

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC149305.8 - phase: 0 /pseudo
(439 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
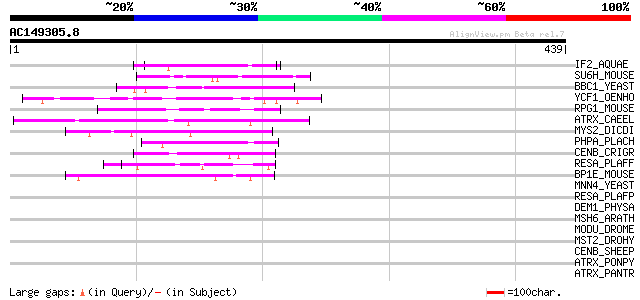
Score E
Sequences producing significant alignments: (bits) Value
IF2_AQUAE (O67825) Translation initiation factor IF-2 52 3e-06
SU6H_MOUSE (Q62383) Suppressor of Ty 6 homolog protein (Chromati... 50 2e-05
BBC1_YEAST (P47068) Myosin tail region-interacting protein MTI1 ... 49 3e-05
YCF1_OENHO (Q9MTH5) Hypothetical 287.9 kDa protein ycf1 48 4e-05
RPG1_MOUSE (Q9EPQ2) X-linked retinitis pigmentosa GTPase regulat... 48 4e-05
ATRX_CAEEL (Q9U7E0) Transcriptional regulator ATRX homolog (X-li... 47 7e-05
MYS2_DICDI (P08799) Myosin II heavy chain, non muscle 47 1e-04
PHPA_PLACH (Q02752) Acidic phosphoprotein precursor (50 kDa anti... 45 4e-04
CENB_CRIGR (P48988) Major centromere autoantigen B (Centromere p... 45 5e-04
RESA_PLAFF (P13830) Ring-infected erythrocyte surface antigen pr... 44 6e-04
BP1E_MOUSE (Q91ZU8) Bullous pemphigoid antigen 1, isoform 5 (BPA... 44 6e-04
MNN4_YEAST (P36044) MNN4 protein 44 0.001
RESA_PLAFP (Q26005) Ring-infected erythrocyte surface antigen (F... 43 0.002
DEM1_PHYSA (P05422) Dermorphin 1 precursor [Contains: Deltorphin... 42 0.003
MSH6_ARATH (O04716) DNA mismatch repair protein MSH6-1 (AtMsh6-1) 42 0.004
MODU_DROME (P13469) DNA-binding protein modulo 42 0.004
MST2_DROHY (Q08696) Axoneme-associated protein mst101(2) 41 0.005
CENB_SHEEP (P49451) Major centromere autoantigen B (Centromere p... 41 0.005
ATRX_PONPY (Q7YQM3) Transcriptional regulator ATRX (X-linked hel... 41 0.005
ATRX_PANTR (Q7YQM4) Transcriptional regulator ATRX (X-linked hel... 41 0.005
>IF2_AQUAE (O67825) Translation initiation factor IF-2
Length = 805
Score = 52.0 bits (123), Expect = 3e-06
Identities = 39/112 (34%), Positives = 60/112 (52%), Gaps = 8/112 (7%)
Query: 107 EEKNDEEKGENVL-EEVIE---ANLIIHDIGNEERKIDEEEGVEEENEEKGNCSSMVKRR 162
EEK +EEK E V+ EEV+E +I+ +I EE+K EEE +EE + K + ++K
Sbjct: 80 EEKKEEEKKEEVIVEEVVEEKKPEVIVEEI--EEKK--EEEEKKEEEKPKKSVEELIKEI 135
Query: 163 REKVMEEARNSVPENGKVMHLVKAFERLLSIKKEKEKNEEEEENDKKNKVMK 214
EK +E E + V+ E +KE++K E++EE K K+ K
Sbjct: 136 LEKKEKEKEKKKVEKERKEEKVRVVEVKKEERKEEKKEEKKEEEKPKIKMSK 187
Score = 47.4 bits (111), Expect = 7e-05
Identities = 29/113 (25%), Positives = 60/113 (52%), Gaps = 2/113 (1%)
Query: 99 EKENFFDGEEKNDEEKGENVLEEVIEANLIIHDIGNEERKIDEEEGVEEENEEKGNCSSM 158
E E + EEK +EEK + +EE+I+ L + E++K+++E E+ +
Sbjct: 108 EIEEKKEEEEKKEEEKPKKSVEELIKEILEKKEKEKEKKKVEKERKEEKVRVVEVKKEER 167
Query: 159 VKRRREKVMEEARNSVPENGKVMHLVKAFERLLSIKKEKEKNEEEEENDKKNK 211
+ ++E+ EE + + + K +++ E +++KEK+K E+ E+ KK +
Sbjct: 168 KEEKKEEKKEEEKPKIKMSKKEREIMRKLEH--AVEKEKKKQEKREKEKKKKE 218
Score = 40.4 bits (93), Expect = 0.009
Identities = 32/114 (28%), Positives = 56/114 (49%), Gaps = 6/114 (5%)
Query: 101 ENFFDGEEKNDEEKGENVLEEVIEANLIIHDIGNEERKIDEEEGVEEENEEKGNCSSMVK 160
E +D +EE+ E V+ E +A + + EE+K EE + EE E+ +V+
Sbjct: 51 EMIYDAFGIKEEEEKEEVVTEQAQAPAEVEEKKEEEKK---EEVIVEEVVEEKKPEVIVE 107
Query: 161 RRREKVMEEARNSVPENGKVMHLVKAFERLLSIKKEKEKNEEEEENDKKNKVMK 214
EK EE + E K V+ + + KKEKEK +++ E ++K + ++
Sbjct: 108 EIEEKKEEEEKK---EEEKPKKSVEELIKEILEKKEKEKEKKKVEKERKEEKVR 158
>SU6H_MOUSE (Q62383) Suppressor of Ty 6 homolog protein (Chromatin
structural protein)
Length = 1726
Score = 49.7 bits (117), Expect = 2e-05
Identities = 42/145 (28%), Positives = 77/145 (52%), Gaps = 15/145 (10%)
Query: 101 ENFFDGEEKNDEEKGENVLEEVIEANLIIHDIGNEERKIDEEEGVEEENEEKGNCSSMV- 159
+ F + E+ ++EE+ EN+ ++ NL D N++ DEEEG E+E + G+ V
Sbjct: 28 KKFVEEEDDDEEEEEENLDDQDERGNL--KDFINDDD--DEEEGEEDEGSDSGDSEDDVG 83
Query: 160 --KRRR----EKVMEEARNSVPENGKVMHLVKAFERLLSIKKEKEKNEEEEENDKKNKVM 213
KR+R +++ ++ + + EN V VK ++ +KK + +E++EE K +
Sbjct: 84 HKKRKRPSFDDRLEDDDFDLIEENLGVK--VKRGQKYRRVKKMSDDDEDDEEEYGKEEHE 141
Query: 214 KWALPGLQFQQPVKDGDEQSEVVSS 238
K A+ G FQ ++G+E E V +
Sbjct: 142 KEAIAGEIFQD--EEGEEGQEAVEA 164
>BBC1_YEAST (P47068) Myosin tail region-interacting protein MTI1
(BBC1 protein)
Length = 1157
Score = 48.5 bits (114), Expect = 3e-05
Identities = 44/153 (28%), Positives = 70/153 (44%), Gaps = 19/153 (12%)
Query: 85 EAYRNLRESQEGF--FEKENFFDG---------EEKNDEEKGENVLEEVIEANLIIHDIG 133
E + RE+ EG EKE F D E + DE +GENV EE G
Sbjct: 340 EITKTSREADEGTNDIEKEQFLDEYTKENQKVEESQADEARGENVAEESEIG------YG 393
