

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC149490.11 + phase: 0
(304 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
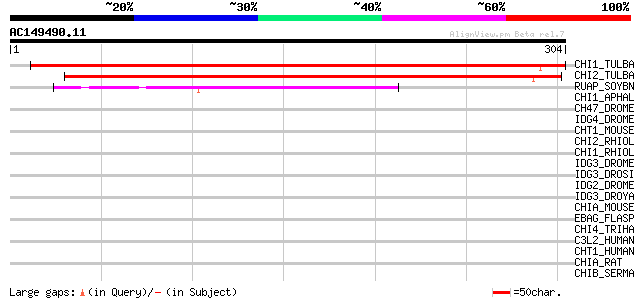
Score E
Sequences producing significant alignments: (bits) Value
CHI1_TULBA (Q9SLP4) Chitinase 1 precursor (EC 3.2.1.14) (Tulip b... 380 e-105
CHI2_TULBA (Q7M443) Chitinase 2 (EC 3.2.1.14) (Tulip bulb chitin... 374 e-103
RUAP_SOYBN (P39657) RuBisCO-associated protein 103 5e-22
CHI1_APHAL (P32470) Chitinase 1 precursor (EC 3.2.1.14) 42 0.002
CH47_DROME (Q23997) Chitinase-like protein DS47 precursor 41 0.004
IDG4_DROME (Q9W303) Chitinase-like protein Idgf4 precursor (Imag... 40 0.007
CHT1_MOUSE (Q9D7Q1) Chitotriosidase 1 precursor (EC 3.2.1.14) (C... 40 0.007
CHI2_RHIOL (P29027) Chitinase 2 precursor (EC 3.2.1.14) 40 0.007
CHI1_RHIOL (P29026) Chitinase 1 precursor (EC 3.2.1.14) 40 0.009
IDG3_DROME (Q8MLZ7) Chitinase-like protein Idgf3 precursor (Imag... 39 0.012
IDG3_DROSI (Q8MX32) Chitinase-like protein Idgf3 precursor (Imag... 39 0.021
IDG2_DROME (Q9V3D4) Chitinase-like protein Idgf2 precursor (Imag... 38 0.027
IDG3_DROYA (Q8MX31) Chitinase-like protein Idgf3 precursor (Imag... 38 0.036
CHIA_MOUSE (Q91XA9) Acidic mammalian chitinase precursor (EC 3.2... 38 0.036
EBAG_FLASP (P80036) Endo-beta-N-acetylglucosaminidase (EC 3.2.1.... 37 0.046
CHI4_TRIHA (P48827) 42 kDa endochitinase precursor (EC 3.2.1.14) 37 0.046
C3L2_HUMAN (Q15782) Chitinase 3-like protein 2 precursor (YKL-39... 37 0.046
CHT1_HUMAN (Q13231) Chitotriosidase 1 precursor (EC 3.2.1.14) (C... 37 0.061
CHIA_RAT (Q6RY07) Acidic mammalian chitinase precursor (EC 3.2.1... 36 0.10
CHIB_SERMA (P11797) Chitinase B precursor (EC 3.2.1.14) 36 0.14
>CHI1_TULBA (Q9SLP4) Chitinase 1 precursor (EC 3.2.1.14) (Tulip bulb
chitinase-1) (TBC-1)
Length = 314
Score = 380 bits (977), Expect = e-105
Identities = 188/295 (63%), Positives = 228/295 (76%), Gaps = 2/295 (0%)
Query: 12 LLILTTSLFASTILAANSNLFREYIGAQFKGVKFSDVPINPNVNFHFILSFAIDYDTSSP 71
LL+L +L + + ++ +FREYIG+QF VKFSDVPINP+V+FHFIL+FAIDY + S
Sbjct: 9 LLLLFFALLSPLLPLTSALVFREYIGSQFNDVKFSDVPINPDVDFHFILAFAIDYTSGSS 68
Query: 72 PSPTNGNFNIFWDTQNLSPSEVSFIKNQNPNVKVALSLGGDTVQGDPVNFIPSSIDSWVS 131
