

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC149489.2 - phase: 0
(365 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
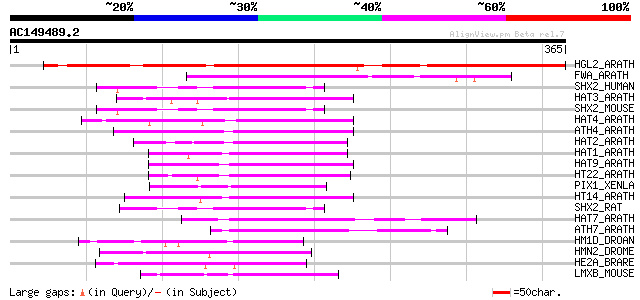
Score E
Sequences producing significant alignments: (bits) Value
HGL2_ARATH (P46607) Homeobox protein GLABRA2 (Homeobox-leucine z... 346 6e-95
FWA_ARATH (Q9FVI6) Homeobox protein FWA 101 3e-21
SHX2_HUMAN (O60902) Short stature homeobox protein 2 (Paired-rel... 74 8e-13
HAT3_ARATH (P46602) Homeobox-leucine zipper protein HAT3 (HD-ZIP... 74 8e-13
SHX2_MOUSE (P70390) Short stature homeobox protein 2 (Homeobox p... 72 2e-12
HAT4_ARATH (Q05466) Homeobox-leucine zipper protein HAT4 (HD-ZIP... 72 2e-12
ATH4_ARATH (P92953) Homeobox-leucine zipper protein ATHB-4 (HD-Z... 71 4e-12
HAT2_ARATH (P46601) Homeobox-leucine zipper protein HAT2 (HD-ZIP... 69 2e-11
HAT1_ARATH (P46600) Homeobox-leucine zipper protein HAT1 (HD-ZIP... 67 7e-11
HAT9_ARATH (P46603) Homeobox-leucine zipper protein HAT9 (Homeod... 67 9e-11
HT22_ARATH (P46604) Homeobox-leucine zipper protein HAT22 (HD-ZI... 65 3e-10
PIX1_XENLA (Q9W751) Pituitary homeobox 1 (X-PITX-1) (xPitx1) 65 4e-10
HT14_ARATH (P46665) Homeobox-leucine zipper protein HAT14 (HD-ZI... 64 5e-10
SHX2_RAT (O35750) Short stature homeobox protein 2 (Paired famil... 64 6e-10
HAT7_ARATH (Q00466) Homeobox-leucine zipper protein HAT7 (HD-ZIP... 64 6e-10
ATH7_ARATH (P46897) Homeobox-leucine zipper protein ATHB-7 (Home... 64 8e-10
HM1D_DROAN (P22544) Homeobox protein B-H1 63 1e-09
HMN2_DROME (P22808) Homeobox protein vnd (Ventral nervous system... 62 2e-09
HE2A_BRARE (P09015) Homeobox protein engrailed-2a (Zf-En-2) (Eng2) 62 2e-09
LMXB_MOUSE (O88609) LIM homeobox transcription factor 1 beta (LI... 61 4e-09
>HGL2_ARATH (P46607) Homeobox protein GLABRA2 (Homeobox-leucine
zipper protein ATHB-10) (HD-ZIP protein ATHB-10)
Length = 745
Score = 346 bits (887), Expect = 6e-95
Identities = 191/346 (55%), Positives = 239/346 (68%), Gaps = 36/346 (10%)
Query: 23 MCADMSNNNPPSTSKAKDFFPSPALSLSLAGIFRHGGGAAAEGEGGSISNMEVEEGEEGS 82
M DMS+ P KDFF SPALSLSLAGIFR+ + E + V++ +
Sbjct: 1 MAVDMSSKQP-----TKDFFSSPALSLSLAGIFRNASSGSTNPEEDFLGRRVVDDED--- 52
Query: 83 TIGGERVEEISSEYSGPAKSKSIDEYEGDELEDDEGEDDEGDGDGDGDGVNKNKKKKRKK 142
