

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC149471.8 + phase: 0
(229 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
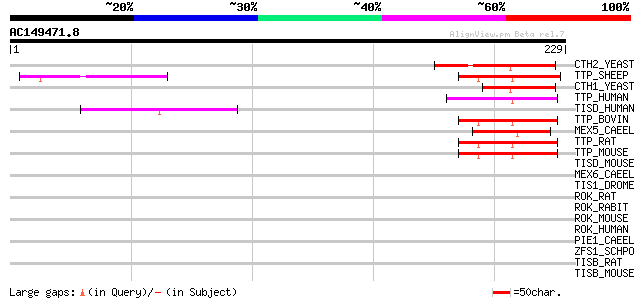
Score E
Sequences producing significant alignments: (bits) Value
CTH2_YEAST (P47977) Zinc finger protein CTH2 (YTIS11 protein) 48 2e-05
TTP_SHEEP (Q6S9E0) Tristetraproline (TTP) (Zinc finger protein 3... 47 5e-05
CTH1_YEAST (P47976) Zinc finger protein CTH1 45 2e-04
TTP_HUMAN (P26651) Tristetraproline (TTP) (Zinc finger protein 3... 44 2e-04
TISD_HUMAN (P47974) Butyrate response factor 2 (TIS11D protein) ... 44 2e-04
TTP_BOVIN (P53781) Tristetraproline (TTP) (Zinc finger protein 3... 44 3e-04
MEX5_CAEEL (Q9XUB2) Zinc finger protein mex-5 44 4e-04
TTP_RAT (P47973) Tristetraproline (TTP) (Zinc finger protein 36)... 43 7e-04
TTP_MOUSE (P22893) Tristetraproline (TTP) (Zinc finger protein 3... 43 7e-04
TISD_MOUSE (P23949) Butyrate response factor 2 (TIS11D protein) 42 0.001
MEX6_CAEEL (Q09436) Zinc finger protein mex-6 42 0.001
TIS1_DROME (P47980) TIS11 protein (dTIS11) 42 0.001
ROK_RAT (P61980) Heterogeneous nuclear ribonucleoprotein K (dC-s... 42 0.001
ROK_RABIT (O19049) Heterogeneous nuclear ribonucleoprotein K (hn... 42 0.001
ROK_MOUSE (P61979) Heterogeneous nuclear ribonucleoprotein K 42 0.001
ROK_HUMAN (P61978) Heterogeneous nuclear ribonucleoprotein K (hn... 42 0.001
PIE1_CAEEL (Q94131) Pharynx and intestine in excess protein 1 (P... 41 0.002
ZFS1_SCHPO (P47979) Zinc-finger protein zfs1 41 0.003
TISB_RAT (P17431) Butyrate response factor 1 (TIS11B protein) (E... 41 0.003
TISB_MOUSE (P23950) Butyrate response factor 1 (TIS11B protein) 41 0.003
>CTH2_YEAST (P47977) Zinc finger protein CTH2 (YTIS11 protein)
Length = 285
Score = 47.8 bits (112), Expect = 2e-05
Identities = 24/51 (47%), Positives = 33/51 (64%), Gaps = 3/51 (5%)
Query: 176 SVSGPPGKNITSQTSAPANNFKTKLCENFT-KGSCTFGERCHFAHGTDELR 225
S + P K+ +T P +KT+LCE+FT KGSC +G +C FAHG EL+
Sbjct: 152 SSAQPKVKSQVQET--PKQLYKTELCESFTLKGSCPYGSKCQFAHGLGELK 200
Score = 37.0 bits (84), Expect = 0.039
Identities = 15/29 (51%), Positives = 16/29 (54%)
Query: 33 KTRLCNKFNTAEGCKFGDKCHFAHGEWEL 61
KT LC F C +G KC FAHG EL
Sbjct: 171 KTELCESFTLKGSCPYGSKCQFAHGLGEL 199
Score = 36.6 bits (83), Expect = 0.050
Identities = 16/29 (55%), Positives = 19/29 (65%), Gaps = 1/29 (3%)
Query: 195 NFKTKLCENFTK-GSCTFGERCHFAHGTD 222
NF+TK C N+ K G C +G RC F HG D
Sbjct: 207 NFRTKPCVNWEKLGYCPYGRRCCFKHGDD 235
>TTP_SHEEP (Q6S9E0) Tristetraproline (TTP) (Zinc finger protein 36
homolog) (Zfp-36)
Length = 325
Score = 46.6 bits (109), Expect = 5e-05
Identities = 21/45 (46%), Positives = 31/45 (68%), Gaps = 3/45 (6%)