Query: 134 NEERKIDEEEGVEEENEEKGNCSSMVKRRREKVMEEARNSVPENGKVMHLVKAFERLLSI 193
+E+R+ D +E EEE+ E+ N + ++ R K+ +R P + S
Sbjct: 394 HEDREGDNDEEKEEEDSEE-NRRAALRERMAKLSGASRFGAPVGFNPFGMASGVGNKPSE 452
Query: 194 K-KEKEKNEEEEENDKKNKVMKWALPGLQFQQP 225
+ K+K+ E+EEE ++ + + A+P + F P
Sbjct: 453 EPKKKQHKEKEEEEPEQLQELPRAIPVMPFVDP 485
>YCF1_OENHO (Q9MTH5) Hypothetical 287.9 kDa protein ycf1
Length = 2434
Score = 48.1 bits (113), Expect = 4e-05
Identities = 67/258 (25%), Positives = 115/258 (43%), Gaps = 59/258 (22%)
Query: 11 IENLNPNEFSNRTP--LQKSLTKGDNNCVNVVVVSPQKRIRQRKFVVAK-KKKNDQSPRK 67
I+N N N N L+K + D+N + Q + R +VA K+K +Q RK
Sbjct: 1982 IQNENENAIQNLIDFFLEKKNSPKDSN----QELHAQAKARIWDALVASLKQKREQKERK 2037
Query: 68 TVLCKCGENDDGSKCVCEAYRNLRESQEGFFEKENFFDGEEKNDEEKGENVLEEVIEANL 127
+K + + R+ + EK ++ +EK EN ++
Sbjct: 2038 ------------NKRIAQLIEKKRQKE---IEKHKRKIEKQMRKKEKIENAKKK------ 2076
Query: 128 IIHDIGNEERKID-EEEGVEEENEEKGNCSSMVKRRREKVMEEARNSVPENGKVMHLVKA 186
I NE++KI+ EEE +E+E +K +K++ K +E+ +N V +N + K
Sbjct: 2077 ----IENEKKKIETEEEKLEKEKRKKERKKEKLKKKVAKNIEKLKNKVAKN-----VAKN 2127
Query: 187 FERLLSIKKEKEKN----EEEEENDKK----------NKVMKWALPGLQFQQPV----KD 228
E+L KK++ KN EEE++ +K NK++ A ++Q+P+ K
Sbjct: 2128 IEKL---KKQRAKNIARLEEEDKKARKKRKRKVQVQENKILYTAFGSDKWQRPIAEYPKS 2184
Query: 229 GDEQSEVVSSCCDDDDFD 246
GD ++ V DDD+ D
Sbjct: 2185 GDIRNFQVILPEDDDEDD 2202
Score = 32.3 bits (72), Expect = 2.5
Identities = 31/153 (20%), Positives = 73/153 (47%), Gaps = 16/153 (10%)
Query: 66 RKTVLCKCGEND---DGSKCVCEAYRNLRESQEGFFEKENFFDGEEKNDEEKGENVLEEV 122
RK V+ END DG + V + E++ +FF E+KN + L
Sbjct: 1961 RKNVIQNENENDPIEDGRQNVIQ-----NENENAIQNLIDFFL-EKKNSPKDSNQELHAQ 2014
Query: 123 IEANLIIHDIGNEERKIDEEEGVEEENEEKGNCSSMVKRRREKVMEEARNSVPENGKVMH 182
+A + + + ++K +++E + + +++++R+K +E+ + + + +
Sbjct: 2015 AKARIWDALVASLKQKREQKE------RKNKRIAQLIEKKRQKEIEKHKRKIEKQMRKKE 2068
Query: 183 LVKAFERLLSIKKEKEKNEEEE-ENDKKNKVMK 214
++ ++ + +K+K + EEE+ E +K+ K K
Sbjct: 2069 KIENAKKKIENEKKKIETEEEKLEKEKRKKERK 2101
>RPG1_MOUSE (Q9EPQ2) X-linked retinitis pigmentosa GTPase
regulator-interacting protein 1 (RPGR-interacting
protein 1)
Length = 1331
Score = 48.1 bits (113), Expect = 4e-05
Identities = 44/146 (30%), Positives = 67/146 (45%), Gaps = 25/146 (17%)
Query: 70 LCKCGENDDGSKCVCEAYRNLRESQEGFFEKE-NFFDGEEKNDEEKGENVLEEVIEANLI 128
L GE +GS V +++ + EGF E +GEEK +E E V EE +E
Sbjct: 872 LTDSGEKSNGSIKVQLDWKSHYLAPEGFQMSEAEKPEGEEKEEEGGEEEVKEEEVE---- 927
Query: 129 IHDIGNEERKIDEEEGVEEENEEKGNCSSMVKRRREKVMEEARNSVPENGKVMHLVKAFE 188
EE + +EEE V+EE EE+ +R E+ EE + E +
Sbjct: 928 -----EEEEEEEEEEEVKEEKEEEEE----EEREEEEEKEEEKEEEEEEDEKEE------ 972
Query: 189 RLLSIKKEKEKNEEEEENDKKNKVMK 214
++E+E+ EEEEE D+ V++
Sbjct: 973 -----EEEEEEEEEEEEEDENKDVLE 993
>ATRX_CAEEL (Q9U7E0) Transcriptional regulator ATRX homolog
(X-linked nuclear protein-1)
Length = 1359
Score = 47.4 bits (111), Expect = 7e-05
Identities = 57/252 (22%), Positives = 103/252 (40%), Gaps = 24/252 (9%)
Query: 4 EKKKPLEIENLNPNEFSNRTPLQKSLTKGDNNCVNVVVVSPQKRIRQRKFVVAKKKKNDQ 63
EK K + + +E S+ QKS K S ++ +RK V K KKN +
Sbjct: 116 EKSKKKRTTSSSEDEDSDEEREQKSKKKSKKTKKQTSSESSEESEEERK--VKKSKKNKE 173
Query: 64 SPRKTVLCKCGENDDGSKCVCEAYRNLRESQEGFFEKENFFDGEEKNDEEKGENVLEEVI 123
K E+D+ K ++ + L++ + E E+ + E K ++K + V+++
Sbjct: 174 KSVKKRAETSEESDEDEKPSKKSKKGLKKKAKSESESESEDEKEVKKSKKKSKKVVKKES 233
Query: 124 EANLIIHDIGNEERKID-------------EEEGVEEENEEKGNCSSMVKRRREKVMEEA 170
E+ D E++K + E E +EE EEK + K++ V + +
Sbjct: 234 ES----EDEAPEKKKTEKRKRSKTSSEESSESEKSDEEEEEKESSPKPKKKKPLAVKKLS 289
Query: 171 RNSVPENGKVMHLVKAFER-----LLSIKKEKEKNEEEEENDKKNKVMKWALPGLQFQQP 225
+ E V L + +R + + EK++ E E +D + KV K + +
Sbjct: 290 SDEESEESDVEVLPQKKKRGAVTLISDSEDEKDQKSESEASDVEEKVSKKKAKKQESSES 349
Query: 226 VKDGDEQSEVVS 237
D E S V+
Sbjct: 350 GSDSSEGSITVN 361
Score = 42.0 bits (97), Expect = 0.003
Identities = 42/202 (20%), Positives = 86/202 (41%), Gaps = 11/202 (5%)
Query: 41 VVSPQKRIRQRKFVVAKKKKNDQSPRKTVLCK----CGENDDGSKCVCEAYRNLRESQEG 96
+ + +K R +K ++++ P+K K E DD + + + +
Sbjct: 27 IENERKEKRAQKLKEKREREGKPPPKKRPAKKRKASSSEEDDDDEEESPRKSSKKSRKRA 86
Query: 97 FFEKENFFDGEEKNDEEKGENVLEEVIEANLIIHDIGNEERKIDEEEGVEEENEEKGNCS 156
E E+ + +E+ D +K ++ ++ ++ E+E +EE E+K
Sbjct: 87 KSESESD-ESDEEEDRKKSKS--KKKVDQKKKEKSKKKRTTSSSEDEDSDEEREQKSKKK 143
Query: 157 SMVKRRREKVMEEARNSVPENGKVMHLVKAFERLLSIKKEKEKNEEEEENDKKNKVMKWA 216