R E+SSE SGP +S+S ++ EG++ +D+E E+++G G K+KRKK
Sbjct: 53 -----RTVEMSSENSGPTRSRSEEDLEGEDHDDEEEEEEDGAAGNKG-----TNKRKRKK 102
Query: 143 YHRHTSEQIRVMEALFKESPHPDEKQRQQLSKQLGLAPRQVKFWFQNRRTQIKAIQERHE 202
YHRHT++QIR MEALFKE+PHPDEKQRQQLSKQLGLAPRQVKFWFQNRRTQIKAIQERHE
Sbjct: 103 YHRHTTDQIRHMEALFKETPHPDEKQRQQLSKQLGLAPRQVKFWFQNRRTQIKAIQERHE 162
Query: 203 NSLLKSEIEKLREKNKTLRETINKA--CCPNCGVPTTNRDGTMATEEQQLRIENAKLKAE 260
NSLLK+E+EKLRE+NK +RE+ +KA CPNCG L +EN+KLKAE
Sbjct: 163 NSLLKAELEKLREENKAMRESFSKANSSCPNCG-----------GGPDDLHLENSKLKAE 211
Query: 261 VERLRAALGKYASGTMSP-SCSTSHDQENIKSSLDFYTGIFCLDESRIMDVVNQAMEELI 319
+++LRAALG+ T P S S DQE+ SLDFYTG+F L++SRI ++ N+A EL
Sbjct: 212 LDKLRAALGR----TPYPLQASCSDDQEHRLGSLDFYTGVFALEKSRIAEISNRATLELQ 267
Query: 320 KMATMGEPMWLRSLETGREILNYDEYMKEFADENSDHGRPKRSIEA 365
KMAT GEPMWLRS+ETGREILNYDEY+KEF + +++IEA
Sbjct: 268 KMATSGEPMWLRSVETGREILNYDEYLKEFPQAQASSFPGRKTIEA 313
>FWA_ARATH (Q9FVI6) Homeobox protein FWA
Length = 686
Score = 101 bits (252), Expect = 3e-21
Identities = 69/224 (30%), Positives = 108/224 (47%), Gaps = 15/224 (6%)
Query: 117 EGEDDEGDGDGDGDGVNKNKKKKRKKYHRHTSEQIRVMEALFKESPHPDEKQRQQLSKQL 176
E E DE D D GVN + ++ HR T+ Q + +E + E+PHP E+QR +L ++L
Sbjct: 18 EAEGDEIDMINDMSGVNDQDGGRMRRTHRRTAYQTQELENFYMENPHPTEEQRYELGQRL 77
Query: 177 GLAPRQVKFWFQNRRTQIKAIQERHENSLLKSEIEKLREKNKTLRETINKACCPNCGVPT 236
+ QVK WFQN+R K + EN L+ E ++L LR + ++ C CG T
Sbjct: 78 NMGVNQVKNWFQNKRNLEKINNDHLENVTLREEHDRLLATQDQLRSAMLRSLCNICGKAT 137
Query: 237 TNRDGTMATEEQQLRIENAKLKAEVERLRAALGKYASGTMSPSCSTSHDQENIKSS---- 292
G E Q+L ENA L+ E+++ + +Y S STS + S+
Sbjct: 138 --NCGDTEYEVQKLMAENANLEREIDQFNS---RYLSHPKQRMVSTSEQAPSSSSNPGIN 192
Query: 293 ----LDFYTGIFCLDE--SRIMDVVNQAMEELIKMATMGEPMWL 330
LDF G ++ S +++ A+ ELI + + P W+
Sbjct: 193 ATPVLDFSGGTRTSEKETSIFLNLAITALRELITLGEVDCPFWM 236
>SHX2_HUMAN (O60902) Short stature homeobox protein 2
(Paired-related homeobox protein SHOT) (Homeobox protein
Og12X)
Length = 331
Score = 73.6 bits (179), Expect = 8e-13
Identities = 49/153 (32%), Positives = 74/153 (48%), Gaps = 30/153 (19%)
Query: 58 GGGAAAEGEGGS---ISNMEVEEGEEGSTIGGERVEEISSEYSGPAKSKSIDEYEGDELE 114
GGG A G GG + +++ E G R+ E+S E D E
Sbjct: 79 GGGGAGGGAGGGRSPVRELDMGAAERSREPGSPRLTEVSPELK-------------DRKE 125
Query: 115 DDEGEDDEGDGDGDGDGVNKNKKKKRKKYHRHTSEQIRVMEALFKESPHPDEKQRQQLSK 174
