

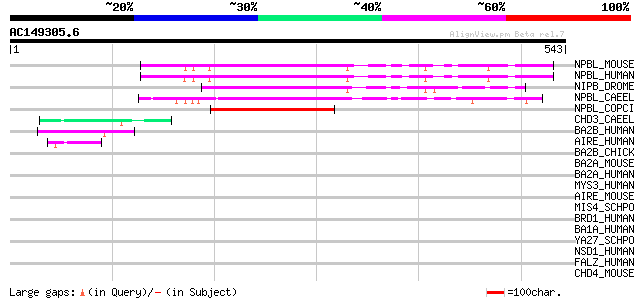
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC149305.6 - phase: 0 /pseudo/partial
(543 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
NPBL_MOUSE (Q6KCD5) Nipped-B-like protein (Delangin homolog) (SC... 119 2e-26
NPBL_HUMAN (Q6KC79) Nipped-B-like protein (Delangin) (SCC2 homolog) 119 2e-26
NIPB_DROME (Q7PLI2) Nipped-B protein (SCC2 homolog) 116 1e-25
NPBL_CAEEL (Q95XZ5) Nipped-B-like protein pqn-85 (Prion-like-(Q/... 98 6e-20
NPBL_COPCI (Q00333) Rad9 protein (SCC2 homolog) 89 4e-17
CHD3_CAEEL (Q22516) Chromodomain helicase-DNA-binding protein 3 ... 47 2e-04
BA2B_HUMAN (Q9UIF8) Bromodomain adjacent to zinc finger domain 2... 46 2e-04
AIRE_HUMAN (O43918) Autoimmune regulator (Autoimmune polyendocri... 44 8e-04
BA2B_CHICK (Q9DE13) Bromodomain adjacent to zinc finger domain 2... 44 0.001
BA2A_MOUSE (Q91YE5) Bromodomain adjacent to zinc finger domain 2... 44 0.001
BA2A_HUMAN (Q9UIF9) Bromodomain adjacent to zinc finger domain 2... 44 0.001
MYS3_HUMAN (Q92794) MYST histone acetyltransferase 3 (Runt-relat... 43 0.002
AIRE_MOUSE (Q9Z0E3) Autoimmune regulator (Autoimmune polyendocri... 42 0.003
MIS4_SCHPO (Q09725) Sister chromatid cohesion protein mis4 (SCC2... 42 0.005
BRD1_HUMAN (O95696) Bromodomain-containing protein 1 (BR140-like... 42 0.005
BA1A_HUMAN (Q9NRL2) Bromodomain adjacent to zinc finger domain p... 41 0.007
YA27_SCHPO (Q09698) Hypothetical protein C2F7.07c in chromosome I 41 0.009
NSD1_HUMAN (Q96L73) Nuclear receptor binding SET domain containi... 41 0.009
FALZ_HUMAN (Q12830) Fetal Alzheimer antigen (Fetal Alz-50-reacti... 39 0.026
CHD4_MOUSE (Q6PDQ2) Chromodomain helicase-DNA-binding protein 4 ... 39 0.026
>NPBL_MOUSE (Q6KCD5) Nipped-B-like protein (Delangin homolog) (SCC2
homolog)
Length = 2798
Score = 119 bits (299), Expect = 2e-26
Identities = 114/453 (25%), Positives = 191/453 (41%), Gaps = 93/453 (20%)
Query: 129 VQQLLLNYFQDVTSADDLHHFICWFYLCSWYKNDPKCQQKPI------------YYFARM 176
+Q+ LL+Y + T D F FY+ W+++ +K + ++ +
Sbjct: 1645 LQKALLDYLDENTETDPSLVFSRKFYIAQWFRDTTLETEKAMKSQKDEESSDATHHAKEL 1704
Query: 177 KS---------------RTIVRDSGSVSSMLTR-------DSIKKITLALGQNSSFCRGF 214
++ R+I++ + S S L D I L F + F
Sbjct: 1705 ETTGQIMHRAENRKKFLRSIIKTTPSQFSTLKMNSDTVDYDDACLIVRYLASMRPFAQSF 1764
Query: 215 DKIFHTLLVSLKENSPVIRAKALRAVSIIVEADPEVLGDKFVQSSVEGRFCDTAISVREA 274
D +L L EN+ +R KA++ +S +V DP +L +Q V GR D + SVREA
Sbjct: 1765 DIYLTQILRVLGENAIAVRTKAMKCLSEVVAVDPSILARLDMQRGVHGRLMDNSTSVREA 1824