Query: 186 TSQTSAP--ANNFKTKLCENFTK-GSCTFGERCHFAHGTDELRKP 227
TS T+ P ++ +KT+LC F++ G C +G +C FAHG ELR+P
Sbjct: 90 TSPTATPTTSSRYKTELCRTFSESGRCRYGAKCQFAHGLGELRQP 134
Score = 42.7 bits (99), Expect = 7e-04
Identities = 24/65 (36%), Positives = 34/65 (51%), Gaps = 6/65 (9%)
Query: 5 GSSPAIP----PIGRNPNVPQSFPDGSSPPVAKTRLCNKFNTAEGCKFGDKCHFAHGEWE 60
G +P P + +P P + P SS KT LC F+ + C++G KC FAHG E
Sbjct: 73 GFAPLAPRPSSELSPSPTSPTATPTTSSR--YKTELCRTFSESGRCRYGAKCQFAHGLGE 130
Query: 61 LGRPT 65
L +P+
Sbjct: 131 LRQPS 135
Score = 39.7 bits (91), Expect = 0.006
Identities = 23/67 (34%), Positives = 27/67 (39%), Gaps = 2/67 (2%)
Query: 27 SSPPVAKTRLCNKFNTAEGCKFGDKCHFAHGEWELGRPTVPAYEDTRAMGQMQSSSVGGR 86
S P KT LC+KF C +G +CHF H E P + S GR
Sbjct: 135 SRHPKYKTELCHKFYLQGRCPYGSRCHFIHNPSE--DLAAPGHPHVLRQSISFSGLPSGR 192
Query: 87 IEPPPPA 93
PPPA
Sbjct: 193 RTSPPPA 199
Score = 37.7 bits (86), Expect = 0.023
Identities = 16/35 (45%), Positives = 23/35 (65%), Gaps = 2/35 (5%)
Query: 196 FKTKLCENF-TKGSCTFGERCHFAHG-TDELRKPG 228
+KT+LC F +G C +G RCHF H +++L PG
Sbjct: 140 YKTELCHKFYLQGRCPYGSRCHFIHNPSEDLAAPG 174
>CTH1_YEAST (P47976) Zinc finger protein CTH1
Length = 325
Score = 44.7 bits (104), Expect = 2e-04
Identities = 18/31 (58%), Positives = 25/31 (80%), Gaps = 1/31 (3%)
Query: 196 FKTKLCENFT-KGSCTFGERCHFAHGTDELR 225
+KT+LCE+FT KG C +G +C FAHG +EL+
Sbjct: 205 YKTELCESFTIKGYCKYGNKCQFAHGLNELK 235
Score = 39.3 bits (90), Expect = 0.008
Identities = 18/42 (42%), Positives = 22/42 (51%)
Query: 20 PQSFPDGSSPPVAKTRLCNKFNTAEGCKFGDKCHFAHGEWEL 61
P P + + KT LC F CK+G+KC FAHG EL
Sbjct: 193 PLQLPQLVNKTLYKTELCESFTIKGYCKYGNKCQFAHGLNEL 234
Score = 39.3 bits (90), Expect = 0.008
Identities = 16/32 (50%), Positives = 24/32 (75%), Gaps = 1/32 (3%)
Query: 193 ANNFKTKLCENFTK-GSCTFGERCHFAHGTDE 223
+NN++TK C N++K G C +G+RC F HG D+
Sbjct: 240 SNNYRTKPCINWSKLGYCPYGKRCCFKHGDDK 271
>TTP_HUMAN (P26651) Tristetraproline (TTP) (Zinc finger protein 36
homolog) (Zfp-36) (TIS11A protein) (TIS11) (Growth
factor-inducible nuclear protein NUP475) (G0/G1 switch
regulatory protein 24)
Length = 326
Score = 44.3 bits (103), Expect = 2e-04
Identities = 19/47 (40%), Positives = 28/47 (59%), Gaps = 1/47 (2%)
Query: 181 PGKNITSQTSAPANNFKTKLCENFTK-GSCTFGERCHFAHGTDELRK 226
P + TS + +KT+LC F++ G C +G +C FAHG ELR+
Sbjct: 89 PSPTSPTATSTTPSRYKTELCRTFSESGRCRYGAKCQFAHGLGELRQ 135
Score = 42.4 bits (98), Expect = 0.001
Identities = 22/59 (37%), Positives = 31/59 (52%), Gaps = 2/59 (3%)
Query: 5 GSSPAIPPIGRNPNVPQSFPDGSS--PPVAKTRLCNKFNTAEGCKFGDKCHFAHGEWEL 61
G +P P +G + + P +S P KT LC F+ + C++G KC FAHG EL
Sbjct: 75 GFAPLAPRLGPELSPSPTSPTATSTTPSRYKTELCRTFSESGRCRYGAKCQFAHGLGEL 133
Score = 40.8 bits (94), Expect = 0.003
