQGTLSRHLFEWSEEGLKPLLWKQRL 692
Query: 127 RIAAGVAKGLEYLHDEMKPPVIYRDLKCSNILLGDDYHPKLSDFGLAKVGPIGDMTHVST 186
+A VA+G+EYLH I+RDLK SNILLGDD K++DFGL ++ P G + + T
Sbjct: 693 TLALDVARGVEYLHGLAHQSFIHRDLKPSNILLGDDMRAKVADFGLVRLAPEGKGS-IET 751
Query: 187 RVMGTYGYCAPDYAMTGQLTSKSDIYSLGVALLELITGRKAFDPSKPAKEQNLVAWAYPL 246
R+ GT+GY AP+YA+TG++T+K D+YS GV L+ELITGRK+ D S+P + +LV+W +
Sbjct: 752 RIAGTFGYLAPEYAVTGRVTTKVDVYSFGVILMELITGRKSLDESQPEESIHLVSWFKRM 811
Query: 247 F-KEQRKFSKMVDPLLE-GQYPARGLYQALAVAAMCVEEQSSMRPVIADVVAAL 298
+ ++ F K +D ++ + ++ +A C + RP + V L
Sbjct: 812 YINKEASFKKAIDTTIDLDEETLASVHTVAELAGHCCAREPYQRPDMGHAVNIL 865
>CRI4_MAIZE (O24585) Putative receptor protein kinase CRINKLY4
precursor (EC 2.7.1.-)
Length = 901
Score = 209 bits (533), Expect = 6e-54
Identities = 122/294 (41%), Positives = 176/294 (59%), Gaps = 7/294 (2%)
Query: 9 AKIFTFDELAAATKSFRVDCFVGEGGFGKVYKGYIKKINQFVAIKQL--DPNGLQGTREF 66
A+ F+++EL AT F D VG+G F V+KG ++ VA+K+ + + ++EF
Sbjct: 490 AQEFSYEELEQATGGFSEDSQVGKGSFSCVFKGILRD-GTVVAVKRAIKASDVKKSSKEF 548
Query: 67 AVEVLTLSLAEHPNLVKLLGFCAEGEQRLLVYEYMPLGSLENHLHDLPPG-KKPLDWNTR 125
E+ LS H +L+ LLG+C +G +RLLVYE+M GSL HLH P KK L+W R
Sbjct: 549 HNELDLLSRLNHAHLLNLLGYCEDGSERLLVYEFMAHGSLYQHLHGKDPNLKKRLNWARR 608
Query: 126 MRIAAGVAKGLEYLHDEMKPPVIYRDLKCSNILLGDDYHPKLSDFGLAKVGPIGDMTHVS 185
+ IA A+G+EYLH PPVI+RD+K SNIL+ +D++ +++DFGL+ +GP T +S
Sbjct: 609 VTIAVQAARGIEYLHGYACPPVIHRDIKSSNILIDEDHNARVADFGLSILGPADSGTPLS 668
Query: 186 TRVMGTYGYCAPDYAMTGQLTSKSDIYSLGVALLELITGRKAFDPSKPAKEQNLVAWAYP 245
GT GY P+Y LT+KSD+YS GV LLE+++GRKA D +E N+V WA P
Sbjct: 669 ELPAGTLGYLDPEYYRLHYLTTKSDVYSFGVVLLEILSGRKAID--MQFEEGNIVEWAVP 726
Query: 246 LFKEQRKFSKMVDPLLEGQYPARGLYQALAVAAMCVEEQSSMRPVIADVVAALD 299
L K F+ ++DP+L L + +VA CV + RP + V AL+
Sbjct: 727 LIKAGDIFA-ILDPVLSPPSDLEALKKIASVACKCVRMRGKDRPSMDKVTTALE 779
>BRI1_ARATH (O22476) BRASSINOSTEROID INSENSITIVE 1 precursor (EC
2.7.1.37) (AtBRI1) (Brassinosteroid LRR receptor kinase)
Length = 1196
Score = 209 bits (533), Expect = 6e-54
Identities = 124/294 (42%), Positives = 175/294 (59%), Gaps = 8/294 (2%)