P+PTNGNF FWDT NLSPS+V+ +K + NVKV+LSLGGD+V G V F PSS+ SWV
Sbjct: 69 PTPTNGNFKPFWDTNNLSPSQVAAVKRTHSNVKVSLSLGGDSVGGKNVFFSPSSVSSWVE 128
Query: 132 NAVSSLTSIIKTYNLDGIDVDYEHFIGDPNTFAECIGRLITTLKNNNVISFASIAPFDDA 191
NAVSSLT IIK Y+LDGID+DYEHF GDPNTFAECIG+L+T LK N V+SF SIAPFDDA
Sbjct: 129 NAVSSLTRIIKQYHLDGIDIDYEHFKGDPNTFAECIGQLVTRLKKNEVVSFVSIAPFDDA 188
Query: 192 EVQNHYLALWKSYSNIIDYVNFQFYAYDKSTTVAQFIDYYNKQSSNYNSEKVLVSFVSDG 251
+VQ+HY ALW+ Y + IDYVNFQFYAY T+V QF+ Y+ +QSSNY+ KVLVSF +D
Sbjct: 189 QVQSHYQALWEKYGHQIDYVNFQFYAYSARTSVEQFLKYFEEQSSNYHGGKVLVSFSTDS 248
Query: 252 STGLGPGNGFLDGCRMLKSQQKLNGIFVYSADDSKADG--FRYEKEAQEVLASPN 304
S GL P NGF C +LK Q KL+GIFV+SADDS FRYE +AQ +LAS N
Sbjct: 249 SGGLKPDNGFFRACSILKKQGKLHGIFVWSADDSLMSNNVFRYEMQAQSMLASVN 303
>CHI2_TULBA (Q7M443) Chitinase 2 (EC 3.2.1.14) (Tulip bulb
chitinase-2) (TBC-2)
Length = 275
Score = 374 bits (960), Expect = e-103
Identities = 184/274 (67%), Positives = 215/274 (78%), Gaps = 2/274 (0%)
Query: 31 LFREYIGAQFKGVKFSDVPINPNVNFHFILSFAIDYDTSSPPSPTNGNFNIFWDTQNLSP 90
LFREYIGAQF VKFSDVPINPNV+FHFIL+FAIDY + S P+PTNGNFN FWDT NLSP
Sbjct: 2 LFREYIGAQFNDVKFSDVPINPNVDFHFILAFAIDYTSGSSPTPTNGNFNPFWDTNNLSP 61
Query: 91 SEVSFIKNQNPNVKVALSLGGDTVQGDPVNFIPSSIDSWVSNAVSSLTSIIKTYNLDGID 150
S+V+ IK NVKV++SLGG++V G+ V F PSS+ SWV NAVSSLT IIK Y+LDGID
Sbjct: 62 SQVAAIKRTYNNVKVSVSLGGNSVGGERVFFNPSSVSSWVDNAVSSLTKIIKQYHLDGID 121
Query: 151 VDYEHFIGDPNTFAECIGRLITTLKNNNVISFASIAPFDDAEVQNHYLALWKSYSNIIDY 210
+DYEHF GDPNTFAECIG+L+T LK N V+SF SIAPFDDA+VQ+HY ALW+ Y + IDY
Sbjct: 122 IDYEHFKGDPNTFAECIGQLVTRLKKNGVVSFVSIAPFDDAQVQSHYQALWEKYGHQIDY 181
Query: 211 VNFQFYAYDKSTTVAQFIDYYNKQSSNYNSEKVLVSFVSDGSTGLGPGNGFLDGCRMLKS 270
VNFQFY Y +V QF+ Y+ Q SNY KVLVSF +D S GL P NGF D C +LK
Sbjct: 182 VNFQFYVYSSRMSVEQFLKYFEMQRSNYPGGKVLVSFSTDNSGGLKPRNGFFDACSILKK 241
Query: 271 QQKLNGIFVYSADDS--KADGFRYEKEAQEVLAS 302
Q KL+GIFV+SADDS D F+YE +AQ +LAS
Sbjct: 242 QGKLHGIFVWSADDSLMSNDVFKYEMQAQSLLAS 275
>RUAP_SOYBN (P39657) RuBisCO-associated protein
Length = 283
Score = 103 bits (257), Expect = 5e-22
Identities = 70/194 (36%), Positives = 96/194 (49%), Gaps = 12/194 (6%)
Query: 25 LAANSNLFREYIGAQFKGVKFSDVPINPNVN-FHFILSFAIDYDTSSPPSPTNGNFNIFW 83