D +G +DEG + K K+R+ T EQ+ +E LF E+ +PD R++LS+
Sbjct: 126 DAKGMEDEG----------QTKIKQRRSRTNFTLEQLNELERLFDETHYPDAFMREELSQ 175
Query: 175 QLGLAPRQVKFWFQNRRTQIKAIQERHENSLLK 207
+LGL+ +V+ WFQNRR + + + EN L K
Sbjct: 176 RLGLSEARVQVWFQNRRAKCR----KQENQLHK 204
>HAT3_ARATH (P46602) Homeobox-leucine zipper protein HAT3 (HD-ZIP
protein 3)
Length = 315
Score = 73.6 bits (179), Expect = 8e-13
Identities = 53/163 (32%), Positives = 79/163 (47%), Gaps = 9/163 (5%)
Query: 71 SNMEVEEGEEGSTIGGERVEEISSEYSGPAKSKSI----DEYEGDELEDDEGEDDE---G 123
S + V+ +EG+ + +SS SG + + G +ED+E E G
Sbjct: 86 STVVVDVEDEGAGVSSPN-STVSSVMSGKKSERELMAAAGAVGGGRVEDNEIERASCSLG 144
Query: 124 DGDGDGDGVNKNKKKKRKKYHRHTSEQIRVMEALFKESPHPDEKQRQQLSKQLGLAPRQV 183
G D DG RKK R + EQ V+E FKE + KQ+ L+KQL L RQV
Sbjct: 145 GGSDDEDGSGNGDDSSRKKL-RLSKEQALVLEETFKEHSTLNPKQKMALAKQLNLRTRQV 203
Query: 184 KFWFQNRRTQIKAIQERHENSLLKSEIEKLREKNKTLRETINK 226
+ WFQNRR + K Q + LK E L ++N+ L++ +++
Sbjct: 204 EVWFQNRRARTKLKQTEVDCEYLKRCCENLTDENRRLQKEVSE 246
>SHX2_MOUSE (P70390) Short stature homeobox protein 2 (Homeobox
protein Og12X) (OG-12) (Paired family homeodomain
protein Prx3)
Length = 331
Score = 72.4 bits (176), Expect = 2e-12
Identities = 48/153 (31%), Positives = 74/153 (47%), Gaps = 30/153 (19%)
Query: 58 GGGAAAEGEGGS---ISNMEVEEGEEGSTIGGERVEEISSEYSGPAKSKSIDEYEGDELE 114
GGG A G GG + +++ E G R+ E+S E D +
Sbjct: 79 GGGGAGGGAGGGRSPVRELDMGAAERSREPGSPRLTEVSPELK-------------DRKD 125
Query: 115 DDEGEDDEGDGDGDGDGVNKNKKKKRKKYHRHTSEQIRVMEALFKESPHPDEKQRQQLSK 174
D +G +DEG + K K+R+ T EQ+ +E LF E+ +PD R++LS+
Sbjct: 126 DAKGMEDEG----------QTKIKQRRSRTNFTLEQLNELERLFDETHYPDAFMREELSQ 175
Query: 175 QLGLAPRQVKFWFQNRRTQIKAIQERHENSLLK 207
+LGL+ +V+ WFQNRR + + + EN L K
Sbjct: 176 RLGLSEARVQVWFQNRRAKCR----KQENQLHK 204
>HAT4_ARATH (Q05466) Homeobox-leucine zipper protein HAT4 (HD-ZIP
protein 4) (HD-ZIP protein ATHB-2)
Length = 284
Score = 72.4 bits (176), Expect = 2e-12
Identities = 58/188 (30%), Positives = 86/188 (44%), Gaps = 17/188 (9%)
Query: 48 SLSLAGIFRHGGGAAAEGEGGSISNMEVEEGEEGSTIGGERVE------EISSEYSGPAK 101
S S G+FR E S+ N + + E + I G V E E +G +
Sbjct: 34 SSSSFGLFRRSSWN--ESFTSSVPNSDSSQKETRTFIRGIDVNRPPSTAEYGDEDAGVSS 91
Query: 102 SKS-IDEYEGDELEDDEGEDDEGDG--DGDGDGVNKNKKKKRKKYHRHTSEQIRVMEALF 158