S K+ +++ E+ E KV K E+ S+KK E +EE +E++K +K K
Sbjct: 144 S--KKTKKQTSSESSEESEEERKVKKSKKNKEK--SVKKRAETSEESDEDEKPSKKSKKG 199
Query: 217 LPGLQFQQPVKDGDEQSEVVSS 238
L + + +++ EV S
Sbjct: 200 LKKKAKSESESESEDEKEVKKS 221
>MYS2_DICDI (P08799) Myosin II heavy chain, non muscle
Length = 2116
Score = 46.6 bits (109), Expect = 1e-04
Identities = 36/174 (20%), Positives = 87/174 (49%), Gaps = 12/174 (6%)
Query: 45 QKRIRQRKFVVAKKKKN--DQSPRKTVLCKCGENDDGSKCVCEAYRNLRESQE---GFFE 99
+K I++++ + + K N D + +K L K ++ + + V + R L+ +E ++
Sbjct: 827 EKEIKEKEREILELKSNLTDSTTQKDKLEKSLKDTESN--VLDLQRQLKAEKETLKAMYD 884
Query: 100 KENFFDGEEKNDEEKGENVLEEVIEANLIIHDIGNEERKIDE-----EEGVEEENEEKGN 154
++ + +++ E + E++ E+ E L + ++ N++R ++E EE ++EE + +
Sbjct: 885 SKDALEAQKRELEIRVEDMESELDEKKLALENLQNQKRSVEEKVRDLEEELQEEQKLRNT 944
Query: 155 CSSMVKRRREKVMEEARNSVPENGKVMHLVKAFERLLSIKKEKEKNEEEEENDK 208
+ K+ E++ E R + ++ + L K + L +E ++ EE DK
Sbjct: 945 LEKLKKKYEEELEEMKRVNDGQSDTISRLEKIKDELQKEVEELTESFSEESKDK 998
Score = 36.6 bits (83), Expect = 0.13
Identities = 34/161 (21%), Positives = 68/161 (42%), Gaps = 8/161 (4%)
Query: 51 RKFVVAKKKKNDQSPRKTVLCKCGENDDGSKCVCEAYRNLRESQEGFFEKENFFDGEEKN 110
+K + +KKK ++S R + D +K E R ++ +++ D + K
Sbjct: 1768 KKQLEDEKKKLNESERAKKRLESENEDFLAKLDAEVKNRSRAEKDRKKYEKDLKDTKYKL 1827
Query: 111 DEEKGENVLEEVIEANLIIHDIGNEERKIDEEEGVEEENEEKGNCSSMVKRRREKVMEEA 170
++E E+ A L E +IDE E+ + K + K+ E ++
Sbjct: 1828 NDEAATKTQTEIGAAKL--------EDQIDELRSKLEQEQAKATQADKSKKTLEGEIDNL 1879
Query: 171 RNSVPENGKVMHLVKAFERLLSIKKEKEKNEEEEENDKKNK 211
R + + GK+ ++ +R L + E+ + EE D K++
Sbjct: 1880 RAQIEDEGKIKMRLEKEKRALEGELEELRETVEEAEDSKSE 1920
Score = 32.0 bits (71), Expect = 3.2
Identities = 47/211 (22%), Positives = 82/211 (38%), Gaps = 17/211 (8%)
Query: 6 KKPLEIENLNPNEFSNRTPLQKS-----LTKGDNNCVNVVVVSPQKRIRQRKFVVAKKKK 60
KK LE E NE +S L K D N ++ ++ K K
Sbjct: 1768 KKQLEDEKKKLNESERAKKRLESENEDFLAKLDAEVKNRSRAEKDRKKYEKDLKDTKYKL 1827
Query: 61 NDQSPRKTVLCKCGENDDGSKCVCEAYRNLRESQEGFFEKENFFDGEEKNDEEKGENVLE 120
ND++ KT + + G+ + + LR E K D +K E + +N+
Sbjct: 1828 NDEAATKT------QTEIGAAKLEDQIDELRSKLEQEQAKATQADKSKKTLEGEIDNLRA 1881
Query: 121 EVIEANLIIHDIGNEERKIDEE-----EGVEEENEEKGNCSSMVKRRREKVMEEARNSVP 175
++ + I + E+R ++ E E VEE + K KR E +E+AR ++
Sbjct: 1882 QIEDEGKIKMRLEKEKRALEGELEELRETVEEAEDSKSEAEQS-KRLVELELEDARRNLQ 1940
Query: 176 ENGKVMHLVKAFERLLSIKKEKEKNEEEEEN 206
+ + + + L + + K EEE+
Sbjct: 1941 KEIDAKEIAEDAKSNLQREIVEAKGRLEEES 1971
>PHPA_PLACH (Q02752) Acidic phosphoprotein precursor (50 kDa
antigen)
Length = 441
Score = 45.1 bits (105), Expect = 4e-04
Identities = 31/114 (27%), Positives = 59/114 (51%), Gaps = 10/114 (8%)
Query: 105 DGEEKNDEEKGENVL------EEVIEANLIIHDIGNEERKIDEEEGVEEENEEKGNCSSM 158
+G +DEE E V E + +L+ +D + E + + ++G E E E+KGN +
Sbjct: 309 EGTANDDEELDEEVASIFDDDEHADDLSLLDYDENSNENQENVKKGNENEGEQKGNENEG 368
Query: 159 VKRRREKVMEEARNSVPENGKVMHLVKAFERLLSIKKEKEKNEEEEENDKKNKV 212
++ ++K +E +N M K ++ +K+K+K E+E++ +KK KV
Sbjct: 369 EQKGKKKKAKEKSKKKVKNKPTMTTKKKKKK----EKKKKKKEKEKKKEKKVKV 418
>CENB_CRIGR (P48988) Major centromere autoantigen B (Centromere
protein B) (CENP-B)
Length = 606
Score = 44.7 bits (104), Expect = 5e-04
Identities = 40/136 (29%), Positives = 62/136 (45%), Gaps = 30/136 (22%)
Query: 99 EKENFFDGEEKNDEEKGENVLEEVIEANLIIHDIGNEERKIDEEEGVEEENEEKGNCSSM 158
E+E + EE+ +EE+GE EE E G EE EE G EEE EE+G+ S
Sbjct: 407 EEEEEEEEEEEEEEEEGEGEEEEEEEEE------GEEEGGEGEEVGEEEEVEEEGDESDE 460
Query: 159 VKRRREKVMEEARN----------------------SVPENGK--VMHLVKAFERLLSIK 194
+ E+ EE+ + SV E + +H ++ E S
Sbjct: 461 EEEEEEEEEEESSSEGLEAEDWAQGVVEASGGFGGYSVQEEAQCPTLHFLEGGEDSDSDS 520
Query: 195 KEKEKNEEEEENDKKN 210
E+E++EEE+E D+++
Sbjct: 521 DEEEEDEEEDEEDEED 536
Score = 38.9 bits (89), Expect = 0.026
Identities = 40/127 (31%), Positives = 62/127 (48%), Gaps = 19/127 (14%)
Query: 92 ESQEGFFEKENFFDGEEKNDEEKGENVLEEVIEANLIIHDIGNEERKIDEEEGVEEENEE 151
E +EG E+E + EE+ +EE GE EEV E ++ E + DEEE EEE EE
Sbjct: 419 EEEEGEGEEEE--EEEEEGEEEGGEG--EEVGEEE----EVEEEGDESDEEEEEEEEEEE 470
Query: 152 KGNCSSMVKRRREKVMEEARN-----SVPENGK--VMHLVKAFERLLSIKKEKEKNEE-- 202
+ + + + + EA SV E + +H ++ E S E+E++EE
Sbjct: 471 ESSSEGLEAEDWAQGVVEASGGFGGYSVQEEAQCPTLHFLEGGEDSDSDSDEEEEDEEED 530
Query: 203 --EEEND 207
+EE+D
Sbjct: 531 EEDEEDD 537
Score = 33.1 bits (74), Expect = 1.5
