

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC149490.13 + phase: 0
(227 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
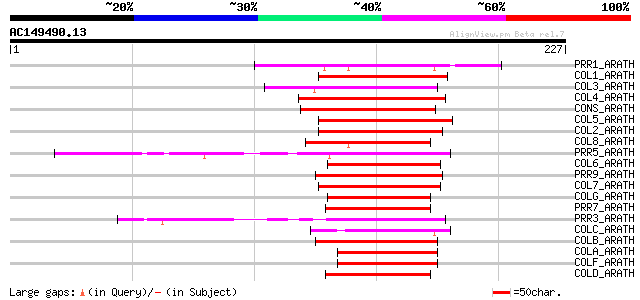
Score E
Sequences producing significant alignments: (bits) Value
PRR1_ARATH (Q9LKL2) Two-component response regulator-like APRR1 ... 62 1e-09
COL1_ARATH (O50055) Zinc finger protein CONSTANS-LIKE 1 60 4e-09
COL3_ARATH (Q9SK53) Zinc finger protein CONSTANS-LIKE 3 60 6e-09
COL4_ARATH (Q940T9) Zinc finger protein CONSTANS-LIKE 4 59 9e-09
CONS_ARATH (Q39057) Zinc finger protein CONSTANS 54 2e-07
COL5_ARATH (Q9FHH8) Zinc finger protein CONSTANS-LIKE 5 54 2e-07
COL2_ARATH (Q96502) Zinc finger protein CONSTANS-LIKE 2 54 3e-07
COL8_ARATH (Q9M9B3) Putative zinc finger protein CONSTANS-LIKE 8 54 4e-07
PRR5_ARATH (Q6LA42) Two-component response regulator-like APRR5 ... 51 2e-06
COL6_ARATH (Q8LG76) Zinc finger protein CONSTANS-LIKE 6 51 3e-06
PRR9_ARATH (Q8L500) Two-component response regulator-like APRR9 ... 50 3e-06
COL7_ARATH (Q9C9A9) Putative zinc finger protein CONSTANS-LIKE 7 50 3e-06
COLG_ARATH (Q8RWD0) Zinc finger protein CONSTANS-LIKE 16 49 7e-06
PRR7_ARATH (Q93WK5) Two-component response regulator-like APRR7 ... 49 1e-05
PRR3_ARATH (Q9LVG4) Two-component response regulator-like APRR3 ... 49 1e-05
COLC_ARATH (Q9LJ44) Putative zinc finger protein CONSTANS-LIKE 12 47 5e-05
COLB_ARATH (O23379) Putative zinc finger protein CONSTANS-LIKE 11 46 8e-05
COLA_ARATH (Q9LUA9) Zinc finger protein CONSTANS-LIKE 10 44 2e-04
COLF_ARATH (Q9C7E8) Zinc finger protein CONSTANS-LIKE 15 43 5e-04
COLD_ARATH (O82256) Putative zinc finger protein CONSTANS-LIKE 13 43 5e-04
>PRR1_ARATH (Q9LKL2) Two-component response regulator-like APRR1
(Pseudo-response regulator 1) (Timing of CAB expression
1) (ABI3-interacting protein 1)
Length = 618
Score = 62.0 bits (149), Expect = 1e-09
Identities = 40/122 (32%), Positives = 66/122 (53%), Gaps = 23/122 (18%)
Query: 101 HHHHVDSPLSSESSMIIEGMSRVCPYS------PEEKKVRIER----------YRIKRNQ 144
+HH +++ L + G + +S P +VR+ + +R KRNQ
Sbjct: 487 YHHPMNTSLQHSQMSLQNGQMSMVHHSWSPAGNPPSNEVRVNKLDRREEALLKFRRKRNQ 546
Query: 145 RNFNKKIKYVCRKTLADRRPRIRGRFAR-----NDEIDKKPIVEWSHIGGGEEEDEEDEK 199
R F+KKI+YV RK LA+RRPR++G+F R N +++ +P + + EEE+EE+E+
Sbjct: 547 RCFDKKIRYVNRKRLAERRPRVKGQFVRKMNGVNVDLNGQP--DSADYDDEEEEEEEEEE 604
Query: 200 WN 201
N
Sbjct: 605 EN 606
>COL1_ARATH (O50055) Zinc finger protein CONSTANS-LIKE 1
Length = 355
Score = 60.1 bits (144), Expect = 4e-09
Identities = 24/53 (45%), Positives = 38/53 (71%)
Query: 127 SPEEKKVRIERYRIKRNQRNFNKKIKYVCRKTLADRRPRIRGRFARNDEIDKK 179
SP +++ R+ RYR K+ R F K I+Y RK A++RPRI+GRFA+ ++D++