++ +FRE+ F + I N+ F LS A DYD ++ TNG F +W
Sbjct: 1 MSTKFKVFREFTSDD----SFLNQVIPENITEFQVTLSLARDYDGNNS---TNGKFIPYW 53
Query: 84 DTQNLSPSEVS-FIKNQNPN---VKVALSLGGDTVQGDPVNFIPSSIDSWVSNAVSSLTS 139
DT+ ++P + F K P VKV +S+G Q S+ ++WVS A +SL S
Sbjct: 54 DTEKVTPEVIKKFKKKYEPTALRVKVLVSIGNKNKQFPFTIGSDSNSEAWVSEATASLKS 113
Query: 140 IIKTYNLDGIDVDYEHFIGDPNTFAECIGRLITTLKNNNVISFASIAPFDDAEVQNHYLA 199
IIKTYNLDGIDV YE + F +G L+ LK N +I+ AS A DA Y
Sbjct: 114 IIKTYNLDGIDVSYEDIAANEADFVNSVGGLVRNLKQNKLITVASFATSADAANNKFYNL 173
Query: 200 LWKSYSNIIDYVNF 213
L+ Y+ D V F
Sbjct: 174 LYAEYATFFDTVVF 187
>CHI1_APHAL (P32470) Chitinase 1 precursor (EC 3.2.1.14)
Length = 423
Score = 42.0 bits (97), Expect = 0.002
Identities = 43/177 (24%), Positives = 78/177 (43%), Gaps = 24/177 (13%)
Query: 92 EVSFIKNQNPNVKVALSLGGDTVQGDPVNFIPSSIDSWVSNA--VSSLTSIIKTYNLDGI 149
++ +K QN N+KV LS+GG T NF P++ S + S +K + DGI
Sbjct: 111 QLYLLKKQNRNMKVMLSIGGWTWS---TNF-PAAASSAATRKTFAQSAVGFMKDWGFDGI 166
Query: 150 DVDYEHFIGDPNTFAECIGRLITTLKNNNVISFAS---------IAPFDDAEVQNHYLAL 200
D+D+E + D + L+ + + S+A+ ++ A N+
Sbjct: 167 DIDWE-YPADATQAQNMV--LLLQAVRSELDSYAAQYAKGHHFLLSIAAPAGPDNYNKLK 223
Query: 201 WKSYSNIIDYVNFQFYAYDKSTTVAQFID---YYNKQSSN---YNSEKVLVSFVSDG 251
+ ++DY+N Y Y S + D Y N Q+ N YN++ + ++++ G
Sbjct: 224 FAELGKVLDYINLMAYDYAGSWSNYTGHDANIYANPQNPNATPYNTDDAVQAYINGG 280
>CH47_DROME (Q23997) Chitinase-like protein DS47 precursor
Length = 452
Score = 40.8 bits (94), Expect = 0.004
Identities = 35/122 (28%), Positives = 57/122 (46%), Gaps = 10/122 (8%)
Query: 93 VSFIKNQNPNVKVALSLGGDT-VQGDP-VNFIPSSIDSWVSNA------VSSLTSIIKTY 144
V+ +K + PNVK+ LS+GGD ++ D +P+ + + V+++ S++KTY
Sbjct: 96 VTRLKRKYPNVKILLSVGGDKDIELDKDAKELPNKYLELLESPTGRTRFVNTVYSLVKTY 155
Query: 145 NLDGIDVDYEHFIGDPNTFAECIGRLITTLKNNNVISFASIAPFDDAEVQNHYLALWKSY 204
DG+DV ++ P IG L K V S SI E + + AL +
Sbjct: 156 GFDGLDVAWQFPKNKPKKVHSGIGSLWKGFK--KVFSGDSIVDEKSEEHKEQFTALLRDV 213
Query: 205 SN 206
N
Sbjct: 214 KN 215
>IDG4_DROME (Q9W303) Chitinase-like protein Idgf4 precursor
(Imaginal disk growth factor protein 4)
Length = 442
Score = 40.0 bits (92), Expect = 0.007
Identities = 28/83 (33%), Positives = 46/83 (54%), Gaps = 7/83 (8%)