Identities = 34/135 (25%), Positives = 56/135 (41%), Gaps = 27/135 (20%)
Query: 133 GNEERKIDEEEGVEEENEEKGNCSSMVKRRREKVMEEARNSVPENGKVMHLVKAFERLLS 192
G EE + +EEE EEE E +G + E+ EE E G+ + + + S
Sbjct: 405 GEEEEEEEEEEEEEEEEEGEGE----EEEEEEEEGEEEGGEGEEVGEEEEVEEEGDE--S 458
Query: 193 IKKEKEKNEEEEENDKKN-KVMKWA--------------------LPGLQFQQPVKDGDE 231
++E+E+ EEEEE+ + + WA P L F + +D D
Sbjct: 459 DEEEEEEEEEEEESSSEGLEAEDWAQGVVEASGGFGGYSVQEEAQCPTLHFLEGGEDSDS 518
Query: 232 QSEVVSSCCDDDDFD 246
S+ ++D+ D
Sbjct: 519 DSDEEEEDEEEDEED 533
>RESA_PLAFF (P13830) Ring-infected erythrocyte surface antigen
precursor
Length = 1073
Score = 44.3 bits (103), Expect = 6e-04
Identities = 41/126 (32%), Positives = 60/126 (47%), Gaps = 24/126 (19%)
Query: 89 NLRESQEGFFE--KENFFDGEEKNDEEKGENVLEEVIEANLIIHDIGNEERKIDEEEGVE 146
N+ E +E E +EN + E+N EE E +EE +E N+ +D EE + EE VE
Sbjct: 958 NVEEVEENVEENVEENVEENVEENVEENVEENVEENVEENVEEYD---EENVEEVEENVE 1014
Query: 147 EENEEKGNCSSMVKRRREKVMEEARNSVPENGKVMHLVKAFERLLSIKKEKEKNEEE--E 204
E EE N V+ E+V E +V EN +++ E+N EE E
Sbjct: 1015 ENVEE--NVEENVEENVEEVEENVEENVEEN---------------VEENVEENVEENVE 1057
Query: 205 ENDKKN 210
E D++N
Sbjct: 1058 EYDEEN 1063
Score = 43.9 bits (102), Expect = 8e-04
Identities = 42/140 (30%), Positives = 64/140 (45%), Gaps = 5/140 (3%)
Query: 75 ENDDGSKCVCEAYRNLRESQEGFFEKENFFDGEEKNDEEKGENVLEEVIEANLIIHDIGN 134
E+D +A N+ E E EN + EE +E ENV E V E + +
Sbjct: 913 EHDAEENVEHDAEENVEHDAEENVE-ENVEEVEENVEENVEENVEENVEEVEENVEENVE 971
Query: 135 EERKIDEEEGVEEENEE--KGNCSSMVKRRREKVMEEARNSVPENGKVMHLVKAFERLLS 192
E + + EE VEE EE + N V+ E+ +EE +V EN + E +
Sbjct: 972 ENVEENVEENVEENVEENVEENVEENVEEYDEENVEEVEENVEENVEENVEENVEENVEE 1031
Query: 193 IKKEKEKNEEE--EENDKKN 210
+++ E+N EE EEN ++N
Sbjct: 1032 VEENVEENVEENVEENVEEN 1051
Score = 37.0 bits (84), Expect = 0.10
Identities = 36/122 (29%), Positives = 52/122 (42%), Gaps = 7/122 (5%)
Query: 89 NLRESQEGFFEKENFFDGEEKNDEEKGENVLEEVIEANLIIHDIGNEERKIDEEEGVEEE 148
N E+ E E+ D EE + + ENV E E N+ N E ++E E EE
Sbjct: 891 NAEENVEHDAEENVEHDAEENVEHDAEENV-EHDAEENVEHDAEENVEENVEEVEENVEE 949
Query: 149 NEEKGNCSSMVKRRREKVMEEARNSVPENGKVMHLVKAFERLLSIKKEKEKNEEEEENDK 208
N E+ N V+ E V E +V EN + + E + E+ E EE D+
Sbjct: 950 NVEE-NVEENVEEVEENVEENVEENVEEN-----VEENVEENVEENVEENVEENVEEYDE 1003
Query: 209 KN 210
+N
Sbjct: 1004 EN 1005
Score = 34.3 bits (77), Expect = 0.65
Identities = 29/91 (31%), Positives = 45/91 (48%), Gaps = 2/91 (2%)
Query: 89 NLRESQEGFFEKENFFDGEEKNDEEKGENVLEEVIEANLIIHDIGNEERKIDEEEGVEEE 148
N+ E+ E E EN + E+ DEE E V EE +E N+ + N E ++E E EE
Sbjct: 981 NVEENVEENVE-ENVEENVEEYDEENVEEV-EENVEENVEENVEENVEENVEEVEENVEE 1038
Query: 149 NEEKGNCSSMVKRRREKVMEEARNSVPENGK 179
N E+ ++ + E V E +V E+ +
Sbjct: 1039 NVEENVEENVEENVEENVEEYDEENVEEHNE 1069
>BP1E_MOUSE (Q91ZU8) Bullous pemphigoid antigen 1, isoform 5 (BPA)
(Hemidesmosomal plaque protein) (Dystonia musculorum
protein) (Dystonin)
Length = 2611
Score = 44.3 bits (103), Expect = 6e-04
Identities = 42/186 (22%), Positives = 84/186 (44%), Gaps = 22/186 (11%)
Query: 45 QKRIRQRKF---VVAKKKKNDQSPRKTVLCKCGENDDGSKCVCEAYRNLRESQEGFFEKE 101
+K +R+ K+ V +K + + + + K E ++ KC+ E ++QE + ++
Sbjct: 1339 EKEMRELKYELSAVQLEKASSEEKARLLKDKLDETNNTLKCLKEDLERKDQAQERYSQQL 1398
Query: 102 NFFDGEEKNDEEKGENVLEEVIEANLIIHDIGNEERKIDEEEG-VEEENEEKGNCSSMVK 160
G+ +K E V +E + I H E + +E+G ++ E + ++ +
Sbjct: 1399 RDLGGQLNQTTDKAEEVRQEANDLKKIKHTYQLELESLHQEKGKLQREVDRVTRAHALAE 1458
Query: 161 R------------RREKVMEEARNSVPENGKVMHLVKAFER-----LLSIKKEKEKNEEE 203
R R EK + E R + + K HL + FER L +I+ EKE N++
Sbjct: 1459 RNIQCLNSQVHASRDEKDLSEERRRLCQR-KSDHLKEEFERSHAQLLQNIQAEKENNDKI 1517
Query: 204 EENDKK 209
++ +K+
Sbjct: 1518 QKLNKE 1523
Score = 32.0 bits (71), Expect = 3.2
Identities = 21/73 (28%), Positives = 41/73 (55%), Gaps = 10/73 (13%)
Query: 133 GNEERKIDEEEGVE-EENEEKGNCSSMVKRRREKVMEEARNSVPENGKVMHLVKAFERLL 191
G+ ++++EEE +EN E G S +++R+R ++ EN K+ + E ++
Sbjct: 1067 GDSLKRLEEEEMKRSKENSEHGAYSDLLQRQRATMV--------ENSKLTGKISELETMV 1118
Query: 192 S-IKKEKEKNEEE 203
+ +KK+K + EEE
Sbjct: 1119 AELKKQKSRVEEE 1131
>MNN4_YEAST (P36044) MNN4 protein
Length = 1178
Score = 43.5 bits (101), Expect = 0.001
Identities = 41/166 (24%), Positives = 75/166 (44%), Gaps = 24/166 (14%)
Query: 46 KRIRQRKFVVAKKKKNDQSPRKTVLCKCGENDDGSKCVCEAYRNLRESQEGFFEKENFFD 105
K + +RK KKK+ ++ +K K + ++ K E + +E +E ++E