Identities = 24/64 (37%), Positives = 27/64 (41%), Gaps = 3/64 (4%)
Query: 30 PVAKTRLCNKFNTAEGCKFGDKCHFAHGEWE-LGRPTVPAYEDTRAMGQMQSSSVGGRIE 88
P KT LC+KF C +G +CHF H E L P P R G R
Sbjct: 140 PKYKTELCHKFYLQGRCPYGSRCHFIHNPSEDLAAPGHPPV--LRQSISFSGLPSGRRTS 197
Query: 89 PPPP 92
PPPP
Sbjct: 198 PPPP 201
Score = 37.7 bits (86), Expect = 0.023
Identities = 16/35 (45%), Positives = 23/35 (65%), Gaps = 2/35 (5%)
Query: 196 FKTKLCENF-TKGSCTFGERCHFAHG-TDELRKPG 228
+KT+LC F +G C +G RCHF H +++L PG
Sbjct: 142 YKTELCHKFYLQGRCPYGSRCHFIHNPSEDLAAPG 176
>TISD_HUMAN (P47974) Butyrate response factor 2 (TIS11D protein)
(EGF-response factor 2) (ERF-2)
Length = 492
Score = 44.3 bits (103), Expect = 2e-04
Identities = 25/66 (37%), Positives = 31/66 (46%), Gaps = 1/66 (1%)
Query: 30 PVAKTRLCNKFNTAEGCKFGDKCHFAHGEWE-LGRPTVPAYEDTRAMGQMQSSSVGGRIE 88
P KT LC F+T C +G +CHF H E P+ A D RA G + +G E
Sbjct: 190 PKYKTELCRTFHTIGFCPYGPRCHFIHNADERRPAPSGGASGDLRAFGTRDALHLGFPRE 249
Query: 89 PPPPAH 94
P P H
Sbjct: 250 PRPKLH 255
Score = 41.2 bits (95), Expect = 0.002
Identities = 17/34 (50%), Positives = 24/34 (70%), Gaps = 1/34 (2%)
Query: 193 ANNFKTKLCENFTK-GSCTFGERCHFAHGTDELR 225
+ +KT+LC F + G+C +GE+C FAHG ELR
Sbjct: 151 STRYKTELCRPFEESGTCKYGEKCQFAHGFHELR 184
Score = 40.0 bits (92), Expect = 0.005
Identities = 17/31 (54%), Positives = 20/31 (63%), Gaps = 1/31 (3%)
Query: 196 FKTKLCENF-TKGSCTFGERCHFAHGTDELR 225
+KT+LC F T G C +G RCHF H DE R
Sbjct: 192 YKTELCRTFHTIGFCPYGPRCHFIHNADERR 222
Score = 39.7 bits (91), Expect = 0.006
Identities = 16/29 (55%), Positives = 19/29 (65%)
Query: 33 KTRLCNKFNTAEGCKFGDKCHFAHGEWEL 61
KT LC F + CK+G+KC FAHG EL
Sbjct: 155 KTELCRPFEESGTCKYGEKCQFAHGFHEL 183
>TTP_BOVIN (P53781) Tristetraproline (TTP) (Zinc finger protein 36
homolog) (Zfp-36) (TIS11A protein) (TIS11)
Length = 324
Score = 43.9 bits (102), Expect = 3e-04
Identities = 20/44 (45%), Positives = 30/44 (67%), Gaps = 3/44 (6%)
Query: 186 TSQTSAP--ANNFKTKLCENFTK-GSCTFGERCHFAHGTDELRK 226
TS T+ P ++ +KT+LC F++ G C +G +C FAHG ELR+
Sbjct: 90 TSPTATPTTSSRYKTELCRTFSESGRCRYGAKCQFAHGLGELRQ 133
Score = 42.0 bits (97), Expect = 0.001
Identities = 23/62 (37%), Positives = 29/62 (46%), Gaps = 11/62 (17%)
Query: 11 PPIGRNPNVPQSFPDGSSPPVA-----------KTRLCNKFNTAEGCKFGDKCHFAHGEW 59
PP G P P+ D S P + KT LC F+ + C++G KC FAHG
Sbjct: 70 PPPGFAPLAPRPSSDWSPSPTSPTATPTTSSRYKTELCRTFSESGRCRYGAKCQFAHGLG 129
Query: 60 EL 61
EL
Sbjct: 130 EL 131
Score = 40.4 bits (93), Expect = 0.003
Identities = 27/78 (34%), Positives = 33/78 (41%), Gaps = 4/78 (5%)
Query: 27 SSPPVAKTRLCNKFNTAEGCKFGDKCHFAHGEWELGRPTVPAYEDTRAMGQMQSSSVGGR 86
S P KT LC+KF C +G +CHF H E P + S GR