Sbjct: 282 SPRDREARVLRYREKKKMRKFEKTIRYASRKAYAEKRPRIKGRFAKKKDVDEE 334
>COL3_ARATH (Q9SK53) Zinc finger protein CONSTANS-LIKE 3
Length = 294
Score = 59.7 bits (143), Expect = 6e-09
Identities = 31/75 (41%), Positives = 42/75 (55%), Gaps = 4/75 (5%)
Query: 105 VDSPLSSESSMIIEGMSRV----CPYSPEEKKVRIERYRIKRNQRNFNKKIKYVCRKTLA 160
+D PL ES + M+ SP E++ R+ RYR KR R F K I+Y RK A
Sbjct: 199 IDVPLVPESGGVTAEMTNTETPAVQLSPAEREARVLRYREKRKNRKFEKTIRYASRKAYA 258
Query: 161 DRRPRIRGRFARNDE 175
+ RPRI+GRFA+ +
Sbjct: 259 EMRPRIKGRFAKRTD 273
>COL4_ARATH (Q940T9) Zinc finger protein CONSTANS-LIKE 4
Length = 362
Score = 58.9 bits (141), Expect = 9e-09
Identities = 26/60 (43%), Positives = 37/60 (61%)
Query: 119 GMSRVCPYSPEEKKVRIERYRIKRNQRNFNKKIKYVCRKTLADRRPRIRGRFARNDEIDK 178
G R P + E++ R+ RYR KR R F K I+Y RK A+ RPRI+GRFA+ + ++
Sbjct: 283 GTQRAVPLTSAEREARVMRYREKRKNRKFEKTIRYASRKAYAEMRPRIKGRFAKRTDTNE 342
>CONS_ARATH (Q39057) Zinc finger protein CONSTANS
Length = 373
Score = 54.3 bits (129), Expect = 2e-07
Identities = 25/55 (45%), Positives = 34/55 (61%)
Query: 120 MSRVCPYSPEEKKVRIERYRIKRNQRNFNKKIKYVCRKTLADRRPRIRGRFARND 174
M V SP +++ R+ RYR KR R F K I+Y RK A+ RPR+ GRFA+ +
Sbjct: 295 MITVTQLSPMDREARVLRYREKRKTRKFEKTIRYASRKAYAEIRPRVNGRFAKRE 349
>COL5_ARATH (Q9FHH8) Zinc finger protein CONSTANS-LIKE 5
Length = 355
Score = 54.3 bits (129), Expect = 2e-07
Identities = 25/55 (45%), Positives = 34/55 (61%)
Query: 127 SPEEKKVRIERYRIKRNQRNFNKKIKYVCRKTLADRRPRIRGRFARNDEIDKKPI 181
S +++ R+ RYR KR R F K I+Y RK A+ RPRI+GRFA+ E + I
Sbjct: 281 SSMDREARVLRYREKRKNRKFEKTIRYASRKAYAESRPRIKGRFAKRTETENDDI 335
>COL2_ARATH (Q96502) Zinc finger protein CONSTANS-LIKE 2
Length = 347
Score = 53.9 bits (128), Expect = 3e-07
Identities = 24/51 (47%), Positives = 35/51 (68%)
Query: 127 SPEEKKVRIERYRIKRNQRNFNKKIKYVCRKTLADRRPRIRGRFARNDEID 177
+P E++ R+ RYR K+ R F+K I+Y RK A+ RPRI+GRFA+ E +
Sbjct: 274 TPMEREARVLRYREKKKTRKFDKTIRYASRKAYAEIRPRIKGRFAKRIETE 324
>COL8_ARATH (Q9M9B3) Putative zinc finger protein CONSTANS-LIKE 8
Length = 313
Score = 53.5 bits (127), Expect = 4e-07
Identities = 29/57 (50%), Positives = 36/57 (62%), Gaps = 6/57 (10%)
Query: 122 RVCPYSPEEKKVRIER------YRIKRNQRNFNKKIKYVCRKTLADRRPRIRGRFAR 172
++ P EEK+VR ER YR KR R F KKI+Y RK AD+RPR++GRF R
Sbjct: 250 QLVPPGIEEKRVRSEREARVWRYRDKRKNRLFEKKIRYEVRKVNADKRPRMKGRFVR 306
>PRR5_ARATH (Q6LA42) Two-component response regulator-like APRR5
(Pseudo-response regulator 5)
Length = 558
Score = 51.2 bits (121), Expect = 2e-06
Identities = 41/167 (24%), Positives = 81/167 (47%), Gaps = 18/167 (10%)
Query: 19 TLSQLNNSELSSGYSSYGGSPSPSTPILMQRSISSHSLQYNNGTHHYPLSAFFADLLDSD 78
T + +NN + ++Y + +P++ S+S H +Y++ H P ++ L D D