Query: 92 EVSFIKNQNPNVKVALSLGGDTVQGDPV-NFIPSSIDSWVSNA----VSSLTSIIKTYNL 146
+V+ +K + P +KV LS+GGD DP N + ++S SNA ++S S++KTY
Sbjct: 91 QVTGLKRKYPALKVLLSVGGDKDTVDPENNKYLTLLES--SNARIPFINSAHSLVKTYGF 148
Query: 147 DGIDVDYEHFIGDPNTFAECIGR 169
DG+D+ ++ P IG+
Sbjct: 149 DGLDLGWQFPKNKPKKVHGSIGK 171
>CHT1_MOUSE (Q9D7Q1) Chitotriosidase 1 precursor (EC 3.2.1.14)
(Chitinase 1)
Length = 396
Score = 40.0 bits (92), Expect = 0.007
Identities = 28/114 (24%), Positives = 51/114 (44%), Gaps = 10/114 (8%)
Query: 41 KGVKFSDVPINPNVNFHFILSFAIDYDTSSPPSPTNGNFNIFWDTQNLSPSEVSFIKNQN 100
+ V+F ++PN+ H I +FA N + L E++ +K +N
Sbjct: 37 EAVRFFPRDVDPNLCTHVIFAFA---------GMDNHQLSTVEHNDELLYQELNSLKTKN 87
Query: 101 PNVKVALSLGGDTVQGDPVNFIPSSIDSWVSNAVSSLTSIIKTYNLDGIDVDYE 154
P +K L++GG T + ++ + V S S ++T DG+D+D+E
Sbjct: 88 PKLKTLLAVGGWTFGTQKFTDMVATASNR-QTFVKSALSFLRTQGFDGLDLDWE 140
>CHI2_RHIOL (P29027) Chitinase 2 precursor (EC 3.2.1.14)
Length = 542
Score = 40.0 bits (92), Expect = 0.007
Identities = 55/187 (29%), Positives = 79/187 (41%), Gaps = 19/187 (10%)
Query: 82 FWDTQNLS-PSEVSFIKN-QNPNVKVALSLGGDT-VQGDPVNFIPSSIDSWVSNAVSSLT 138
F +TQ LS P+ + IK Q+ VKV LSLGG V G + + N
Sbjct: 88 FPNTQLLSCPAVGADIKKCQDKGVKVILSLGGAAGVYGFTSDAQGQQFAQTIWNLFGGGN 147
Query: 139 SIIKTYN---LDGIDVDYEHFIGDPNTFAECIGRLITTLKNNNVISFASIAPFDDAEVQN 195
S + + +DG+D+D E G + + L +N +I A PF DA + +
Sbjct: 148 SDTRPFGDAVIDGVDLDIEG--GSSTGYVAFVNALRQKFSSNFLIGAAPQCPFPDAILGS 205
Query: 196 HYLALWKSYSNIIDYVNFQFYAYDKSTTVAQF----IDYYNKQSSNYNSEKVLVSFVSDG 251
S DYVN QFY S T + F D + K +S + K++ + V
Sbjct: 206 ------VLNSASFDYVNVQFYNNYCSATGSSFNFDTWDNWAKTTSPNKNVKIMFT-VPGS 258
Query: 252 STGLGPG 258
ST G G
Sbjct: 259 STAAGSG 265
>CHI1_RHIOL (P29026) Chitinase 1 precursor (EC 3.2.1.14)
Length = 540
Score = 39.7 bits (91), Expect = 0.009
Identities = 56/186 (30%), Positives = 80/186 (42%), Gaps = 17/186 (9%)
Query: 82 FWDTQNLS-PSEVSFIKN-QNPNVKVALSLGGDT-VQGDPVNFIPSSIDSWVSNAVSSLT 138
F +TQ LS P+ + IK Q+ VKV LSLGG V G + + N +
Sbjct: 88 FPNTQLLSCPAVGADIKKCQDKGVKVILSLGGAAGVYGFTSDAQGQQFAQTIWNLFGGGS 147
Query: 139 SIIKTYN---LDGIDVDYEHFIGDPNTFAECIGRLITTLKNNNVISFASIAPFDDAEVQN 195