Query: 6 EKVAKIFTFDELAAATKSFRVDCFVGEGGFGKVYKGYIKKINQFVAIKQLDPNGLQGTRE 65
EK + TF +L AT F D +G GGFG VYK +K VAIK+L QG RE
Sbjct: 865 EKPLRKLTFADLLQATNGFHNDSLIGSGGFGDVYKAILKD-GSAVAIKKLIHVSGQGDRE 923
Query: 66 FAVEVLTLSLAEHPNLVKLLGFCAEGEQRLLVYEYMPLGSLENHLHDLPPGKKPLDWNTR 125
F E+ T+ +H NLV LLG+C G++RLLVYE+M GSLE+ LHD L+W+TR
Sbjct: 924 FMAEMETIGKIKHRNLVPLLGYCKVGDERLLVYEFMKYGSLEDVLHDPKKAGVKLNWSTR 983
Query: 126 MRIAAGVAKGLEYLHDEMKPPVIYRDLKCSNILLGDDYHPKLSDFGLAKVGPIGDMTHVS 185
+IA G A+GL +LH P +I+RD+K SN+LL ++ ++SDFG+A++ D TH+S
Sbjct: 984 RKIAIGSARGLAFLHHNCSPHIIHRDMKSSNVLLDENLEARVSDFGMARLMSAMD-THLS 1042
Query: 186 TRVM-GTYGYCAPDYAMTGQLTSKSDIYSLGVALLELITGRKAFDPSKPAKEQNLVAWAY 244
+ GT GY P+Y + + ++K D+YS GV LLEL+TG++ D S + NLV W
Sbjct: 1043 VSTLAGTPGYVPPEYYQSFRCSTKGDVYSYGVVLLELLTGKRPTD-SPDFGDNNLVGWVK 1101
Query: 245 PLFKEQRKFSKMVDPLLEGQYPAR--GLYQALAVAAMCVEEQSSMRPVIADVVA 296
K + S + DP L + PA L Q L VA C+++++ RP + V+A
Sbjct: 1102 QHAK--LRISDVFDPELMKEDPALEIELLQHLKVAVACLDDRAWRRPTMVQVMA 1153
>BRI1_LYCPE (Q8L899) Systemin receptor SR160 precursor (EC 2.7.1.37)
(Brassinosteroid LRR receptor kinase)
Length = 1207
Score = 201 bits (510), Expect = 3e-51
Identities = 123/294 (41%), Positives = 169/294 (56%), Gaps = 8/294 (2%)
Query: 6 EKVAKIFTFDELAAATKSFRVDCFVGEGGFGKVYKGYIKKINQFVAIKQLDPNGLQGTRE 65
EK + TF +L AT F D VG GGFG VYK +K VAIK+L QG RE
Sbjct: 870 EKPLRKLTFADLLEATNGFHNDSLVGSGGFGDVYKAQLKD-GSVVAIKKLIHVSGQGDRE 928
Query: 66 FAVEVLTLSLAEHPNLVKLLGFCAEGEQRLLVYEYMPLGSLENHLHDLPPGKKPLDWNTR 125
F E+ T+ +H NLV LLG+C GE+RLLVYEYM GSLE+ LHD L+W R
Sbjct: 929 FTAEMETIGKIKHRNLVPLLGYCKVGEERLLVYEYMKYGSLEDVLHDRKKTGIKLNWPAR 988
Query: 126 MRIAAGVAKGLEYLHDEMKPPVIYRDLKCSNILLGDDYHPKLSDFGLAKVGPIGDMTHVS 185
+IA G A+GL +LH P +I+RD+K SN+LL ++ ++SDFG+A++ D TH+S
Sbjct: 989 RKIAIGAARGLAFLHHNCIPHIIHRDMKSSNVLLDENLEARVSDFGMARLMSAMD-THLS 1047
Query: 186 TRVM-GTYGYCAPDYAMTGQLTSKSDIYSLGVALLELITGRKAFDPSKPAKEQNLVAWAY 244
+ GT GY P+Y + + ++K D+YS GV LLEL+TG++ D S + NLV W
Sbjct: 1048 VSTLAGTPGYVPPEYYQSFRCSTKGDVYSYGVVLLELLTGKQPTD-SADFGDNNLVGWV- 1105