Sbjct: 135 SRHPKYKTELCHKFYLQGRCPYGSRCHFIHNPSE--DLAAPGHPHVLRQSISFSGLPSGR 192
Query: 87 IEPPPPAHGAAAGFGVSA 104
PPPA + AG VS+
Sbjct: 193 RTSPPPA--SLAGPSVSS 208
Score = 37.7 bits (86), Expect = 0.023
Identities = 16/35 (45%), Positives = 23/35 (65%), Gaps = 2/35 (5%)
Query: 196 FKTKLCENF-TKGSCTFGERCHFAHG-TDELRKPG 228
+KT+LC F +G C +G RCHF H +++L PG
Sbjct: 140 YKTELCHKFYLQGRCPYGSRCHFIHNPSEDLAAPG 174
>MEX5_CAEEL (Q9XUB2) Zinc finger protein mex-5
Length = 468
Score = 43.5 bits (101), Expect = 4e-04
Identities = 18/35 (51%), Positives = 23/35 (65%), Gaps = 3/35 (8%)
Query: 192 PANNFKTKLCENFTKGS---CTFGERCHFAHGTDE 223
P N +KTKLC+NF +G C +G RC F H TD+
Sbjct: 311 PNNKYKTKLCKNFARGGTGFCPYGLRCEFVHPTDK 345
Score = 39.3 bits (90), Expect = 0.008
Identities = 38/142 (26%), Positives = 54/142 (37%), Gaps = 7/142 (4%)
Query: 90 PPPAHGAAAGFGVSATATVSINAT--LAGAIIGKNDVNSKQICHITGAKLSIREHDSDP- 146
PP G +S+T T + ++ L ++ + + +I + EHD P
Sbjct: 162 PPQQQPRQVGVEISSTRTAPLTSSTPLPTSLEYETVQRDNRNRNIQFRYHRVMEHDELPI 221
Query: 147 -NLRNIELEGSFDQIKQASAMVHDLILNVSSVSGPPGKNITSQTSAPANNFKTKLCENFT 205
+ I L+ D A H + G G I S N+KT+LC
Sbjct: 222 DEISKITLDNHNDDTMSAEKENH-FHEHRGEKFGRRGFPIPETDSQQPPNYKTRLCMMHA 280
Query: 206 KG--SCTFGERCHFAHGTDELR 225
G C G RC FAHG ELR
Sbjct: 281 SGIKPCDMGARCKFAHGLKELR 302
Score = 35.4 bits (80), Expect = 0.11
Identities = 19/45 (42%), Positives = 22/45 (48%), Gaps = 1/45 (2%)
Query: 25 DGSSPPVAKTRLCNKFNTA-EGCKFGDKCHFAHGEWELGRPTVPA 68
D PP KTRLC + + C G +C FAHG EL PA
Sbjct: 264 DSQQPPNYKTRLCMMHASGIKPCDMGARCKFAHGLKELRATDAPA 308
>TTP_RAT (P47973) Tristetraproline (TTP) (Zinc finger protein 36)
(Zfp-36) (TIS11A protein) (TIS11)
Length = 320
Score = 42.7 bits (99), Expect = 7e-04
Identities = 19/44 (43%), Positives = 30/44 (68%), Gaps = 3/44 (6%)
Query: 186 TSQTSAP--ANNFKTKLCENFTK-GSCTFGERCHFAHGTDELRK 226
TS T+ P ++ +KT+LC +++ G C +G +C FAHG ELR+
Sbjct: 85 TSPTATPTTSSRYKTELCRTYSESGRCRYGAKCQFAHGPGELRQ 128
Score = 42.0 bits (97), Expect = 0.001
Identities = 22/62 (35%), Positives = 30/62 (47%), Gaps = 11/62 (17%)
Query: 11 PPIGRNPNVPQSFPDGSSPPVA-----------KTRLCNKFNTAEGCKFGDKCHFAHGEW 59
PP G P P+ P+ S P + KT LC ++ + C++G KC FAHG
Sbjct: 65 PPPGFAPLAPRPGPELSPSPTSPTATPTTSSRYKTELCRTYSESGRCRYGAKCQFAHGPG 124
Query: 60 EL 61
EL
Sbjct: 125 EL 126
Score = 41.2 bits (95), Expect = 0.002
Identities = 25/74 (33%), Positives = 29/74 (38%), Gaps = 3/74 (4%)
Query: 20 PQSFPDGSSPPVAKTRLCNKFNTAEGCKFGDKCHFAHGEWE-LGRPTVPAYEDTRAMGQM 78
P + P KT LC+KF C +G +CHF H E L P P R
Sbjct: 123 PGELRQANRHPKYKTELCHKFYLQGRCPYGSRCHFIHNPTEDLALPGQP--HVLRQSISF 180
Query: 79 QSSSVGGRIEPPPP 92
G R PPPP
Sbjct: 181 SGLPSGRRTSPPPP 194
Score = 38.9 bits (89), Expect = 0.010