S + G E +E D +G D DG N KK + K +Q ++E F
Sbjct: 92 PNSTVSSSTGKRSEREEDTDPQGSRGISDDEDGDNSRKKLRLSK------DQSAILEETF 145
Query: 159 KESPHPDEKQRQQLSKQLGLAPRQVKFWFQNRRTQIKAIQERHENSLLKSEIEKLREKNK 218
K+ + KQ+Q L+KQLGL RQV+ WFQNRR + K Q + L+ E L E+N+
Sbjct: 146 KDHSTLNPKQKQALAKQLGLRARQVEVWFQNRRARTKLKQTEVDCEFLRRCCENLTEENR 205
Query: 219 TLRETINK 226
L++ + +
Sbjct: 206 RLQKEVTE 213
>ATH4_ARATH (P92953) Homeobox-leucine zipper protein ATHB-4 (HD-ZIP
protein ATHB-4)
Length = 318
Score = 71.2 bits (173), Expect = 4e-12
Identities = 47/158 (29%), Positives = 77/158 (47%), Gaps = 5/158 (3%)
Query: 69 SISNMEVEEGEEGSTIGGERVEEISSEYSGPAKSKSIDEYEGDELEDDEGEDDEGDGDGD 128
S++ +++EE + V +S A ++ DE E + G G D D
Sbjct: 95 SVAVVDLEEEAAVVSSPNSAVSSLSGNKRDLAVARGGDENEAERASCSRGGGSGGSDDED 154
Query: 129 GDGVNKNKKKKRKKYHRHTSEQIRVMEALFKESPHPDEKQRQQLSKQLGLAPRQVKFWFQ 188
G + ++KK R + +Q V+E FKE + KQ+ L+KQL L RQV+ WFQ
Sbjct: 155 GGNGDGSRKKLRL-----SKDQALVLEETFKEHSTLNPKQKLALAKQLNLRARQVEVWFQ 209
Query: 189 NRRTQIKAIQERHENSLLKSEIEKLREKNKTLRETINK 226
NRR + K Q + LK + L E+N+ L++ +++
Sbjct: 210 NRRARTKLKQTEVDCEYLKRCCDNLTEENRRLQKEVSE 247
>HAT2_ARATH (P46601) Homeobox-leucine zipper protein HAT2 (HD-ZIP
protein 2)
Length = 283
Score = 68.9 bits (167), Expect = 2e-11
Identities = 48/141 (34%), Positives = 70/141 (49%), Gaps = 11/141 (7%)
Query: 82 STIGGERVEEISSEYSGPAKSKSIDEYEGDELEDDEGEDDEGDGDGDGDGVNKNKKKKRK 141
STI G+R E +G + DE+ D G G D + DG ++KK R
Sbjct: 81 STISGKRSEREGISGTGVGSGD-----DHDEITPDRGYS-RGTSDEEEDGGETSRKKLRL 134
Query: 142 KYHRHTSEQIRVMEALFKESPHPDEKQRQQLSKQLGLAPRQVKFWFQNRRTQIKAIQERH 201
+ +Q +E FKE + KQ+ L+K+L L RQV+ WFQNRR + K Q
Sbjct: 135 -----SKDQSAFLEETFKEHNTLNPKQKLALAKKLNLTARQVEVWFQNRRARTKLKQTEV 189
Query: 202 ENSLLKSEIEKLREKNKTLRE 222
+ LK +EKL E+N+ L++
Sbjct: 190 DCEYLKRCVEKLTEENRRLQK 210
>HAT1_ARATH (P46600) Homeobox-leucine zipper protein HAT1 (HD-ZIP
protein 1)
Length = 282
Score = 67.0 bits (162), Expect = 7e-11
Identities = 47/136 (34%), Positives = 67/136 (48%), Gaps = 9/136 (6%)
Query: 92 ISSEYSGPAKSKSIDEYEGDELEDD-----EGEDDEGDGDGDGDGVNKNKKKKRKKYHRH 146
ISS SG +S + G DD + G D + D + +KK R
Sbjct: 84 ISSTVSGKRRSTEREGTSGGGCGDDLDITLDRSSSRGTSDEEEDYGGETCRKKL----RL 139
Query: 147 TSEQIRVMEALFKESPHPDEKQRQQLSKQLGLAPRQVKFWFQNRRTQIKAIQERHENSLL 206