Query: 245 PLFKEQRKFSKMVDP--LLEGQYPARGLYQALAVAAMCVEEQSSMRPVIADVVA 296
+ K + + D L E L Q L VA C++++ RP + V+A
Sbjct: 1106 -KLHAKGKITDVFDRELLKEDASIEIELLQHLKVACACLDDRHWKRPTMIQVMA 1158
>BRI1_LYCES (Q8GUQ5) Brassinosteroid LRR receptor kinase precursor (EC
2.7.1.37) (tBRI1) (Altered brassinolide sensitivity 1)
(Systemin receptor SR160)
Length = 1207
Score = 200 bits (508), Expect = 4e-51
Identities = 123/294 (41%), Positives = 169/294 (56%), Gaps = 8/294 (2%)
Query: 6 EKVAKIFTFDELAAATKSFRVDCFVGEGGFGKVYKGYIKKINQFVAIKQLDPNGLQGTRE 65
EK + TF +L AT F D VG GGFG VYK +K VAIK+L QG RE
Sbjct: 870 EKPLRKLTFADLLEATNGFHNDSLVGSGGFGDVYKAQLKD-GSVVAIKKLIHVSGQGDRE 928
Query: 66 FAVEVLTLSLAEHPNLVKLLGFCAEGEQRLLVYEYMPLGSLENHLHDLPPGKKPLDWNTR 125
F E+ T+ +H NLV LLG+C GE+RLLVYEYM GSLE+ LHD L+W R
Sbjct: 929 FTAEMETIGKIKHRNLVPLLGYCKVGEERLLVYEYMKYGSLEDVLHDRKKIGIKLNWPAR 988
Query: 126 MRIAAGVAKGLEYLHDEMKPPVIYRDLKCSNILLGDDYHPKLSDFGLAKVGPIGDMTHVS 185
+IA G A+GL +LH P +I+RD+K SN+LL ++ ++SDFG+A++ D TH+S
Sbjct: 989 RKIAIGAARGLAFLHHNCIPHIIHRDMKSSNVLLDENLEARVSDFGMARLMSAMD-THLS 1047
Query: 186 TRVM-GTYGYCAPDYAMTGQLTSKSDIYSLGVALLELITGRKAFDPSKPAKEQNLVAWAY 244
+ GT GY P+Y + + ++K D+YS GV LLEL+TG++ D S + NLV W
Sbjct: 1048 VSTLAGTPGYVPPEYYQSFRCSTKGDVYSYGVVLLELLTGKQPTD-SADFGDNNLVGWV- 1105
Query: 245 PLFKEQRKFSKMVDP--LLEGQYPARGLYQALAVAAMCVEEQSSMRPVIADVVA 296
+ K + + D L E L Q L VA C++++ RP + V+A
Sbjct: 1106 -KLHAKGKITDVFDRELLKEDASIEIELLQHLKVACACLDDRHWKRPTMIQVMA 1158
>SRK6_BRAOL (Q09092) Putative serine/threonine-protein kinase
receptor precursor (EC 2.7.1.37) (S-receptor kinase)
(SRK)
Length = 849
Score = 190 bits (483), Expect = 3e-48
Identities = 106/298 (35%), Positives = 180/298 (59%), Gaps = 10/298 (3%)
Query: 5 DEKVAKIFTFDELAAATKSFRVDCFVGEGGFGKVYKGYIKKINQFVAIKQLDPNGLQGTR 64
+E + + + AT++F +G+GGFG VYKG + + +A+K+L +QGT
Sbjct: 509 EELELPLIEMETVVKATENFSSCNKLGQGGFGIVYKGRLLDGKE-IAVKRLSKTSVQGTD 567
Query: 65 EFAVEVLTLSLAEHPNLVKLLGFCAEGEQRLLVYEYMPLGSLENHLHDLPPGKKPLDWNT 124
EF EV ++ +H NLV++LG C EG++++L+YEY+ SL+++L K L+WN
Sbjct: 568 EFMNEVTLIARLQHINLVQVLGCCIEGDEKMLIYEYLENLSLDSYLFGKTRRSK-LNWNE 626