S + + +DG+D+D E G +A + L +N +I A PF DA + +
Sbjct: 148 SDTRPFGDAVIDGVDLDIEG--GASTGYAAFVNALRQKFSSNFLIGAAPQCPFPDAILGS 205
Query: 196 HYLALWKSYSNIIDYVNFQFYAYDKSTTVAQF-IDYYNKQSSNYNSEK-VLVSFVSDGS- 252
S DYVN QFY S T + F D ++ + + K V + F GS
Sbjct: 206 ------VLNSASFDYVNVQFYNNYCSATGSSFNFDTWDNWAKTTSPNKNVKIMFTIPGSP 259
Query: 253 TGLGPG 258
T G G
Sbjct: 260 TAAGSG 265
>IDG3_DROME (Q8MLZ7) Chitinase-like protein Idgf3 precursor
(Imaginal disk growth factor protein 3)
Length = 441
Score = 39.3 bits (90), Expect = 0.012
Identities = 37/150 (24%), Positives = 74/150 (48%), Gaps = 13/150 (8%)
Query: 10 FILLILTTSLFASTILAANSNL--FREYIGAQFKGV-KFSDVPINPNVNF--HFILSFA- 63
++ L L+ ++ A ++A NL F + G+Q +G+ +FS + I + F H + +A
Sbjct: 6 WLSLALSLAVLAQFKVSAAPNLVCFYDSQGSQRQGLAQFSMIDIELALQFCTHLVYGYAG 65
Query: 64 IDYDTSSPPSPTNGNFNIFWDTQNLSPSEVSFIKNQNPNVKVALSLGG--DTVQGDPVNF 121
++ D S N D + ++++ +K + P++K LS+GG DT +G+
Sbjct: 66 VNADNYEMQS-----INKRLDLEQRHLAQITSMKERYPHIKFLLSVGGDADTNEGNQYIK 120
Query: 122 IPSSIDSWVSNAVSSLTSIIKTYNLDGIDV 151
+ S + S +++ YN DG+D+
Sbjct: 121 LLESGQQGHRRFIESARDLVRRYNFDGLDL 150
>IDG3_DROSI (Q8MX32) Chitinase-like protein Idgf3 precursor
(Imaginal disk growth factor protein 3)
Length = 441
Score = 38.5 bits (88), Expect = 0.021
Identities = 38/150 (25%), Positives = 73/150 (48%), Gaps = 13/150 (8%)
Query: 10 FILLILTTSLFASTILAANSNL--FREYIGAQFKGV-KFSDVPINPNVNF--HFILSFA- 63
++ L L+ ++ A ++A NL F + G+Q +G+ +FS I + F H + +A
Sbjct: 6 WLSLALSLAVLAQFKVSAAPNLVCFYDSQGSQRQGLAQFSITDIELALQFCTHLVYGYAG 65
Query: 64 IDYDTSSPPSPTNGNFNIFWDTQNLSPSEVSFIKNQNPNVKVALSLGG--DTVQGDPVNF 121
++ D S N D + ++V+ +K + P++K LS+GG DT +G+
Sbjct: 66 VNADNFEMQS-----INKRLDLEQRHLAQVTSMKERYPHIKFLLSVGGDADTNEGNQYIK 120
Query: 122 IPSSIDSWVSNAVSSLTSIIKTYNLDGIDV 151
+ S + S +++ YN DG+D+
Sbjct: 121 LLESGQQGHRRFIESARDLVRRYNFDGLDL 150
>IDG2_DROME (Q9V3D4) Chitinase-like protein Idgf2 precursor
(Imaginal disk growth factor protein 2)
Length = 440
Score = 38.1 bits (87), Expect = 0.027
Identities = 22/68 (32%), Positives = 38/68 (55%), Gaps = 4/68 (5%)
Query: 91 SEVSFIKNQNPNVKVALSLGGD-TVQGDPVNFIPSSIDSWVSNAVSSLTS---IIKTYNL 146
SEV+ +K + P++KV LS+GGD + D N ++ + + S ++KTY