+ +Q V+E FKE + KQ+ L+K+LGL RQV+ WFQNRR + K Q + L
Sbjct: 140 SKDQSAVLEDTFKEHNTLNPKQKLALAKKLGLTARQVEVWFQNRRARTKLKQTEVDCEYL 199
Query: 207 KSEIEKLREKNKTLRE 222
K +EKL E+N+ L +
Sbjct: 200 KRCVEKLTEENRRLEK 215
>HAT9_ARATH (P46603) Homeobox-leucine zipper protein HAT9
(Homeodomain-leucine zipper protein HAT9) (Homeodomain
transcription factor HAT9) (HD-ZIP protein 9)
Length = 274
Score = 66.6 bits (161), Expect = 9e-11
Identities = 45/135 (33%), Positives = 68/135 (50%), Gaps = 6/135 (4%)
Query: 92 ISSEYSGPAKSKSIDEYEGDELEDDEGEDDEGDGDGDGDGVNKNKKKKRKKYHRHTSEQI 151
+SS SG + D E E++ E D D +G++ KK R T +Q
Sbjct: 69 VSSFSSGRVVKRERDGGEESPEEEEMTERVISDYHEDEEGISARKKL------RLTKQQS 122
Query: 152 RVMEALFKESPHPDEKQRQQLSKQLGLAPRQVKFWFQNRRTQIKAIQERHENSLLKSEIE 211
++E FK+ + KQ+Q L++QL L PRQV+ WFQNRR + K Q + LK E
Sbjct: 123 ALLEESFKDHSTLNPKQKQVLARQLNLRPRQVEVWFQNRRARTKLKQTEVDCEFLKKCCE 182
Query: 212 KLREKNKTLRETINK 226
L ++N L++ I +
Sbjct: 183 TLADENIRLQKEIQE 197
>HT22_ARATH (P46604) Homeobox-leucine zipper protein HAT22 (HD-ZIP
protein 22)
Length = 278
Score = 65.1 bits (157), Expect = 3e-10
Identities = 48/140 (34%), Positives = 70/140 (49%), Gaps = 15/140 (10%)
Query: 92 ISSEYSGPAKSKSIDEYEGDELEDDEGEDDE-------GDGDGDGDGVNKNKKKKRKKYH 144
ISS SG K + E G + E++ E E D D +GV+ KK
Sbjct: 77 ISSFSSGRVKRER--EISGGDGEEEAEETTERVVCSRVSDDHDDEEGVSARKKL------ 128
Query: 145 RHTSEQIRVMEALFKESPHPDEKQRQQLSKQLGLAPRQVKFWFQNRRTQIKAIQERHENS 204
R T +Q ++E FK + KQ+Q L++QL L PRQV+ WFQNRR + K Q +
Sbjct: 129 RLTKQQSALLEDNFKLHSTLNPKQKQALARQLNLRPRQVEVWFQNRRARTKLKQTEVDCE 188
Query: 205 LLKSEIEKLREKNKTLRETI 224
LK E L ++N+ L++ +
Sbjct: 189 FLKKCCETLTDENRRLQKEL 208
>PIX1_XENLA (Q9W751) Pituitary homeobox 1 (X-PITX-1) (xPitx1)
Length = 305
Score = 64.7 bits (156), Expect = 4e-10
Identities = 33/116 (28%), Positives = 67/116 (57%), Gaps = 2/116 (1%)
Query: 93 SSEYSGPAKSKSIDEYEGDELEDDEGEDDEGDGDGDGDGVNKNKKKKRKKYHRHTSEQIR 152
SSE P + + + + + E + + +G+ DG+GD +K KK++R++ H TS+Q++
Sbjct: 34 SSEARDPMDNSASESSDTEIAEKERTGEPKGE-DGNGDDPSKKKKQRRQRTH-FTSQQLQ 91
Query: 153 VMEALFKESPHPDEKQRQQLSKQLGLAPRQVKFWFQNRRTQIKAIQERHENSLLKS 208
+EA F+ + +PD R++++ L +V+ WF+NRR + + + + L K+
Sbjct: 92 ELEATFQRNRYPDMSMREEIAVWTNLTEARVRVWFKNRRAKWRKRERNQQMDLCKN 147