Sbjct: 1032 KLLEERKRREKKKKEEEEKKKKEEEEKKKKEEEEKKKKEEEEKKKKEEEEKKKKEE---- 1087
Query: 106 GEEKNDEEKGENVLEEVIEANLIIHDIGNEERKIDEEEGVEEENEEKGNCSSMVKRRREK 165
EEK +E+ E +E EE K +EEG + +NE++ N + + +++
Sbjct: 1088 -EEKKKQEEEEKKKKE-------------EEEKKKQEEGEKMKNEDEENKKNEDEEKKKN 1133
Query: 166 VMEEARNSVPENGKVMHLVKAFERLLSIKKEKEKNEEEEENDKKNK 211
EE + +N K K + +E+EK + EEE KK +
Sbjct: 1134 EEEEKKKQEEKNKKNEDEEKKKQ------EEEEKKKNEEEEKKKQE 1173
Score = 43.1 bits (100), Expect = 0.001
Identities = 38/133 (28%), Positives = 63/133 (46%), Gaps = 18/133 (13%)
Query: 86 AYRNLRESQEGFFEKENFFDGEEKNDEEKGENVLEEVIEANLIIHDIGNEERKIDEEEGV 145
AY L E ++ +K+ + ++K +EE+ + EE E++K +EEE
Sbjct: 1029 AYAKLLEERKRREKKKKEEEEKKKKEEEEKKKKEEE-------------EKKKKEEEEKK 1075
Query: 146 EEENEEKGNCSSMVKRRRE----KVMEEARNSVPENGKVMHLVKAFERLLSIKKEKEKNE 201
++E EEK K+++E K EE E G+ M E + +EK+KNE
Sbjct: 1076 KKEEEEKKKKEEEEKKKQEEEEKKKKEEEEKKKQEEGEKMKNEDE-ENKKNEDEEKKKNE 1134
Query: 202 EEEENDKKNKVMK 214
EEE+ ++ K K
Sbjct: 1135 EEEKKKQEEKNKK 1147
Score = 41.2 bits (95), Expect = 0.005
Identities = 38/129 (29%), Positives = 63/129 (48%), Gaps = 6/129 (4%)
Query: 88 RNLRESQEGFFEKENFFDGEEKNDEEKGENVLEEVIEANLIIHDIGNEERKIDEEEGVEE 147
R RE ++ E++ + EEK +E+ E +E E + E++K +EEE ++
Sbjct: 1037 RKRREKKKKEEEEKKKKEEEEKKKKEEEEKKKKEEEEKKKKEEE---EKKKKEEEEKKKQ 1093
Query: 148 ENEEKGNCSSMVKRRREKVMEEARNSVPENGKVMHLVKAFERLLSIKKEKEKN--EEEEE 205
E EEK K+++E+ E+ +N EN K K KK++EKN E+EE
Sbjct: 1094 EEEEKKKKEEEEKKKQEEG-EKMKNEDEENKKNEDEEKKKNEEEEKKKQEEKNKKNEDEE 1152
Query: 206 NDKKNKVMK 214
K+ + K
Sbjct: 1153 KKKQEEEEK 1161
Score = 39.3 bits (90), Expect = 0.020
Identities = 25/91 (27%), Positives = 46/91 (50%)
Query: 118 VLEEVIEANLIIHDIGNEERKIDEEEGVEEENEEKGNCSSMVKRRREKVMEEARNSVPEN 177
V E+ A L+ E++K +EEE ++E EEK K+++E+ ++ + +
Sbjct: 1024 VYEDYAYAKLLEERKRREKKKKEEEEKKKKEEEEKKKKEEEEKKKKEEEEKKKKEEEEKK 1083
Query: 178 GKVMHLVKAFERLLSIKKEKEKNEEEEENDK 208
K K E KKE+E+ +++EE +K
Sbjct: 1084 KKEEEEKKKQEEEEKKKKEEEEKKKQEEGEK 1114
>RESA_PLAFP (Q26005) Ring-infected erythrocyte surface antigen
(Fragment)
Length = 304
Score = 42.7 bits (99), Expect = 0.002
Identities = 38/118 (32%), Positives = 61/118 (51%), Gaps = 9/118 (7%)
Query: 100 KENFFDGEEKNDEEKGENV---LEEVIEANLIIHDIGNEERKIDEEEGVEEENEEKGNCS 156
+EN + E+N EE ENV +EE +E N+ + N E ++E E EEN E+
Sbjct: 162 EENAEENVEENVEEVEENVEENVEENVEENVEENVEENVEENVEEVEENVEENVEENVEE 221
Query: 157 SMVKRRREKVMEEARNSVPENGKVMHLVKAF--ERLLSIKKEKEKNEEE--EENDKKN 210
++ + E V E +V EN V V+ + E + +++ E+N EE EEN ++N
Sbjct: 222 NVEENVEENVEENVEENVEEN--VEENVEEYDEENVEEVEENVEENVEENVEENVEEN 277
Score = 37.7 bits (86), Expect = 0.059
Identities = 34/108 (31%), Positives = 49/108 (44%), Gaps = 6/108 (5%)
Query: 107 EEKNDEEKGENVLEEVIEANLIIHDIGNEERKIDEEEGVEEENEEKG--NCSSMVKRRRE 164
E KN E ++ EAN + HD E + D EE VE + EE N V+ E
Sbjct: 121 EMKNQNENVPEHVQHNAEAN-VEHD-AEENVEHDAEENVEHDAEENAEENVEENVEEVEE 178
Query: 165 KVMEEARNSVPENGKVMHLVKAFERLLSIKKEKEKNEEE--EENDKKN 210
V E +V EN + E + +++ E+N EE EEN ++N
Sbjct: 179 NVEENVEENVEENVEENVEENVEENVEEVEENVEENVEENVEENVEEN 226
Score = 37.7 bits (86), Expect = 0.059
Identities = 34/111 (30%), Positives = 52/111 (46%), Gaps = 10/111 (9%)
Query: 100 KENFFDGEEKNDEEKGENVLEEVIEANLIIHDIGNEERKIDEEEGVEEENEEKGNCSSMV 159
+EN E+N E E EE +E N+ EE + + EE VEE EE N V
Sbjct: 146 EENVEHDAEENVEHDAEENAEENVEENV-------EEVEENVEENVEENVEE--NVEENV 196
Query: 160 KRRREKVMEEARNSVPENGKVMHLVKAFERLLSIKKEKEKNEEEEENDKKN 210
+ E+ +EE +V EN + ++ + E + E+ E EEN ++N
Sbjct: 197 EENVEENVEEVEENVEENVE-ENVEENVEENVEENVEENVEENVEENVEEN 246
Score = 36.6 bits (83), Expect = 0.13
Identities = 38/124 (30%), Positives = 57/124 (45%), Gaps = 31/124 (25%)
Query: 89 NLRESQEGFFEKENFFDGEEKNDEEKGENVLEEVIEANLIIHDIGNEERKIDEEEGVEEE 148
N+ E +E +EN + E+N EE E +EE +E N+ EE + EE VEE
Sbjct: 203 NVEEVEENV--EENVEENVEENVEENVEENVEENVEENV-------EE---NVEENVEEY 250
Query: 149 NEEKGNCSSMVKRRREKVMEEARNSVPENGKVMHLVKAFERLLSIKKEKEKNEEE--EEN 206
+EE N + + E V E +V EN +++ E+N EE EE
Sbjct: 251 DEE--NVEEVEENVEENVEENVEENVEEN---------------VEENVEENVEENVEEY 293
Query: 207 DKKN 210
D++N
Sbjct: 294 DEEN 297
Score = 35.4 bits (80), Expect = 0.29
Identities = 37/123 (30%), Positives = 54/123 (43%), Gaps = 19/123 (15%)
Query: 92 ESQEGFFEKENFFDGEEKNDEEKGENV--LEEVIEANLIIHDIGNEERKIDE--EEGVEE 147
E+ E E+ D EE +E ENV +EE +E N+ E ++E EE VEE
Sbjct: 147 ENVEHDAEENVEHDAEENAEENVEENVEEVEENVEENV--------EENVEENVEENVEE 198