Sbjct: 85 SEVTSLKRKYPHLKVLLSVGGDHDIDPDHPNKYIDLLEGEKVRQIGFIRSAYDLVKTYGF 144
Query: 147 DGIDVDYE 154
DG+D+ Y+
Sbjct: 145 DGLDLAYQ 152
>IDG3_DROYA (Q8MX31) Chitinase-like protein Idgf3 precursor
(Imaginal disk growth factor protein 3)
Length = 441
Score = 37.7 bits (86), Expect = 0.036
Identities = 38/150 (25%), Positives = 72/150 (47%), Gaps = 13/150 (8%)
Query: 10 FILLILTTSLFASTILAANSNL--FREYIGAQFKGV-KFSDVPINPNVNF--HFILSFA- 63
++ L L+ ++ A ++A NL F + G Q +G+ +FS + + F H + +A
Sbjct: 6 WLSLALSLAVLAQFKVSAAPNLVCFYDSQGFQRQGLAQFSMTDMELALQFCTHLVYGYAG 65
Query: 64 IDYDTSSPPSPTNGNFNIFWDTQNLSPSEVSFIKNQNPNVKVALSLGG--DTVQGDPVNF 121
++ D S N D + ++VS +K + P++K LS+GG DT +G+
Sbjct: 66 VNADNYEMQS-----INKRLDLEQRHLAQVSSLKERYPHIKFLLSVGGDADTNEGNQYIK 120
Query: 122 IPSSIDSWVSNAVSSLTSIIKTYNLDGIDV 151
+ S + S +++ YN DG+D+
Sbjct: 121 LLESGQQGHRRFIESARDLVRRYNFDGLDL 150
>CHIA_MOUSE (Q91XA9) Acidic mammalian chitinase precursor (EC
3.2.1.14) (AMCase) (YNL)
Length = 473
Score = 37.7 bits (86), Expect = 0.036
Identities = 37/148 (25%), Positives = 68/148 (45%), Gaps = 13/148 (8%)
Query: 11 ILLILTTSLFASTILAANSNLFREYIG-AQFK-GV-KFSDVPINPNVNFHFILSFAIDYD 67
+LL+ +L + L + NL + AQ++ G+ F INP + H I +FA
Sbjct: 4 LLLVTGLALLLNAQLGSAYNLICYFTNWAQYRPGLGSFKPDDINPCLCTHLIYAFA---- 59
Query: 68 TSSPPSPTNGNFNIFWDTQNLSPSEVSFIKNQNPNVKVALSLGGDTVQGDPVNFIPSSID 127
N I W+ L + + +KN+N +K L++GG P + S+
Sbjct: 60 ----GMQNNEITTIEWNDVTLYKA-FNDLKNRNSKLKTLLAIGGWNFGTAPFTTMVSTSQ 114
Query: 128 SWVSNAVSSLTSIIKTYNLDGIDVDYEH 155
+ ++S+ ++ Y DG+D+D+E+
Sbjct: 115 NR-QTFITSVIKFLRQYGFDGLDLDWEY 141
>EBAG_FLASP (P80036) Endo-beta-N-acetylglucosaminidase (EC 3.2.1.96)
(Mannosyl-glycoprotein
endo-beta-N-acetyl-glucosaminidase)
(DI-N-acetylchitobiosyl beta-N-acetylglucosaminidase)
Length = 267
Score = 37.4 bits (85), Expect = 0.046
Identities = 46/184 (25%), Positives = 74/184 (40%), Gaps = 34/184 (18%)
Query: 76 NGNFNIFWDTQN----LSPSEVSFIKNQNPNVKVALSLGGDTVQGDPVNFIPSSIDSWVS 131
NG+ + ++ +N L + Q +KV+LS+ G+ NF +
Sbjct: 49 NGSKAVLYNNENVQATLDDAATQIRPLQAKGIKVSLSILGNHQGAGIANFPTQAA---AE 105
Query: 132 NAVSSLTSIIKTYNLDGIDVDYEH----FIGDPNTFAECIGRLITTLKNN---NVISFAS 184
+ + +++ + Y LDG+D+D E+ G P + IG LI+ L+ + +ISF