>HT14_ARATH (P46665) Homeobox-leucine zipper protein HAT14 (HD-ZIP
protein 14)
Length = 225
Score = 64.3 bits (155), Expect = 5e-10
Identities = 49/157 (31%), Positives = 71/157 (45%), Gaps = 10/157 (6%)
Query: 76 EEGEEGSTIGGERVEEISSEYSGPAKSKSIDEYEGDELEDDEGEDDEGD------GDGDG 129
EE EE + V S S I Y + + DDE + + D
Sbjct: 11 EEEEEEEAVPSMSVSPPDSVTSSFQLDFGIKSYGYERRSNKRDIDDEVERSASRASNEDN 70
Query: 130 DGVNKNKKKKRKKYHRHTSEQIRVMEALFKESPHPDEKQRQQLSKQLGLAPRQVKFWFQN 189
D N + +KK R + +Q +E FKE + KQ+ L+KQL L PRQV+ WFQN
Sbjct: 71 DDENGSTRKKL----RLSKDQSAFLEDSFKEHSTLNPKQKIALAKQLNLRPRQVEVWFQN 126
Query: 190 RRTQIKAIQERHENSLLKSEIEKLREKNKTLRETINK 226
RR + K Q + LK E L E+N+ L++ + +
Sbjct: 127 RRARTKLKQTEVDCEYLKRCCESLTEENRRLQKEVKE 163
>SHX2_RAT (O35750) Short stature homeobox protein 2 (Paired family
homeodomain protein Prx3) (Fragment)
Length = 237
Score = 63.9 bits (154), Expect = 6e-10
Identities = 42/135 (31%), Positives = 66/135 (48%), Gaps = 27/135 (20%)
Query: 73 MEVEEGEEGSTIGGERVEEISSEYSGPAKSKSIDEYEGDELEDDEGEDDEGDGDGDGDGV 132
+++ E G R+ E+S E D ED +G +DEG
Sbjct: 3 LDMGAAERSREPGSPRLTEVSPELK-------------DRKEDAKGMEDEG--------- 40
Query: 133 NKNKKKKRKKYHRHTSEQIRVMEALFKESPHPDEKQRQQLSKQLGLAPRQVKFWFQNRRT 192
+ K K+R+ T EQ+ +E LF E+ +PD R++LS++LGL+ +V+ WFQNRR
Sbjct: 41 -QTKIKQRRSRTNFTLEQLNELERLFDETHYPDAFMREELSQRLGLSEARVQVWFQNRRA 99
Query: 193 QIKAIQERHENSLLK 207
+ + + EN L K
Sbjct: 100 KCR----KQENQLHK 110
>HAT7_ARATH (Q00466) Homeobox-leucine zipper protein HAT7 (HD-ZIP
protein 7) (HD-ZIP protein ATHB-3)
Length = 251
Score = 63.9 bits (154), Expect = 6e-10
Identities = 53/194 (27%), Positives = 84/194 (42%), Gaps = 29/194 (14%)
Query: 114 EDDEGEDDEGDGDGDGDGVNKNKKKKRKKYHRHTSEQIRVMEALFKESPHPDEKQRQQLS 173
+D GE+D DG + + KK R EQ+R +E F+ + +++ QL+
Sbjct: 31 QDQVGEEDNLSDDGSHMMLGEKKK-------RLNLEQVRALEKSFELGNKLEPERKMQLA 83
Query: 174 KQLGLAPRQVKFWFQNRRTQIKAIQERHENSLLKSEIEKLREKNKTLRETINKACCPNCG 233
K LGL PRQ+ WFQNRR + K Q + LK + + L+ N +L K
Sbjct: 84 KALGLQPRQIAIWFQNRRARWKTKQLERDYDSLKKQFDVLKSDNDSLLAHNKKL------ 137
Query: 234 VPTTNRDGTMATEEQQLRIENAKLKAEVERLRAALGKYASGTMSPSCSTSHDQENIKSSL 293
+ ++ R E+AK+K E +A + S + ST ++ N S
Sbjct: 138 ------HAELVALKKHDRKESAKIKRE----------FAEASWSNNGSTENNHNNNSSDA 181
Query: 294 DFYTGIFCLDESRI 307
+ + I L S I