Identities = 17/35 (48%), Positives = 23/35 (65%), Gaps = 2/35 (5%)
Query: 196 FKTKLCENF-TKGSCTFGERCHFAHG-TDELRKPG 228
+KT+LC F +G C +G RCHF H T++L PG
Sbjct: 135 YKTELCHKFYLQGRCPYGSRCHFIHNPTEDLALPG 169
>TTP_MOUSE (P22893) Tristetraproline (TTP) (Zinc finger protein 36)
(Zfp-36) (TIS11A protein) (TIS11) (Growth
factor-inducible nuclear protein NUP475) (TPA induced
sequence 11)
Length = 319
Score = 42.7 bits (99), Expect = 7e-04
Identities = 19/44 (43%), Positives = 30/44 (68%), Gaps = 3/44 (6%)
Query: 186 TSQTSAP--ANNFKTKLCENFTK-GSCTFGERCHFAHGTDELRK 226
TS T+ P ++ +KT+LC +++ G C +G +C FAHG ELR+
Sbjct: 84 TSPTATPTTSSRYKTELCRTYSESGRCRYGAKCQFAHGLGELRQ 127
Score = 41.2 bits (95), Expect = 0.002
Identities = 22/62 (35%), Positives = 30/62 (47%), Gaps = 11/62 (17%)
Query: 11 PPIGRNPNVPQSFPDGSSPPVA-----------KTRLCNKFNTAEGCKFGDKCHFAHGEW 59
PP G P P+ P+ S P + KT LC ++ + C++G KC FAHG
Sbjct: 64 PPPGFAPLAPRPGPELSPSPTSPTATPTTSSRYKTELCRTYSESGRCRYGAKCQFAHGLG 123
Query: 60 EL 61
EL
Sbjct: 124 EL 125
Score = 40.4 bits (93), Expect = 0.003
Identities = 24/64 (37%), Positives = 27/64 (41%), Gaps = 3/64 (4%)
Query: 30 PVAKTRLCNKFNTAEGCKFGDKCHFAHGEWE-LGRPTVPAYEDTRAMGQMQSSSVGGRIE 88
P KT LC+KF C +G +CHF H E L P P R G R
Sbjct: 132 PKYKTELCHKFYLQGRCPYGSRCHFIHNPTEDLALPGQP--HVLRQSISFSGLPSGRRSS 189
Query: 89 PPPP 92
PPPP
Sbjct: 190 PPPP 193
Score = 38.9 bits (89), Expect = 0.010
Identities = 17/35 (48%), Positives = 23/35 (65%), Gaps = 2/35 (5%)
Query: 196 FKTKLCENF-TKGSCTFGERCHFAHG-TDELRKPG 228
+KT+LC F +G C +G RCHF H T++L PG
Sbjct: 134 YKTELCHKFYLQGRCPYGSRCHFIHNPTEDLALPG 168
>TISD_MOUSE (P23949) Butyrate response factor 2 (TIS11D protein)
Length = 367
Score = 42.4 bits (98), Expect = 0.001
Identities = 26/70 (37%), Positives = 31/70 (44%), Gaps = 7/70 (10%)
Query: 30 PVAKTRLCNKFNTAEGCKFGDKCHFAHGEWELGRPTVP-----AYEDTRAMGQMQSSSVG 84
P KT LC F+T C +G +CHF H E R P A D RA G + +G
Sbjct: 163 PKYKTELCRTFHTIGFCPYGPRCHFIHNADE--RRPAPSGGGGASGDLRAFGARDALHLG 220
Query: 85 GRIEPPPPAH 94
EP P H
Sbjct: 221 FAREPRPKLH 230
Score = 41.2 bits (95), Expect = 0.002
Identities = 17/34 (50%), Positives = 24/34 (70%), Gaps = 1/34 (2%)
Query: 193 ANNFKTKLCENFTK-GSCTFGERCHFAHGTDELR 225
+ +KT+LC F + G+C +GE+C FAHG ELR
Sbjct: 124 STRYKTELCRPFEESGTCKYGEKCQFAHGFHELR 157
Score = 40.0 bits (92), Expect = 0.005
Identities = 17/31 (54%), Positives = 20/31 (63%), Gaps = 1/31 (3%)
Query: 196 FKTKLCENF-TKGSCTFGERCHFAHGTDELR 225
+KT+LC F T G C +G RCHF H DE R
Sbjct: 165 YKTELCRTFHTIGFCPYGPRCHFIHNADERR 195
Score = 39.7 bits (91), Expect = 0.006
Identities = 16/29 (55%), Positives = 19/29 (65%)
Query: 33 KTRLCNKFNTAEGCKFGDKCHFAHGEWEL 61
KT LC F + CK+G+KC FAHG EL
Sbjct: 128 KTELCRPFEESGTCKYGEKCQFAHGFHEL 156