Sbjct: 405 TPTPINNIQFRDPNTAYTSAMAPASLSPSPSSVSPH--EYSSMFH--PFNSKPEGLQDRD 460
Query: 79 ---DAPVRKVCSTGDLQRINGMQHNHHHHVDSPLSSESSMIIEGMSRVCPYSPE--EKKV 133
D R+ S+ G +H+D + ++ +G S + +++
Sbjct: 461 CSMDVDERRYVSSATEHSAIG------NHIDQLIEKKNE---DGYSLSVGKIQQSLQREA 511
Query: 134 RIERYRIKRNQRNFNKKIKYVCRKTLADRRPRIRGRFARNDEIDKKP 180
+ ++R+KR R + KK++Y RK LA++RPRI+G+F R + + P
Sbjct: 512 ALTKFRMKRKDRCYEKKVRYESRKKLAEQRPRIKGQFVRQVQSTQAP 558
>COL6_ARATH (Q8LG76) Zinc finger protein CONSTANS-LIKE 6
Length = 406
Score = 50.8 bits (120), Expect = 3e-06
Identities = 22/46 (47%), Positives = 32/46 (68%)
Query: 131 KKVRIERYRIKRNQRNFNKKIKYVCRKTLADRRPRIRGRFARNDEI 176
++ R+ RYR KR R F+KKI+Y RK A++RPR++GRF + I
Sbjct: 357 REARVSRYREKRRTRLFSKKIRYEVRKLNAEKRPRMKGRFVKRSSI 402
>PRR9_ARATH (Q8L500) Two-component response regulator-like APRR9
(Pseudo-response regulator 9)
Length = 468
Score = 50.4 bits (119), Expect = 3e-06
Identities = 20/52 (38%), Positives = 36/52 (68%)
Query: 126 YSPEEKKVRIERYRIKRNQRNFNKKIKYVCRKTLADRRPRIRGRFARNDEID 177
+S +++ + ++R+KR R F+KK++Y RK LA++RPR++G+F R D
Sbjct: 412 WSRSQREAALMKFRLKRKDRCFDKKVRYQSRKKLAEQRPRVKGQFVRTVNSD 463
>COL7_ARATH (Q9C9A9) Putative zinc finger protein CONSTANS-LIKE 7
Length = 392
Score = 50.4 bits (119), Expect = 3e-06
Identities = 23/50 (46%), Positives = 34/50 (68%)
Query: 127 SPEEKKVRIERYRIKRNQRNFNKKIKYVCRKTLADRRPRIRGRFARNDEI 176
S E++ R+ RY+ KR R F+KKI+Y RK A++RPRI+GRF + +
Sbjct: 341 SDGEREARVLRYKEKRRTRLFSKKIRYEVRKLNAEQRPRIKGRFVKRTSL 390
>COLG_ARATH (Q8RWD0) Zinc finger protein CONSTANS-LIKE 16
Length = 417
Score = 49.3 bits (116), Expect = 7e-06
Identities = 21/42 (50%), Positives = 31/42 (73%)
Query: 131 KKVRIERYRIKRNQRNFNKKIKYVCRKTLADRRPRIRGRFAR 172
++ R+ RYR KR R F+KKI+Y RK A++RPR++GRF +
Sbjct: 361 REARVSRYREKRRTRLFSKKIRYEVRKLNAEKRPRMKGRFVK 402
>PRR7_ARATH (Q93WK5) Two-component response regulator-like APRR7
(Pseudo-response regulator 7)
Length = 727
Score = 48.5 bits (114), Expect = 1e-05
Identities = 19/43 (44%), Positives = 32/43 (74%)
Query: 130 EKKVRIERYRIKRNQRNFNKKIKYVCRKTLADRRPRIRGRFAR 172
+++ + ++R KR +R F KK++Y RK LA++RPR+RG+F R
Sbjct: 668 QREAALTKFRQKRKERCFRKKVRYQSRKKLAEQRPRVRGQFVR 710
>PRR3_ARATH (Q9LVG4) Two-component response regulator-like APRR3
(Pseudo-response regulator 3)
Length = 495
Score = 48.5 bits (114), Expect = 1e-05
Identities = 36/135 (26%), Positives = 64/135 (46%), Gaps = 24/135 (17%)
Query: 45 ILMQRSISSHSLQYNNG-THHYPLSAFFADLLDSDDAPVRKVCSTGDLQRINGMQHNHHH 103
+L ++S+ S +YNNG T + D+P+ K+ +
Sbjct: 378 VLRHSNLSAFS-KYNNGATSAKKAPEENVESCSPHDSPIAKLLGSSSSS----------- 425
Query: 104 HVDSPLSSESSMIIEGMSRVCPYSPEEKKVRIERYRIKRNQRNFNKKIKYVCRKTLADRR 163
D+PL +SS G R +++ + ++R+KR +R F KK++Y RK LA++R