Sbjct: 106 DFAAQVSATVSKYGLDGVDLDDEYSDYGTNGTPQPNQQSIGGLISALRADVPGKLISFYD 165
Query: 185 IAPFDDAEVQNHYLALWKSYSNI---IDYVNFQFY---------AYDKSTTVAQFIDYYN 232
I P AL S S I +DY +Y DKS A +D N
Sbjct: 166 IGPASS--------ALSSSSSTIGSKLDYAWNPYYGTYSAPSIPGLDKSRLSAAAVDVQN 217
Query: 233 KQSS 236
S
Sbjct: 218 TPQS 221
>CHI4_TRIHA (P48827) 42 kDa endochitinase precursor (EC 3.2.1.14)
Length = 423
Score = 37.4 bits (85), Expect = 0.046
Identities = 41/172 (23%), Positives = 72/172 (41%), Gaps = 22/172 (12%)
Query: 96 IKNQNPNVKVALSLGGDTVQGDPVNFIPS--SIDSWVSNAVSSLTSIIKTYNLDGIDVDY 153
+K N +KV LS+GG T NF PS S D+ N + + +K + DGID+D+
Sbjct: 115 VKKANRGLKVLLSIGGWTWS---TNF-PSAASTDANRKNFAKTAITFMKDWGFDGIDIDW 170
Query: 154 EHFIGDPNTFAECIGRLITTLKNNNVISFASIAP--------FDDAEVQNHYLALWKSYS 205
E+ T A + L+ +++ A AP A N+
Sbjct: 171 EY--PADATQASNMILLLKEVRSQRDAYAAQYAPGYHFLLTIAAPAGKDNYSKLRLADLG 228
Query: 206 NIIDYVNFQFYAYDKSTT------VAQFIDYYNKQSSNYNSEKVLVSFVSDG 251
++DY+N Y Y S + F + N ++ +N++ + +++ G
Sbjct: 229 QVLDYINLMAYDYAGSFSPLTGHDANLFNNPSNPNATPFNTDSAVKDYINGG 280
>C3L2_HUMAN (Q15782) Chitinase 3-like protein 2 precursor (YKL-39)
(Chondrocyte protein 39)
Length = 390
Score = 37.4 bits (85), Expect = 0.046
Identities = 31/112 (27%), Positives = 53/112 (46%), Gaps = 14/112 (12%)
Query: 44 KFSDVPINPNVNFHFILSFAIDYDTSSPPSPTNGNFNIFWDTQNLSPSEVSFIKNQNPNV 103
KF+ I+P + H I SFA S N I ++ + ++ +K +NP +
Sbjct: 45 KFTPENIDPFLCSHLIYSFA---------SIENNKVIIKDKSEVMLYQTINSLKTKNPKL 95
Query: 104 KVALSLGGDTVQGDPVNFIPSSIDSWVSNA--VSSLTSIIKTYNLDGIDVDY 153
K+ LS+GG F P +DS S ++S+ ++ +N DG+DV +
Sbjct: 96 KILLSIGGYLFGSK--GFHP-MVDSSTSRLEFINSIILFLRNHNFDGLDVSW 144
>CHT1_HUMAN (Q13231) Chitotriosidase 1 precursor (EC 3.2.1.14)
(Chitinase 1)
Length = 466
Score = 37.0 bits (84), Expect = 0.061
Identities = 36/142 (25%), Positives = 60/142 (41%), Gaps = 26/142 (18%)
Query: 44 KFSDVPINPNVNFHFILSFAIDYDTSSPPSPTNGNFNIF-WDTQNLSPSEVSFIKNQNPN 102
+F ++P++ H I +FA TN + W+ + L E + +K NP
Sbjct: 40 RFLPKDLDPSLCTHLIYAFA---------GMTNHQLSTTEWNDETLY-QEFNGLKKMNPK 89
Query: 103 VKVALSLGGDTVQGDPVNFIPSSIDSWVSNA------VSSLTSIIKTYNLDGIDVDYEH- 155
+K L++GG NF V+ A V+S ++ Y+ DG+D+D+E+
Sbjct: 90 LKTLLAIGG-------WNFGTQKFTDMVATANNRQTFVNSAIRFLRKYSFDGLDLDWEYP 142