Query: 125 RMRIAAGVAKGLEYLHDEMKPPVIYRDLKCSNILLGDDYHPKLSDFGLAKVGPIGDMTHV 184
R I GVA+GL YLH + + +I+RDLK SNILL + PK+SDFG+A++ +
Sbjct: 627 RFDITNGVARGLLYLHQDSRFRIIHRDLKVSNILLDKNMIPKISDFGMARIFERDETEAN 686
Query: 185 STRVMGTYGYCAPDYAMTGQLTSKSDIYSLGVALLELITGRKAFDPSKPAKEQNLVAWAY 244
+ +V+GTYGY +P+YAM G + KSD++S GV +LE+++G+K E +L+++ +
Sbjct: 687 TMKVVGTYGYMSPEYAMYGIFSEKSDVFSFGVIVLEIVSGKKNRGFYNLDYENDLLSYVW 746
Query: 245 PLFKEQRKFSKMVDPLLEGQ-------YPARGLYQALAVAAMCVEEQSSMRPVIADVV 295
+KE R ++VDP++ + + + + + + +CV+E + RP ++ VV
Sbjct: 747 SRWKEGRAL-EIVDPVIVDSLSSQPSIFQPQEVLKCIQIGLLCVQELAEHRPAMSSVV 803
>SIRK_ARATH (O64483) Senescence-induced receptor-like
serine/threonine kinase precursor (FLG22-induced
receptor-like kinase 1)
Length = 876
Score = 182 bits (463), Expect = 7e-46
Identities = 108/291 (37%), Positives = 159/291 (54%), Gaps = 12/291 (4%)
Query: 10 KIFTFDELAAATKSFRVDCFVGEGGFGKVYKGYIKKINQFVAIKQLDPNGLQGTREFAVE 69
+ F + E+ T +F + +G+GGFGKVY G I + VA+K L QG +EF E
Sbjct: 562 RYFKYSEVVNITNNF--ERVIGKGGFGKVYHGVIN--GEQVAVKVLSEESAQGYKEFRAE 617
Query: 70 VLTLSLAEHPNLVKLLGFCAEGEQRLLVYEYMPLGSLENHLHDLPPGKKP--LDWNTRMR 127
V L H NL L+G+C E +L+YEYM +L ++L GK+ L W R++
Sbjct: 618 VDLLMRVHHTNLTSLVGYCNEINHMVLIYEYMANENLGDYL----AGKRSFILSWEERLK 673
Query: 128 IAAGVAKGLEYLHDEMKPPVIYRDLKCSNILLGDDYHPKLSDFGLAKVGPIGDMTHVSTR 187
I+ A+GLEYLH+ KPP+++RD+K +NILL + K++DFGL++ + +ST
Sbjct: 674 ISLDAAQGLEYLHNGCKPPIVHRDVKPTNILLNEKLQAKMADFGLSRSFSVEGSGQISTV 733
Query: 188 VMGTYGYCAPDYAMTGQLTSKSDIYSLGVALLELITGRKAFDPSKPAKEQNLVAWAYPLF 247
V G+ GY P+Y T Q+ KSD+YSLGV LLE+ITG+ A SK K ++
Sbjct: 734 VAGSIGYLDPEYYSTRQMNEKSDVYSLGVVLLEVITGQPAIASSKTEKVH--ISDHVRSI 791
Query: 248 KEQRKFSKMVDPLLEGQYPARGLYQALAVAAMCVEEQSSMRPVIADVVAAL 298
+VD L +Y ++ +A C E S+ RP ++ VV L
Sbjct: 792 LANGDIRGIVDQRLRERYDVGSAWKMSEIALACTEHTSAQRPTMSQVVMEL 842
>RPK1_IPONI (P93194) Receptor-like protein kinase precursor (EC
2.7.1.37)
Length = 1109
Score = 182 bits (461), Expect = 1e-45
Identities = 111/301 (36%), Positives = 165/301 (53%), Gaps = 8/301 (2%)
Query: 15 DELAAATKSFRVDCFVGEGGFGKVYKGYIKKINQFVAIKQLDPNGLQ-GTREFAVEVLTL 73