Sbjct: 182 NHVSMIKDLFPSSI 195
>ATH7_ARATH (P46897) Homeobox-leucine zipper protein ATHB-7
(Homeodomain transcription factor ATHB-7) (HD-ZIP
protein ATHB-7)
Length = 258
Score = 63.5 bits (153), Expect = 8e-10
Identities = 44/156 (28%), Positives = 74/156 (47%), Gaps = 25/156 (16%)
Query: 133 NKNKKKKRKKYHRHTSEQIRVMEALFKESPHPDEKQRQQLSKQLGLAPRQVKFWFQNRRT 192
N NK +R R + EQI+ +E +F+ + +++ QL+++LGL PRQV WFQN+R
Sbjct: 27 NHNKNNQR----RFSDEQIKSLEMMFESETRLEPRKKVQLARELGLQPRQVAIWFQNKRA 82
Query: 193 QIKAIQERHENSLLKSEIEKLREKNKTLRETINKACCPNCGVPTTNRDGTMATEEQQLRI 252
+ K+ Q E ++L+ + L + ++L++ A + R+
Sbjct: 83 RWKSKQLETEYNILRQNYDNLASQFESLKKE------------------KQALVSELQRL 124
Query: 253 ENAKLKAEVERLRAALGKYASGTMSPSCSTSHDQEN 288
+ A K E R G A +S ST H+ EN
Sbjct: 125 KEATQKKTQEEERQCSGDQAVVALS---STHHESEN 157
>HM1D_DROAN (P22544) Homeobox protein B-H1
Length = 606
Score = 62.8 bits (151), Expect = 1e-09
Identities = 44/155 (28%), Positives = 78/155 (49%), Gaps = 18/155 (11%)
Query: 46 ALSLSLAGIFRHGGGAAAEGEGGSISNMEVEEGEEGSTIGGERVEEISSEYSGPAK---- 101
A S + AG GGG G GG+ + E+++ + E E+ S+ G A
Sbjct: 242 AASAAAAG----GGGGGGLGVGGAPAGAELDDSSDYH----EENEDCDSDEGGSAGGGGG 293
Query: 102 -SKSIDEYE--GDELEDDEGEDDEGDGDGDGDGVNKNKKKKRKKYHRHTSEQIRVMEALF 158
S +D++ + +DD+G + D G++K ++K R + T Q++ +E F
Sbjct: 294 GSNHMDDHSVCSNGGKDDDGNSIKSGSTSDMSGLSKKQRKARTAF---TDHQLQTLEKSF 350
Query: 159 KESPHPDEKQRQQLSKQLGLAPRQVKFWFQNRRTQ 193
+ + ++RQ+L+ +L L+ QVK W+QNRRT+
Sbjct: 351 ERQKYLSVQERQELAHKLDLSDCQVKTWYQNRRTK 385
>HMN2_DROME (P22808) Homeobox protein vnd (Ventral nervous system
defective protein) (Homeobox protein NK-2)
Length = 723
Score = 62.4 bits (150), Expect = 2e-09
Identities = 40/141 (28%), Positives = 60/141 (42%), Gaps = 3/141 (2%)
Query: 60 GAAAEGEGGSISNMEVEEGEEGSTIGGERVEEISSEYSGPAKSKSIDEYEGDELEDDEGE 119
G++ G G +T + +E A S D D +E+D +
Sbjct: 465 GSSGNGSAGGAPTAHALHNNNNNTTNNNN-HSLKAEGINGAGSGHDDSLNEDGIEEDIDD 523
Query: 120 DDEGDGDGDGD--GVNKNKKKKRKKYHRHTSEQIRVMEALFKESPHPDEKQRQQLSKQLG 177
D+ DG G GD G + KKRK+ T Q +E F++ + +R+ L+ +
Sbjct: 524 VDDADGSGGGDANGSDGLPNKKRKRRVLFTKAQTYELERRFRQQRYLSAPEREHLASLIR 583
Query: 178 LAPRQVKFWFQNRRTQIKAIQ 198
L P QVK WFQN R + K Q
Sbjct: 584 LTPTQVKIWFQNHRYKTKRAQ 604
>HE2A_BRARE (P09015) Homeobox protein engrailed-2a (Zf-En-2) (Eng2)