Query: 275 ALELVGRHIASHPDVGFKYFEKITERIKDTGVSVRKRAIKIIRDMCCSDGNFLRV----Y 330
A+EL+GR + P + +Y++ + ERI DTG+SVRKR IKI+RD+C F ++
Sbjct: 1825 AVELLGRFVLCRPQLAEQYYDMLIERILDTGISVRKRVIKILRDICIEQPTFPKITEMCV 1884
Query: 331 KSLHRDNFTCY***IEYPAYSASDLQDLVCKTFYEFWFEEPSTPQTQVFKDGSTVPLEVA 390
K + R N ++ LV +TF + WF TP K+ T
Sbjct: 1885 KMIRRVN-------------DEEGIKKLVNETFQKLWF----TPTPHNDKEAMT------ 1921
Query: 391 KKTEQIVEMLKRLPN------NQLLVTVIKRCLTLDFLPQSAKASGVNPVSLVTVRKRCE 444
+K I +++ + QLL ++K S + S P V+K C
Sbjct: 1922 RKILNITDVVAACRDTGYDWFEQLLQNLLK----------SEEDSSYKP-----VKKACT 1966
Query: 445 LMCKCLLEKILQVDEMNSNELEK-----HALPYVLVLHAFCLVDPTLCAPASNPSQFVLT 499
+ L+E IL+ +E ++ K + + L F + P L + +T
Sbjct: 1967 QLVDNLVEHILKYEESLADSDNKGVNSGRLVACITTLFLFSKIRPQLMV------KHAMT 2020
Query: 500 LQPYLKTQVKTNLKTYILGSLQCYYALCIELLE 532
+QPYL T+ T ++ ++ L + L+E
Sbjct: 2021 MQPYLTTKCSTQNDFMVICNVAKILELVVPLME 2053
>NPBL_HUMAN (Q6KC79) Nipped-B-like protein (Delangin) (SCC2 homolog)
Length = 2804
Score = 119 bits (299), Expect = 2e-26
Identities = 116/453 (25%), Positives = 189/453 (41%), Gaps = 93/453 (20%)
Query: 129 VQQLLLNYFQDVTSADDLHHFICWFYLCSWYKNDPKCQQKP------------------- 169
+Q+ LL+Y + T D F FY+ W+++ +K
Sbjct: 1651 LQKALLDYLDENTETDPSLVFSRKFYIAQWFRDTTLETEKAMKSQKDEESSEGTHHAKEI 1710
Query: 170 -----IYYFARMKS---RTIVRDSGSVSSMLTR-------DSIKKITLALGQNSSFCRGF 214
I + A + R+I++ + S S L D I L F + F
Sbjct: 1711 ETTGQIMHRAENRKKFLRSIIKTTPSQFSTLKMNSDTVDYDDACLIVRYLASMRPFAQSF 1770
Query: 215 DKIFHTLLVSLKENSPVIRAKALRAVSIIVEADPEVLGDKFVQSSVEGRFCDTAISVREA 274
D +L L EN+ +R KA++ +S +V DP +L +Q V GR D + SVREA
Sbjct: 1771 DIYLTQILRVLGENAIAVRTKAMKCLSEVVAVDPSILARLDMQRGVHGRLMDNSTSVREA 1830
Query: 275 ALELVGRHIASHPDVGFKYFEKITERIKDTGVSVRKRAIKIIRDMCCSDGNFLRV----Y 330
A+EL+GR + P + +Y++ + ERI DTG+SVRKR IKI+RD+C F ++
Sbjct: 1831 AVELLGRFVLCRPQLAEQYYDMLIERILDTGISVRKRVIKILRDICIEQPTFPKITEMCV 1890
Query: 331 KSLHRDNFTCY***IEYPAYSASDLQDLVCKTFYEFWFEEPSTPQTQVFKDGSTVPLEVA 390
K + R N ++ LV +TF + WF TP K+ T
Sbjct: 1891 KMIRRVN-------------DEEGIKKLVNETFQKLWF----TPTPHNDKEAMT------ 1927
Query: 391 KKTEQIVEMLKRLPN------NQLLVTVIKRCLTLDFLPQSAKASGVNPVSLVTVRKRCE 444
+K I +++ + QLL ++K S + S P V+K C
Sbjct: 1928 RKILNITDVVAACRDTGYDWFEQLLQNLLK----------SEEDSSYKP-----VKKACT 1972
Query: 445 LMCKCLLEKILQVDEMNSNELEK-----HALPYVLVLHAFCLVDPTLCAPASNPSQFVLT 499
+ L+E IL+ +E ++ K + + L F + P L + +T
Sbjct: 1973 QLVDNLVEHILKYEESLADSDNKGVNSGRLVACITTLFLFSKIRPQLMV------KHAMT 2026
Query: 500 LQPYLKTQVKTNLKTYILGSLQCYYALCIELLE 532
+QPYL T+ T ++ ++ L + L+E
Sbjct: 2027 MQPYLTTKCSTQNDFMVICNVAKILELVVPLME 2059