Sbjct: 426 --DNPLKQQSS----GSDRWA-----QREAALMKFRLKRKERCFEKKVRYHSRKKLAEQR 474
Query: 164 PRIRGRFARNDEIDK 178
P ++G+F R + K
Sbjct: 475 PHVKGQFIRKRDDHK 489
>COLC_ARATH (Q9LJ44) Putative zinc finger protein CONSTANS-LIKE 12
Length = 337
Score = 46.6 bits (109), Expect = 5e-05
Identities = 24/58 (41%), Positives = 35/58 (59%), Gaps = 4/58 (6%)
Query: 124 CPYSPEEKKVRIERYRIKRNQRNFNKKIKYVCRKTLADRRPRIRGRFAR-NDEIDKKP 180
CP + E K+R Y+ K+ +R+F K+I+Y RK AD R R++GRF + D D P
Sbjct: 276 CPQARNEAKLR---YKEKKLKRSFGKQIRYASRKARADTRKRVKGRFVKAGDSYDYDP 330
>COLB_ARATH (O23379) Putative zinc finger protein CONSTANS-LIKE 11
Length = 330
Score = 45.8 bits (107), Expect = 8e-05
Identities = 19/50 (38%), Positives = 34/50 (68%)
Query: 126 YSPEEKKVRIERYRIKRNQRNFNKKIKYVCRKTLADRRPRIRGRFARNDE 175
++P+ + +RY+ K+++R F K+I+Y RK AD R R++GRF ++ E
Sbjct: 271 FNPKLRDEAKKRYKQKKSKRMFGKQIRYASRKARADTRKRVKGRFVKSGE 320
>COLA_ARATH (Q9LUA9) Zinc finger protein CONSTANS-LIKE 10
Length = 373
Score = 44.3 bits (103), Expect = 2e-04
Identities = 18/41 (43%), Positives = 29/41 (69%)
Query: 135 IERYRIKRNQRNFNKKIKYVCRKTLADRRPRIRGRFARNDE 175
+ RY+ K+ R F+K+++YV RK AD R R++GRF ++ E
Sbjct: 320 VMRYKEKKKARKFDKRVRYVSRKERADVRRRVKGRFVKSGE 360
>COLF_ARATH (Q9C7E8) Zinc finger protein CONSTANS-LIKE 15
Length = 433
Score = 43.1 bits (100), Expect = 5e-04
Identities = 19/41 (46%), Positives = 27/41 (65%)
Query: 135 IERYRIKRNQRNFNKKIKYVCRKTLADRRPRIRGRFARNDE 175
++RY+ KR R ++K I+Y RK AD R R+RGRF + E
Sbjct: 389 MQRYKEKRKTRRYDKTIRYESRKARADTRLRVRGRFVKASE 429
>COLD_ARATH (O82256) Putative zinc finger protein CONSTANS-LIKE 13
Length = 332
Score = 43.1 bits (100), Expect = 5e-04
Identities = 19/43 (44%), Positives = 27/43 (62%)
Query: 130 EKKVRIERYRIKRNQRNFNKKIKYVCRKTLADRRPRIRGRFAR 172
E+ + RY+ K+ R + K I+Y RK A+ R RIRGRFA+
Sbjct: 286 ERNSALSRYKEKKKSRRYEKHIRYESRKVRAESRTRIRGRFAK 328
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.314 0.132 0.387
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 28,647,838
Number of Sequences: 164201
Number of extensions: 1246795
Number of successful extensions: 3425
Number of sequences better than 10.0: 47
Number of HSP's better than 10.0 without gapping: 24
Number of HSP's successfully gapped in prelim test: 23
Number of HSP's that attempted gapping in prelim test: 3390
Number of HSP's gapped (non-prelim): 52
length of query: 227
length of database: 59,974,054
effective HSP length: 106
effective length of query: 121
effective length of database: 42,568,748
effective search space: 5150818508
effective search space used: 5150818508
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (22.0 bits)
S2: 64 (29.3 bits)
Medicago: description of AC149490.13