Length = 265
Score = 62.4 bits (150), Expect = 2e-09
Identities = 44/161 (27%), Positives = 73/161 (45%), Gaps = 24/161 (14%)
Query: 57 HGGGAAAEGEGGSISNMEVEEGEEGSTIGGERVEEISSEYSGPAKSKSIDEYEGDELEDD 116
H + G+ GS + EE T G + EI SE + +++D+ G E +
Sbjct: 74 HNPTGPSTGQVGS--TVPAEEASTTHTSSGGKEAEIESEEPLKPRGENVDQCLGSESDSS 131
Query: 117 EGEDDEGDGDG-----------------DGDGVNKNKKKKRKKYHRH-----TSEQIRVM 154
+ + G G G K KKK K + T+EQ++ +
Sbjct: 132 QSNSNGQTGQGMLWPAWVYCTRYSDRPSSGPRSRKPKKKAASKEDKRPRTAFTAEQLQRL 191
Query: 155 EALFKESPHPDEKQRQQLSKQLGLAPRQVKFWFQNRRTQIK 195
+A F+ + + E++RQ L+++LGL Q+K WFQN+R +IK
Sbjct: 192 KAEFQTNRYLTEQRRQSLAQELGLNESQIKIWFQNKRAKIK 232
>LMXB_MOUSE (O88609) LIM homeobox transcription factor 1 beta
(LIM/homeobox protein LMX1B) (LIM-homeobox protein 1.2)
(LMX-1.2)
Length = 372
Score = 61.2 bits (147), Expect = 4e-09
Identities = 38/131 (29%), Positives = 73/131 (55%), Gaps = 5/131 (3%)
Query: 87 ERVEEISSEYSGPAKSKSI-DEYEGDELEDDEGEDDEGDGDGDGDGVNKNKKKKRKKYHR 145
E+ +++ S S P +S S+ E E +++ +G+ + G GD DG + + K+ +
Sbjct: 147 EKEKDLLSSVS-PDESDSVKSEDEDGDMKPAKGQGSQSKGSGD-DGKDPRRPKRPRTIL- 203
Query: 146 HTSEQIRVMEALFKESPHPDEKQRQQLSKQLGLAPRQVKFWFQNRRTQIKAIQERHENSL 205
T++Q R +A F+ S P K R+ L+ + GL+ R V+ WFQN+R ++K + RH+
Sbjct: 204 -TTQQRRAFKASFEVSSKPCRKVRETLAAETGLSVRVVQVWFQNQRAKMKKLARRHQQQQ 262
Query: 206 LKSEIEKLREK 216
+ ++L ++
Sbjct: 263 EQQNSQRLGQE 273
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.309 0.129 0.360
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 44,161,452
Number of Sequences: 164201
Number of extensions: 2010497
Number of successful extensions: 14340
Number of sequences better than 10.0: 1439
Number of HSP's better than 10.0 without gapping: 1034
Number of HSP's successfully gapped in prelim test: 426
Number of HSP's that attempted gapping in prelim test: 10378
Number of HSP's gapped (non-prelim): 2696
length of query: 365
length of database: 59,974,054
effective HSP length: 112
effective length of query: 253
effective length of database: 41,583,542
effective search space: 10520636126
effective search space used: 10520636126
T: 11
A: 40
X1: 16 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.7 bits)
S2: 66 (30.0 bits)
Medicago: description of AC149489.2