>MEX6_CAEEL (Q09436) Zinc finger protein mex-6
Length = 467
Score = 42.4 bits (98), Expect = 0.001
Identities = 18/37 (48%), Positives = 23/37 (61%), Gaps = 3/37 (8%)
Query: 189 TSAPANNFKTKLCENFTKGS---CTFGERCHFAHGTD 222
T P N +KTKLC+NF +G C +G RC F H +D
Sbjct: 311 TRYPNNKYKTKLCKNFARGGSGVCPYGLRCEFVHPSD 347
Score = 38.9 bits (89), Expect = 0.010
Identities = 18/34 (52%), Positives = 20/34 (57%), Gaps = 2/34 (5%)
Query: 194 NNFKTKLCENFTKG--SCTFGERCHFAHGTDELR 225
+NFKT+LC G C G RC FAHG ELR
Sbjct: 272 HNFKTRLCMTHAAGINPCALGARCKFAHGLKELR 305
Score = 30.0 bits (66), Expect = 4.7
Identities = 17/44 (38%), Positives = 19/44 (42%), Gaps = 1/44 (2%)
Query: 25 DGSSPPVAKTRLCNKFNTA-EGCKFGDKCHFAHGEWELGRPTVP 67
D P KTRLC C G +C FAHG EL +P
Sbjct: 267 DSQLPHNFKTRLCMTHAAGINPCALGARCKFAHGLKELRASDIP 310
>TIS1_DROME (P47980) TIS11 protein (dTIS11)
Length = 437
Score = 42.0 bits (97), Expect = 0.001
Identities = 17/29 (58%), Positives = 19/29 (64%)
Query: 33 KTRLCNKFNTAEGCKFGDKCHFAHGEWEL 61
KT LC F A CK+G+KC FAHG EL
Sbjct: 138 KTELCRPFEEAGECKYGEKCQFAHGSHEL 166
Score = 42.0 bits (97), Expect = 0.001
Identities = 17/33 (51%), Positives = 24/33 (72%), Gaps = 1/33 (3%)
Query: 194 NNFKTKLCENFTK-GSCTFGERCHFAHGTDELR 225
+ +KT+LC F + G C +GE+C FAHG+ ELR
Sbjct: 135 SRYKTELCRPFEEAGECKYGEKCQFAHGSHELR 167
Score = 37.7 bits (86), Expect = 0.023
Identities = 15/31 (48%), Positives = 19/31 (60%), Gaps = 1/31 (3%)
Query: 196 FKTKLCENF-TKGSCTFGERCHFAHGTDELR 225
+KT+ C F + G C +G RCHF H DE R
Sbjct: 175 YKTEYCRTFHSVGFCPYGPRCHFVHNADEAR 205
Score = 35.8 bits (81), Expect = 0.086
Identities = 18/55 (32%), Positives = 25/55 (44%), Gaps = 2/55 (3%)
Query: 30 PVAKTRLCNKFNTAEGCKFGDKCHFAHG--EWELGRPTVPAYEDTRAMGQMQSSS 82
P KT C F++ C +G +CHF H E + A T++ Q Q SS
Sbjct: 173 PKYKTEYCRTFHSVGFCPYGPRCHFVHNADEARAQQAAQAAKSSTQSQSQSQQSS 227
>ROK_RAT (P61980) Heterogeneous nuclear ribonucleoprotein K
(dC-stretch binding protein) (CSBP)
Length = 463
Score = 42.0 bits (97), Expect = 0.001
Identities = 27/75 (36%), Positives = 38/75 (50%)
Query: 101 GVSATATVSINATLAGAIIGKNDVNSKQICHITGAKLSIREHDSDPNLRNIELEGSFDQI 160
G T V+I LAG+IIGK KQI H +GA + I E R I + G+ DQI
Sbjct: 385 GPIITTQVTIPKDLAGSIIGKGGQRIKQIRHESGASIKIDEPLEGSEDRIITITGTQDQI 444
Query: 161 KQASAMVHDLILNVS 175
+ A ++ + + S
Sbjct: 445 QNAQYLLQNSVKQYS 459
>ROK_RABIT (O19049) Heterogeneous nuclear ribonucleoprotein K (hnRNP
K)
Length = 463
Score = 42.0 bits (97), Expect = 0.001
Identities = 27/75 (36%), Positives = 38/75 (50%)
Query: 101 GVSATATVSINATLAGAIIGKNDVNSKQICHITGAKLSIREHDSDPNLRNIELEGSFDQI 160
G T V+I LAG+IIGK KQI H +GA + I E R I + G+ DQI
Sbjct: 385 GPIITTQVTIPKDLAGSIIGKGGQRIKQIRHESGASIKIDEPLEGSEDRIITITGTQDQI 444