Query: 156 -FIGDPNTFAECIGRLITTLKN 176
G P E L+ L N
Sbjct: 143 GSQGSPAVDKERFTTLVQDLAN 164
>CHIA_RAT (Q6RY07) Acidic mammalian chitinase precursor (EC
3.2.1.14) (AMCase)
Length = 473
Score = 36.2 bits (82), Expect = 0.10
Identities = 28/111 (25%), Positives = 50/111 (44%), Gaps = 10/111 (9%)
Query: 45 FSDVPINPNVNFHFILSFAIDYDTSSPPSPTNGNFNIFWDTQNLSPSEVSFIKNQNPNVK 104
F INP + H I +FA N I W+ L + + +KN+N +K
Sbjct: 41 FKPDDINPCLCTHLIYAFA--------GMQNNQITTIEWNDVTLYKA-FNDLKNRNSKLK 91
Query: 105 VALSLGGDTVQGDPVNFIPSSIDSWVSNAVSSLTSIIKTYNLDGIDVDYEH 155
L++GG P + S+ + ++S+ ++ Y DG+D+D+E+
Sbjct: 92 TLLAIGGWNFGTAPFTTMVSTSQNR-QTFITSVIKFLRQYGFDGLDLDWEY 141
>CHIB_SERMA (P11797) Chitinase B precursor (EC 3.2.1.14)
Length = 499
Score = 35.8 bits (81), Expect = 0.14
Identities = 33/160 (20%), Positives = 62/160 (38%), Gaps = 19/160 (11%)
Query: 76 NGNFNIFWD--TQNLSPSEV----SFIKNQNPNVKVALSLGGDTVQGD----PVNFIPS- 124
N N WD T + +V + +K NP++++ S+GG D N++ +
Sbjct: 55 NSNLECAWDPATNDAKARDVVNRLTALKAHNPSLRIMFSIGGWYYSNDLGVSHANYVNAV 114
Query: 125 SIDSWVSNAVSSLTSIIKTYNLDGIDVDYEH-FIGDPNTFAECIGRLITTLKNNNVISFA 183
+ + S I+K Y DG+D+D+E+ + + F + + T L +
Sbjct: 115 KTPAARTKFAQSCVRIMKDYGFDGVDIDWEYPQAAEVDGFIAALQEIRTLLNQQTIADGR 174
Query: 184 SIAPFD-------DAEVQNHYLALWKSYSNIIDYVNFQFY 216
P+ A + Y + +DY+N Y
Sbjct: 175 QALPYQLTIAGAGGAFFLSRYYSKLAQIVAPLDYINLMTY 214
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.318 0.135 0.398
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 37,094,719
Number of Sequences: 164201
Number of extensions: 1597201
Number of successful extensions: 4172
Number of sequences better than 10.0: 59
Number of HSP's better than 10.0 without gapping: 6
Number of HSP's successfully gapped in prelim test: 53
Number of HSP's that attempted gapping in prelim test: 4137
Number of HSP's gapped (non-prelim): 63
length of query: 304
length of database: 59,974,054
effective HSP length: 110
effective length of query: 194
effective length of database: 41,911,944
effective search space: 8130917136
effective search space used: 8130917136
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 65 (29.6 bits)
Medicago: description of AC149490.11