+++ AT++ +G+G G +YK + ++ A+K+L G++ G+ E+ T+
Sbjct: 807 NKVLEATENLNDKYVIGKGAHGTIYKATLSP-DKVYAVKKLVFTGIKNGSVSMVREIETI 865
Query: 74 SLAEHPNLVKLLGFCAEGEQRLLVYEYMPLGSLENHLHDLPPGKKPLDWNTRMRIAAGVA 133
H NL+KL F E L++Y YM GSL + LH+ P KPLDW+TR IA G A
Sbjct: 866 GKVRHRNLIKLEEFWLRKEYGLILYTYMENGSLHDILHETNP-PKPLDWSTRHNIAVGTA 924
Query: 134 KGLEYLHDEMKPPVIYRDLKCSNILLGDDYHPKLSDFGLAKVGPIGDMTHVSTRVMGTYG 193
GL YLH + P +++RD+K NILL D P +SDFG+AK+ + S V GT G
Sbjct: 925 HGLAYLHFDCDPAIVHRDIKPMNILLDSDLEPHISDFGIAKLLDQSATSIPSNTVQGTIG 984
Query: 194 YCAPDYAMTGQLTSKSDIYSLGVALLELITGRKAFDPSKPAKEQNLVAWAYPLFKEQRKF 253
Y AP+ A T + +SD+YS GV LLELIT +KA DPS E ++V W ++ + +
Sbjct: 985 YMAPENAFTTVKSRESDVYSYGVVLLELITRKKALDPSFNG-ETDIVGWVRSVWTQTGEI 1043
Query: 254 SKMVDPLLEGQY----PARGLYQALAVAAMCVEEQSSMRPVIADVVAALDFIASHKYDPQ 309
K+VDP L + + +AL++A C E++ RP + DVV L + Y
Sbjct: 1044 QKIVDPSLLDELIDSSVMEQVTEALSLALRCAEKEVDKRPTMRDVVKQLTRWSIRSYSSS 1103
Query: 310 V 310
V
Sbjct: 1104 V 1104
>CLV1_ARATH (Q9SYQ8) Receptor protein kinase CLAVATA1 precursor (EC
2.7.1.-)
Length = 980
Score = 181 bits (460), Expect = 2e-45
Identities = 111/276 (40%), Positives = 157/276 (56%), Gaps = 13/276 (4%)
Query: 30 VGEGGFGKVYKGYIKKINQFVAIKQLDPNGL-QGTREFAVEVLTLSLAEHPNLVKLLGFC 88
+G+GG G VY+G + N VAIK+L G + F E+ TL H ++V+LLG+
Sbjct: 698 IGKGGAGIVYRGSMPN-NVDVAIKRLVGRGTGRSDHGFTAEIQTLGRIRHRHIVRLLGYV 756
Query: 89 AEGEQRLLVYEYMPLGSLENHLHDLPPGKKPLDWNTRMRIAAGVAKGLEYLHDEMKPPVI 148
A + LL+YEYMP GSL LH G L W TR R+A AKGL YLH + P ++
Sbjct: 757 ANKDTNLLLYEYMPNGSLGELLHGSKGGH--LQWETRHRVAVEAAKGLCYLHHDCSPLIL 814
Query: 149 YRDLKCSNILLGDDYHPKLSDFGLAKVGPIGDMTHVSTRVMGTYGYCAPDYAMTGQLTSK 208
+RD+K +NILL D+ ++DFGLAK G + + + G+YGY AP+YA T ++ K
Sbjct: 815 HRDVKSNNILLDSDFEAHVADFGLAKFLVDGAASECMSSIAGSYGYIAPEYAYTLKVDEK 874
Query: 209 SDIYSLGVALLELITGRKAFDPSKPAKEQNLVAWAYPLFKEQRKFS------KMVDPLLE 262
SD+YS GV LLELI G+K + + ++V W +E + S +VDP L
Sbjct: 875 SDVYSFGVVLLELIAGKKPV--GEFGEGVDIVRWVRNTEEEITQPSDAAIVVAIVDPRLT 932
Query: 263 GQYPARGLYQALAVAAMCVEEQSSMRPVIADVVAAL 298
G YP + +A MCVEE+++ RP + +VV L