Query: 148 ENEEKGNCSSMVKRRREKVMEEARNSVPENGKVMHLVKAFERLLSIKKEKEKNEEEEEND 207
EE N + + E V E +V EN + + E + E+ E EE D
Sbjct: 199 NVEE--NVEEVEENVEENVEENVEENVEEN-----VEENVEENVEENVEENVEENVEEYD 251
Query: 208 KKN 210
++N
Sbjct: 252 EEN 254
>DEM1_PHYSA (P05422) Dermorphin 1 precursor [Contains: Deltorphin
(Dermenkephalin); Dermorphin]
Length = 197
Score = 42.0 bits (97), Expect = 0.003
Identities = 40/154 (25%), Positives = 70/154 (44%), Gaps = 24/154 (15%)
Query: 100 KENFFDGEEKNDEEKGENVLEEVIEANLIIHDIGNEERKIDEEEGVEEENEEKGNCSSMV 159
KE + EE+N+ E+ E+ + H + E +K D EE EEN E+G S +
Sbjct: 23 KEEKRETEEENENEENHEEGSEM--KRYMFHLMDGEAKKRDSEENEIEENHEEG---SEM 77
Query: 160 KR----------RREKVMEEARNSVPENGKVMHLVKAFE------RLLSIKKEKEKNEEE 203
KR ++ K + E N EN + +K + IK+E E+ +E
Sbjct: 78 KRYAFGYPSGEAKKIKRVSEEENENEENHEEGSEMKRYAFGYPSGEAKKIKRESEEEKEI 137
Query: 204 EENDKKNKVMK---WALPGLQFQQPVKDGDEQSE 234
EEN ++ MK + P + ++ ++ +E++E
Sbjct: 138 EENHEEGSEMKRYAFGYPSGEAKKIKRESEEENE 171
Score = 37.7 bits (86), Expect = 0.059
Identities = 30/130 (23%), Positives = 62/130 (47%), Gaps = 19/130 (14%)
Query: 122 VIEANLIIHDIGNEERKIDEEEGVEEENEEKGNCSSMVKRRREKVME-EAR------NSV 174
++ L+ + EE++ EEE EEN E+G S +KR +M+ EA+ N +
Sbjct: 11 ILFLGLVSLSVCKEEKRETEEENENEENHEEG---SEMKRYMFHLMDGEAKKRDSEENEI 67
Query: 175 PENGKVMHLVKAF---------ERLLSIKKEKEKNEEEEENDKKNKVMKWALPGLQFQQP 225
EN + +K + +++ + +E+ +NEE E + K + P + ++
Sbjct: 68 EENHEEGSEMKRYAFGYPSGEAKKIKRVSEEENENEENHEEGSEMKRYAFGYPSGEAKKI 127
Query: 226 VKDGDEQSEV 235
++ +E+ E+
Sbjct: 128 KRESEEEKEI 137
Score = 36.2 bits (82), Expect = 0.17
Identities = 45/174 (25%), Positives = 70/174 (39%), Gaps = 47/174 (27%)
Query: 80 SKCVCEAYRNLRESQEGFFEKENFFDGEEK--------NDEEKGENVLEEVIEANLIIHD 131
S VC+ + RE++E +EN +G E + E K + E IE N H+
Sbjct: 18 SLSVCKEEK--RETEEENENEENHEEGSEMKRYMFHLMDGEAKKRDSEENEIEEN---HE 72
Query: 132 IGNEERK---------------IDEEEGVEEENEEKGNCSSMVKR----------RREKV 166
G+E ++ + EEE EEN E+G S +KR ++ K
Sbjct: 73 EGSEMKRYAFGYPSGEAKKIKRVSEEENENEENHEEG---SEMKRYAFGYPSGEAKKIKR 129
Query: 167 MEEARNSVPENGKVMHLVKAFE------RLLSIKKEKEKNEEEEENDKKNKVMK 214
E + EN + +K + IK+E E+ E EEN ++ MK
Sbjct: 130 ESEEEKEIEENHEEGSEMKRYAFGYPSGEAKKIKRESEEENENEENHEEGSEMK 183
>MSH6_ARATH (O04716) DNA mismatch repair protein MSH6-1 (AtMsh6-1)
Length = 1324
Score = 41.6 bits (96), Expect = 0.004
Identities = 28/82 (34%), Positives = 48/82 (58%), Gaps = 3/82 (3%)
Query: 100 KENFFDGEEKNDEEKGENVLEEVIEANLIIHDIGNEERKIDEEEGVEEENEE--KGNCSS 157
+E+ DG++ +DE+ G+NV +EV E+ ++ +E ++DEEE VEE++EE K N S
Sbjct: 212 EEDKSDGDDSSDEDWGKNVGKEVCESEEDDVEL-VDENEMDEEELVEEKDEETSKVNRVS 270
Query: 158 MVKRRREKVMEEARNSVPENGK 179
R+ K E ++ + K
Sbjct: 271 KTDSRKRKTSEVTKSGGEKKSK 292
>MODU_DROME (P13469) DNA-binding protein modulo
Length = 542
Score = 41.6 bits (96), Expect = 0.004
Identities = 38/123 (30%), Positives = 57/123 (45%), Gaps = 14/123 (11%)
Query: 98 FEKENFFDGEEKNDEEKGENVLEEVIEANLIIHDIG--NEERKIDEEEGVEEENEEKGNC 155
F +E+ D EE+NDE+ G++ E EA +I D +EE D+EE +++ E G
Sbjct: 58 FSEEDESDVEEQNDEQPGDDSDFETEEAAGLIDDEAEEDEEYNSDDEEDDDDDELEPGEV 117
Query: 156 SSMVKRRREKVMEEARNSVPENGKVMHLVKAFERLLSIKKEKEKNEEEEENDKKNKVMKW 215
S + E E + E V E+ +S K EK +E+ EEN KV
Sbjct: 118 S-----KSEGADEVDESDDDEEAPV-------EKPVSKKSEKANSEKSEENRGIPKVKVG 165
Query: 216 ALP 218
+P
Sbjct: 166 KIP 168
>MST2_DROHY (Q08696) Axoneme-associated protein mst101(2)
Length = 1391
Score = 41.2 bits (95), Expect = 0.005
Identities = 37/162 (22%), Positives = 71/162 (42%), Gaps = 8/162 (4%)
Query: 50 QRKFVVAKKKKNDQSPRKTVLCKCGENDDGSKCVCEAYRNLRESQEGFFEKENFFDGEEK 109
++K A KK+ + + RK KC E K E + + +++ ++N + K
Sbjct: 700 KKKCEEAAKKEKEAAERK----KCEELAKKIKKAAEKKKCKKLAKKKKAGEKN----KLK 751
Query: 110 NDEEKGENVLEEVIEANLIIHDIGNEERKIDEEEGVEEENEEKGNCSSMVKRRREKVMEE 169
+KG+ L+E + + E++K E E+E EK C K+R+E+ ++
Sbjct: 752 KGNKKGKKALKEKKKCRELAKKKAAEKKKCKEAAKKEKEAAEKKKCEKTAKKRKEEAEKK 811
Query: 170 ARNSVPENGKVMHLVKAFERLLSIKKEKEKNEEEEENDKKNK 211
+ K K E+ +KE+ + ++ E+ KK K
Sbjct: 812 KCEKTAKKRKEAAEKKKCEKAAKKRKEEAEKKKCEKTAKKRK 853
Score = 40.4 bits (93), Expect = 0.009
Identities = 42/171 (24%), Positives = 71/171 (40%), Gaps = 14/171 (8%)
Query: 56 AKKKKNDQSPRKTVLCKCGENDDGSKCVCEAYRNLRESQEGFFEKENFFDGEEKNDEEKG 115
AKK+K + +K + K CE R+ + EK+ EK +++
Sbjct: 801 AKKRKEEAEKKKCEKTAKKRKEAAEKKKCEKAAKKRKEEA---EKKKC----EKTAKKRK 853