Query: 161 KQASAMVHDLILNVS 175
+ A ++ + + S
Sbjct: 445 QNAQYLLQNSVKQYS 459
>ROK_MOUSE (P61979) Heterogeneous nuclear ribonucleoprotein K
Length = 463
Score = 42.0 bits (97), Expect = 0.001
Identities = 27/75 (36%), Positives = 38/75 (50%)
Query: 101 GVSATATVSINATLAGAIIGKNDVNSKQICHITGAKLSIREHDSDPNLRNIELEGSFDQI 160
G T V+I LAG+IIGK KQI H +GA + I E R I + G+ DQI
Sbjct: 385 GPIITTQVTIPKDLAGSIIGKGGQRIKQIRHESGASIKIDEPLEGSEDRIITITGTQDQI 444
Query: 161 KQASAMVHDLILNVS 175
+ A ++ + + S
Sbjct: 445 QNAQYLLQNSVKQYS 459
>ROK_HUMAN (P61978) Heterogeneous nuclear ribonucleoprotein K (hnRNP
K) (Transformation up-regulated nuclear protein) (TUNP)
Length = 463
Score = 42.0 bits (97), Expect = 0.001
Identities = 27/75 (36%), Positives = 38/75 (50%)
Query: 101 GVSATATVSINATLAGAIIGKNDVNSKQICHITGAKLSIREHDSDPNLRNIELEGSFDQI 160
G T V+I LAG+IIGK KQI H +GA + I E R I + G+ DQI
Sbjct: 385 GPIITTQVTIPKDLAGSIIGKGGQRIKQIRHESGASIKIDEPLEGSEDRIITITGTQDQI 444
Query: 161 KQASAMVHDLILNVS 175
+ A ++ + + S
Sbjct: 445 QNAQYLLQNSVKQYS 459
>PIE1_CAEEL (Q94131) Pharynx and intestine in excess protein 1
(Pie-1 protein)
Length = 335
Score = 41.2 bits (95), Expect = 0.002
Identities = 19/58 (32%), Positives = 29/58 (49%), Gaps = 1/58 (1%)
Query: 171 ILNVSSVSGPPGKNITSQTSAPANNFKTKLCENFTK-GSCTFGERCHFAHGTDELRKP 227
I ++ + P S +KT+LC+ F + G C + + C +AHG DELR P
Sbjct: 74 ITPLAQIDEAPATKRHSSAKDKHTEYKTRLCDAFRREGYCPYNDNCTYAHGQDELRVP 131
Score = 35.8 bits (81), Expect = 0.086
Identities = 15/32 (46%), Positives = 19/32 (58%)
Query: 33 KTRLCNKFNTAEGCKFGDKCHFAHGEWELGRP 64
KTRLC+ F C + D C +AHG+ EL P
Sbjct: 100 KTRLCDAFRREGYCPYNDNCTYAHGQDELRVP 131
Score = 35.8 bits (81), Expect = 0.086
Identities = 18/55 (32%), Positives = 28/55 (50%), Gaps = 7/55 (12%)
Query: 169 DLILNVSSVSGPPGKNITSQTSAPANNF----KTKLCENFTKGSCTFGERCHFAH 219
D +N SS S + + P+NN + ++C NF +G+C +G RC F H
Sbjct: 157 DTTINRSSSSASKHHD---ENRRPSNNHGSSNRRQICHNFERGNCRYGPRCRFIH 208
>ZFS1_SCHPO (P47979) Zinc-finger protein zfs1
Length = 404
Score = 40.8 bits (94), Expect = 0.003
Identities = 22/56 (39%), Positives = 32/56 (56%), Gaps = 5/56 (8%)
Query: 176 SVSGPPGKNITSQTSAPANN---FKTKLCENFT-KGSCTFGERCHFAHGTDELRKP 227
S S P G +S AP +KT+ C+N+ G+C +G +C FAHG EL++P
Sbjct: 305 SASHPHGSG-SSNGVAPNGKRALYKTEPCKNWQISGTCRYGSKCQFAHGNQELKEP 359
Score = 37.7 bits (86), Expect = 0.023
Identities = 18/47 (38%), Positives = 23/47 (48%), Gaps = 1/47 (2%)
Query: 33 KTRLCNKFNTAEGCKFGDKCHFAHGEWELGR-PTVPAYEDTRAMGQM 78
KT C + + C++G KC FAHG EL P P Y+ R M
Sbjct: 328 KTEPCKNWQISGTCRYGSKCQFAHGNQELKEPPRHPKYKSERCRSFM 374
>TISB_RAT (P17431) Butyrate response factor 1 (TIS11B protein)
(EGF-inducible protein CMG1)
Length = 338