Sbjct: 933 G-YPLTSVIHVFKIAMMCVEEEAAARPTMREVVHML 967
>IRA4_MOUSE (Q8R4K2) Interleukin-1 receptor-associated kinase-4 (EC
2.7.1.37) (IRAK-4)
Length = 459
Score = 173 bits (439), Expect = 4e-43
Identities = 115/295 (38%), Positives = 165/295 (54%), Gaps = 24/295 (8%)
Query: 12 FTFDELAAATKSFRVDCF------VGEGGFGKVYKGYIKKINQFVAIKQLDPNGLQGTRE 65
F+F EL + T +F +GEGGFG VYKG + N VA+K+L T E
Sbjct: 168 FSFHELKSITNNFDEQPASAGGNRMGEGGFGVVYKGCVN--NTIVAVKKLGAMVEISTEE 225
Query: 66 ----FAVEVLTLSLAEHPNLVKLLGFCAEGEQRLLVYEYMPLGSLENHLHDLPPGKKPLD 121
F E+ ++ +H NLV+LLGF ++ + LVY YMP GSL + L L G PL
Sbjct: 226 LKQQFDQEIKVMATCQHENLVELLGFSSDSDNLCLVYAYMPNGSLLDRLSCLD-GTPPLS 284
Query: 122 WNTRMRIAAGVAKGLEYLHDEMKPPVIYRDLKCSNILLGDDYHPKLSDFGLAKVGPIGDM 181
W+TR ++A G A G+ +LH+ I+RD+K +NILL D+ K+SDFGLA+
Sbjct: 285 WHTRCKVAQGTANGIRFLHENHH---IHRDIKSANILLDKDFTAKISDFGLARASARLAQ 341
Query: 182 THVSTRVMGTYGYCAPDYAMTGQLTSKSDIYSLGVALLELITGRKAFDPSKPAKEQNLVA 241
T +++R++GT Y AP+ A+ G++T KSDIYS GV LLELITG A D ++ + Q L+
Sbjct: 342 TVMTSRIVGTTAYMAPE-ALRGEITPKSDIYSFGVVLLELITGLAAVDENR--EPQLLLD 398
Query: 242 WAYPLFKEQRKFSKMVDPLLEGQYPA--RGLYQALAVAAMCVEEQSSMRPVIADV 294
+ E++ D + PA +Y A A+ C+ E+ + RP IA V
Sbjct: 399 IKEEIEDEEKTIEDYTDEKMSDADPASVEAMYSA---ASQCLHEKKNRRPDIAKV 450
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.320 0.138 0.410
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 38,003,437
Number of Sequences: 164201
Number of extensions: 1608060
Number of successful extensions: 7155
Number of sequences better than 10.0: 1591
Number of HSP's better than 10.0 without gapping: 1073
Number of HSP's successfully gapped in prelim test: 518
Number of HSP's that attempted gapping in prelim test: 3473
Number of HSP's gapped (non-prelim): 1782
length of query: 317
length of database: 59,974,054
effective HSP length: 110
effective length of query: 207
effective length of database: 41,911,944
effective search space: 8675772408
effective search space used: 8675772408
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 66 (30.0 bits)
Medicago: description of AC149490.2