Query: 116 ENVLEEVIEANLIIHDIGNEERKIDEEEGVEEENEEKGNCSSMVKRRRE----KVMEEA- 170
E ++ E E++K ++ +E EK C+ K+ +E K EEA
Sbjct: 854 ETAEKKKCEKAAKKRKQAAEKKKCEKAAKKRKEAAEKKKCAEAAKKEKELAEKKKCEEAA 913
Query: 171 --RNSVPENGKVMHLVKAFERLLSIKKEKEKNEEEEENDKKNKVMKWALPG 219
V E K L K ++ KK K+ ++E++ +KNK+ K A G
Sbjct: 914 KKEKEVAERKKCEELAKKIKKAAEKKKCKKLAKKEKKAGEKNKLKKKAGKG 964
Score = 35.0 bits (79), Expect = 0.38
Identities = 40/179 (22%), Positives = 75/179 (41%), Gaps = 22/179 (12%)
Query: 45 QKRIRQRKFVVAKKKKNDQSPRKTVLCKCGENDDGSKCVCEAYRNLRESQEGFFEKENFF 104
++R ++ K KK+ +++ ++ + + ++ +K + EA + + EKE
Sbjct: 1065 EERAKKLKEAAEKKQCEERAKKEKEAAEKKQCEERAKKLKEAAEKKQCEERAKKEKEA-- 1122
Query: 105 DGEEKNDEEKGENVLEEVIEANLIIHDIGNEERKIDEEEGVEEENEEKGNCSSMVKRRRE 164
E+K EE + E E++K E E+E EK C+ K+ +E
Sbjct: 1123 -AEKKRCEEAAKREKEAA------------EKKKCAEAAKKEKEATEKQKCAEAAKKEKE 1169
Query: 165 -----KVMEEARNS--VPENGKVMHLVKAFERLLSIKKEKEKNEEEEENDKKNKVMKWA 216
K E A+ + K L K + +KK +E ++E+E +K K K A
Sbjct: 1170 AAEKKKCAEAAKREKEAAQKKKCADLAKKEQEPAEMKKCEEAAKKEKEAAEKQKCAKAA 1228
Score = 34.7 bits (78), Expect = 0.50
Identities = 41/172 (23%), Positives = 76/172 (43%), Gaps = 17/172 (9%)
Query: 45 QKRIRQRKFVVAKKKKNDQSPRKTVLCKCGENDDGSKCVCEAYRNLRESQEGFFEKENFF 104
+K ++RK A+KKK +++ +K E + KC E + ++++ E KE
Sbjct: 576 EKAAKERK-EAAEKKKCEEAAKKEK-----EVAERKKCE-ELAKKIKKAAEKKKCKEAAK 628
Query: 105 DGEEKNDEEKGENVLEEVIEANLIIHDIGNEERKIDEEEGVEEENEEKGNCSSMVKRRRE 164
+E + EK + +++ +A E++K + E+E EK C K+R+E
Sbjct: 629 KEKEAAEREKCGELAKKIKKAA--------EKKKCKKLAKKEKETAEKKKCEKAAKKRKE 680
Query: 165 KVMEEARNSVPENGKVMHLVKAFERLLSIKKEKEKNEEEEENDKKNKVMKWA 216
++ + K K E + KKEKE E ++ + K+ K A
Sbjct: 681 AAEKKKCAEAAKKEKEAAEKKKCEE--AAKKEKEAAERKKCEELAKKIKKAA 730
Score = 33.5 bits (75), Expect = 1.1
Identities = 36/164 (21%), Positives = 73/164 (43%), Gaps = 14/164 (8%)
Query: 56 AKKKKNDQSPRKTVLCKCGENDDGSKCVCEAYRNLRESQEGFFEKENFFDGEEKNDEEKG 115
A+KKK ++ +K E + KC E + ++++ E +E G+E + +K
Sbjct: 442 AEKKKCKEAAKKEK-----EAAERKKCE-ELAKKIKKAAEKKKCEETAKKGKEVAERKKC 495
Query: 116 ENVLEEVIEANLI--IHDIGNEERKIDEEEGVEE------ENEEKGNCSSMVKRRREKVM 167
E + +++ +A + + +E++ E++ E+ E EK C K+R+E
Sbjct: 496 EELAKKIKKAEIKKKCKKLAKKEKETAEKKKCEKAAKKRKEAAEKKKCEKAAKKRKEAAE 555
Query: 168 EEARNSVPENGKVMHLVKAFERLLSIKKEKEKNEEEEENDKKNK 211
++ + K K E+ +KE + ++ EE KK K
Sbjct: 556 KKKCEKSAKKRKEAAEKKKCEKAAKERKEAAEKKKCEEAAKKEK 599
Score = 31.2 bits (69), Expect = 5.5
Identities = 47/230 (20%), Positives = 90/230 (38%), Gaps = 40/230 (17%)
Query: 4 EKKKPLEIENLNPNEFSNRTPLQKSLTKGDNNCVNVVVVSPQKRIRQRKFVV----AKKK 59
EK K E +P E ++ +K TKG + N + + R ++ K +V +K
Sbjct: 164 EKSKEKECNQNSPAE-GDKDRTKKGKTKGKSGGGNKKRSTKENRAKKGKKLVKNRFTQKL 222
Query: 60 KNDQSPRKTVLCKCGEN--DDGSKCVCEAYRNLRESQEGFFEKENFFD------------ 105
++ +C+C +N +D K + +Y+ + + K +
Sbjct: 223 EHCIKSEWADVCECRQNFTEDERKRLAASYKCMGTKIKSICRKRVIAEMCEAAGYVKSSE 282
Query: 106 ----GEEKNDEEKGENVLEEVIEANLIIHDIGNEERKIDEEEGVEEENEEKGNCSSMVKR 161
G++K ++EK E LE I +E EEE + +G + K+
Sbjct: 283 PKKKGKKKKNDEKKEKELEREIL-----------------KEQAEEEAKIRGVVKEVKKK 325
Query: 162 RREKVMEEARNSVPENGKVMHLVKAFERLLSIKKEKEKNEEEEENDKKNK 211
+EK +++ + K K L +KE+++ + +E KK K
Sbjct: 326 CKEKALKKKCKDLGRKMKEEAEKKKCAALAKKQKEEDEKKACKELAKKKK 375
Score = 31.2 bits (69), Expect = 5.5
Identities = 39/189 (20%), Positives = 73/189 (37%), Gaps = 27/189 (14%)
Query: 25 LQKSLTKGDNNCVNVVVVSPQKRIRQRKFV--VAKKKKNDQSPRKTVLCKCGENDDGSKC 82
L K G+ N + +K ++++K +AKKK ++ K K E + KC
Sbjct: 738 LAKKKKAGEKNKLKKGNKKGKKALKEKKKCRELAKKKAAEKKKCKEAAKKEKEAAEKKKC 797
Query: 83 VCEAYRNLRESQEGFFEKENFFDGEEKNDEEKGENVLEEVIEANLIIHDIGNEERKIDEE 142
A + +E+ +++K E ++ EA E++K ++
Sbjct: 798 EKTAKKR-----------------KEEAEKKKCEKTAKKRKEAA--------EKKKCEKA 832
Query: 143 EGVEEENEEKGNCSSMVKRRREKVMEEARNSVPENGKVMHLVKAFERLLSIKKEKEKNEE 202
+E EK C K+R+E ++ + K K E+ +KE + ++
Sbjct: 833 AKKRKEEAEKKKCEKTAKKRKETAEKKKCEKAAKKRKQAAEKKKCEKAAKKRKEAAEKKK 892
Query: 203 EEENDKKNK 211
E KK K
Sbjct: 893 CAEAAKKEK 901
Score = 31.2 bits (69), Expect = 5.5
Identities = 38/176 (21%), Positives = 69/176 (38%), Gaps = 20/176 (11%)
Query: 45 QKRIRQRKFVVAKKKKNDQSPRKTVLCKCGENDDGSKCVCEAYRNLRESQEGFFEKENFF 104
++R ++ K KK+ + + R+ + + + +K EA + ++ EKE
Sbjct: 1113 EERAKKEKEAAEKKRCEEAAKREKEAAEKKKCAEAAKKEKEATEKQKCAEAAKKEKEA-- 1170
Query: 105 DGEEKNDEEKGENVLEEVIEANLIIHDIGNEE------RKIDEEEGVEEENEEKGNCSSM 158