Score = 40.8 bits (94), Expect = 0.003
Identities = 17/29 (58%), Positives = 18/29 (61%)
Query: 33 KTRLCNKFNTAEGCKFGDKCHFAHGEWEL 61
KT LC F CK+GDKC FAHG EL
Sbjct: 116 KTELCRPFEENGACKYGDKCQFAHGIHEL 144
Score = 40.8 bits (94), Expect = 0.003
Identities = 16/34 (47%), Positives = 25/34 (73%), Gaps = 1/34 (2%)
Query: 193 ANNFKTKLCENFTK-GSCTFGERCHFAHGTDELR 225
++ +KT+LC F + G+C +G++C FAHG ELR
Sbjct: 112 SSRYKTELCRPFEENGACKYGDKCQFAHGIHELR 145
Score = 38.5 bits (88), Expect = 0.013
Identities = 16/31 (51%), Positives = 20/31 (63%), Gaps = 1/31 (3%)
Query: 196 FKTKLCENF-TKGSCTFGERCHFAHGTDELR 225
+KT+LC F T G C +G RCHF H +E R
Sbjct: 153 YKTELCRTFHTIGFCPYGPRCHFIHNAEERR 183
Score = 35.4 bits (80), Expect = 0.11
Identities = 14/31 (45%), Positives = 17/31 (54%)
Query: 30 PVAKTRLCNKFNTAEGCKFGDKCHFAHGEWE 60
P KT LC F+T C +G +CHF H E
Sbjct: 151 PKYKTELCRTFHTIGFCPYGPRCHFIHNAEE 181
>TISB_MOUSE (P23950) Butyrate response factor 1 (TIS11B protein)
Length = 338
Score = 40.8 bits (94), Expect = 0.003
Identities = 17/29 (58%), Positives = 18/29 (61%)
Query: 33 KTRLCNKFNTAEGCKFGDKCHFAHGEWEL 61
KT LC F CK+GDKC FAHG EL
Sbjct: 116 KTELCRPFEENGACKYGDKCQFAHGIHEL 144
Score = 40.8 bits (94), Expect = 0.003
Identities = 16/34 (47%), Positives = 25/34 (73%), Gaps = 1/34 (2%)
Query: 193 ANNFKTKLCENFTK-GSCTFGERCHFAHGTDELR 225
++ +KT+LC F + G+C +G++C FAHG ELR
Sbjct: 112 SSRYKTELCRPFEENGACKYGDKCQFAHGIHELR 145
Score = 38.5 bits (88), Expect = 0.013
Identities = 16/31 (51%), Positives = 20/31 (63%), Gaps = 1/31 (3%)
Query: 196 FKTKLCENF-TKGSCTFGERCHFAHGTDELR 225
+KT+LC F T G C +G RCHF H +E R
Sbjct: 153 YKTELCRTFHTIGFCPYGPRCHFIHNAEERR 183
Score = 35.4 bits (80), Expect = 0.11
Identities = 14/31 (45%), Positives = 17/31 (54%)
Query: 30 PVAKTRLCNKFNTAEGCKFGDKCHFAHGEWE 60
P KT LC F+T C +G +CHF H E
Sbjct: 151 PKYKTELCRTFHTIGFCPYGPRCHFIHNAEE 181
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.314 0.132 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 29,593,907
Number of Sequences: 164201
Number of extensions: 1230977
Number of successful extensions: 2832
Number of sequences better than 10.0: 81
Number of HSP's better than 10.0 without gapping: 45
Number of HSP's successfully gapped in prelim test: 36
Number of HSP's that attempted gapping in prelim test: 2614
Number of HSP's gapped (non-prelim): 244
length of query: 229
length of database: 59,974,054
effective HSP length: 107
effective length of query: 122
effective length of database: 42,404,547
effective search space: 5173354734
effective search space used: 5173354734
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (22.0 bits)
S2: 64 (29.3 bits)
Medicago: description of AC149471.8