E+K E + E + D+ +E +K +E E+E EK C+
Sbjct: 1171 -AEKKKCAEAAKREKEAAQKKKCA--DLAKKEQEPAEMKKCEEAAKKEKEAAEKQKCAKA 1227
Query: 159 VKRRREKVMEEARNSVPENGKVMHLVKAFERLLSIKKEKEKNEEEEENDKKNKVMK 214
K+ +E E K K + KK E ++E+E +KK K K
Sbjct: 1228 AKKEKEAA---------EKKKCAEAAKKEQEAAEKKKCAEAAKKEKEAEKKRKCEK 1274
>CENB_SHEEP (P49451) Major centromere autoantigen B (Centromere
protein B) (CENP-B) (Fragment)
Length = 239
Score = 41.2 bits (95), Expect = 0.005
Identities = 33/123 (26%), Positives = 63/123 (50%), Gaps = 9/123 (7%)
Query: 92 ESQEGFFEKENFFDGEEKNDEEKGENV-LEEVIEANLIIHDIGNEERKIDE--EEGVEEE 148
E +EG E+E DGEE+ + +GE + EE +E + + EE + +E EG+E E
Sbjct: 57 EEEEGEGEEEEEEDGEEEEEAGEGEELGEEEEVEEEGDVDTVEEEEEEEEESSSEGLEAE 116
Query: 149 NEEKGNCSSMVKRRREKVMEEARNSVPENGKVMHLVKAFERLLSIKKEKEKNEEEEENDK 208
+ +G + EEA+ +H ++ E S +E+E++++E+E+D+
Sbjct: 117 DWAQGVVEAGGSFGGYGAQEEAQ------CPTLHFLEGEEDSESDSEEEEEDDDEDEDDE 170
Query: 209 KNK 211
++
Sbjct: 171 DDE 173
Score = 32.7 bits (73), Expect = 1.9
Identities = 34/132 (25%), Positives = 50/132 (37%), Gaps = 34/132 (25%)
Query: 135 EERKIDEEEGVEEENEEKGNCSSMVKRRREKVMEEARNSVPENGKVMHLVKAFERLLSIK 194
EE + +EEEG EE EE+ E++ EE V E G V +
Sbjct: 52 EEEEEEEEEGEGEEEEEEDGEEEEEAGEGEELGEE--EEVEEEGDVDTV----------- 98
Query: 195 KEKEKNEEEEENDKKNKVMKWA--------------------LPGLQFQQPVKDGDEQSE 234
E+E+ EEEE + + + WA P L F + +D + SE
Sbjct: 99 -EEEEEEEEESSSEGLEAEDWAQGVVEAGGSFGGYGAQEEAQCPTLHFLEGEEDSESDSE 157
Query: 235 VVSSCCDDDDFD 246
D+D+ D
Sbjct: 158 EEEEDDDEDEDD 169
>ATRX_PONPY (Q7YQM3) Transcriptional regulator ATRX (X-linked helicase
II) (X-linked nuclear protein) (XNP)
Length = 2492
Score = 41.2 bits (95), Expect = 0.005
Identities = 53/232 (22%), Positives = 91/232 (38%), Gaps = 29/232 (12%)
Query: 16 PNEFSNRTPLQKSLTKGDNNCVNVVVVSPQKRIRQRKFVVAKKKKNDQSPRKTVLCKCGE 75
P E RT Q GD N V + K K + + R + GE
Sbjct: 1296 PEEGKKRTGKQNEENPGDEEAKNQVNSESDSDSEESK----KPRYRHRLLRHKLTVSDGE 1351
Query: 76 NDDGSKCVCEAYRNLRESQEGFFEKENFFDGEEKNDEEKGENVLEEVIEANLIIHDIGNE 135
+ + K + ++ ++ E D E+ + +E G V EEV E+ +E
Sbjct: 1352 SGEEKKTKPKEHKEVKGRNRRKVSSE---DSEDSDFQESG--VSEEVSESE-------DE 1399
Query: 136 ERKIDEEEGVEEENEEKGNCSSMVKRRREKVMEEARNSVPENGKVMHLVKAFERLLSIKK 195
+R E E + + KRRR KV E++ + N + K E ++
Sbjct: 1400 QRPRTRSAKKAELEENQRSYKQKKKRRRIKVQEDSSSENKSNSEEEEEEKEEE---EEEE 1456
Query: 196 EKEKNEEEEEND----------KKNKVMKWALPGLQFQQPVKDGDEQSEVVS 237
E+E+ EEE+END K K++K + Q +K+ +E+ + ++
Sbjct: 1457 EEEEEEEEDENDDSKSPGKGRKKIRKILKDDKLRTETQNALKEEEERRKRIA 1508
>ATRX_PANTR (Q7YQM4) Transcriptional regulator ATRX (X-linked helicase
II) (X-linked nuclear protein) (XNP)
Length = 2492
Score = 41.2 bits (95), Expect = 0.005
Identities = 53/232 (22%), Positives = 91/232 (38%), Gaps = 29/232 (12%)
Query: 16 PNEFSNRTPLQKSLTKGDNNCVNVVVVSPQKRIRQRKFVVAKKKKNDQSPRKTVLCKCGE 75
P E RT Q GD N V + K K + + R + GE
Sbjct: 1296 PEEGKKRTGKQNEENPGDEEAKNQVNSESDSDSEESK----KPRYRHRLLRHKLTVSDGE 1351
Query: 76 NDDGSKCVCEAYRNLRESQEGFFEKENFFDGEEKNDEEKGENVLEEVIEANLIIHDIGNE 135
+ + K + ++ ++ E D E+ + +E G V EEV E+ +E
Sbjct: 1352 SGEEKKTKPKEHKEVKGRNRRKVSSE---DSEDSDFQESG--VSEEVSESE-------DE 1399
Query: 136 ERKIDEEEGVEEENEEKGNCSSMVKRRREKVMEEARNSVPENGKVMHLVKAFERLLSIKK 195
+R E E + + KRRR KV E++ + N + K E ++
Sbjct: 1400 QRPRTRSAKKAELEENQRSYKQKKKRRRIKVQEDSSSENKSNSEEEEEEKEEE---EEEE 1456
Query: 196 EKEKNEEEEEND----------KKNKVMKWALPGLQFQQPVKDGDEQSEVVS 237
E+E+ EEE+END K K++K + Q +K+ +E+ + ++
Sbjct: 1457 EEEEEEEEDENDDSKSPGKGRKKIRKILKDDKLRTETQNALKEEEERRKRIA 1508
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.339 0.149 0.490
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 50,915,517
Number of Sequences: 164201
Number of extensions: 2256872
Number of successful extensions: 22054
Number of sequences better than 10.0: 499
Number of HSP's better than 10.0 without gapping: 64
Number of HSP's successfully gapped in prelim test: 459
Number of HSP's that attempted gapping in prelim test: 17659
Number of HSP's gapped (non-prelim): 1886
length of query: 439
length of database: 59,974,054
effective HSP length: 113
effective length of query: 326
effective length of database: 41,419,341
effective search space: 13502705166
effective search space used: 13502705166
T: 11
A: 40
X1: 15 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 39 (21.8 bits)
S2: 67 (30.4 bits)
Medicago: description of AC149305.8