>NIPB_DROME (Q7PLI2) Nipped-B protein (SCC2 homolog)
Length = 2077
Score = 116 bits (291), Expect = 1e-25
Identities = 87/331 (26%), Positives = 164/331 (49%), Gaps = 50/331 (15%)
Query: 188 VSSMLTRDSIKKITLALGQNSSFCRGFDKIFHTLLVSLKENSPVIRAKALRAVSIIVEAD 247
+ + + ++ + I L F + FD +++ + E S +R +A++ ++ IVE D
Sbjct: 1101 IKTYIDYNNAQLIAQYLATKRPFSQSFDGCLKKIILVVNEPSIAVRTRAMKCLANIVEVD 1160
Query: 248 PEVLGDKFVQSSVEGRFCDTAISVREAALELVGRHIASHPDVGFKYFEKITERIKDTGVS 307
P VL K +Q V +F DTAISVREAA++LVG+ + S+ D+ +Y++ ++ RI DTGVS
Sbjct: 1161 PLVLKRKDMQMGVNQKFLDTAISVREAAVDLVGKFVLSNQDLIDQYYDMLSTRILDTGVS 1220
Query: 308 VRKRAIKIIRDMCCSDGNFLRV----YKSLHRDNFTCY***IEYPAYSASDLQDLVCKTF 363
VRKR IKI+RD+C +F ++ K + R + +Q LV + F
Sbjct: 1221 VRKRVIKILRDICLEYPDFSKIPEICVKMIRR-------------VHDEEGIQKLVTEVF 1267
Query: 364 YEFWFEEPSTPQTQVFKDGSTVPLEVAKKTEQIVEMLKRLPN------NQLLVTVIK--- 414
+ WF TP T+ K G + +K I++++ + LL+++ K
Sbjct: 1268 MKMWF----TPCTKNDKIG------IQRKINHIIDVVNTAHDTGTTWLEGLLMSIFKPRD 1317
Query: 415 -RCLTLDFLPQSAKASGVNPVSLVTVRKRCELMCKCLLEKILQVDEMNSNELEKHALPYV 473
+ + + K + P+ +V C+ + L+++++++++ +++ + L +
Sbjct: 1318 NMLRSEGCVQEFIKKNSEPPMDIVIA---CQQLADGLVDRLIELEDTDNSRM----LGCI 1370
Query: 474 LVLHAFCLVDPTLCAPASNPSQFVLTLQPYL 504
LH V P L + +T++PYL
Sbjct: 1371 TTLHLLAKVRPQLLV------KHAITIEPYL 1395
>NPBL_CAEEL (Q95XZ5) Nipped-B-like protein pqn-85
(Prion-like-(Q/n-rich)-domain-bearing protein 85) (SCC2
homolog)
Length = 2203
Score = 97.8 bits (242), Expect = 6e-20
Identities = 110/446 (24%), Positives = 188/446 (41%), Gaps = 83/446 (18%)
Query: 127 EIVQQLLLNYFQDVTSADDLHHFICWFYLCSWYKN----------------DPKCQQKPI 170
++++ L++Y +T++ D+ + C FY+ WYK D +K +
Sbjct: 1184 KVLETSLIDYLV-ITNSSDIIVYACNFYVGEWYKEVAEDLESARSKLKQTVDTNESEKDV 1242
Query: 171 -----------YYFARMK--------SRTIVR--DSGSVSSMLTRDSIKKITLALGQNSS 209
Y A MK + I R + + ML D+ + L Q+
Sbjct: 1243 KKAERKYEKIQYRGAEMKVFLSKILDKKEIKRRLEKSNKVKMLDSDAFWAVKF-LAQSRE 1301
Query: 210 FCRGFDKIF-HTLLVSLKENSPVIRAKALRAVSIIVEADPEVLGDKFVQSSVEGRFCDTA 268
F FD H + + E +R+KAL+ +S I+EAD VL + VQ +V R D+
Sbjct: 1302 FTHSFDTYLKHIVFGAGSETIVALRSKALKCLSSIIEADSSVLILEDVQQAVHTRMVDSH 1361
Query: 269 ISVREAALELVGRHIASHPDVGFKYFEKITERIKDTGVSVRKRAIKIIRDMCCSDGNFLR 328
VRE+A+EL+GR + + KY+ +I ERI DTGV+VRKR I+I+R++C F
Sbjct: 1362 AQVRESAVELIGRFVLYDEEYVRKYYSQIAERILDTGVAVRKRVIRIMREICEKFPTFEM 1421
Query: 329 VYKSLHRDNFTCY***IEYPAYSASDLQDLVCKTFYEFWFEEPSTPQTQVFKDGSTVPLE 388
+ L R + ++ LV +TF WF+ T+++ + V +
Sbjct: 1422 IPDMLAR---------MIRRVTDEEGVKKLVFETFTTLWFQ---PVDTRIYT--NAVATK 1467
Query: 389 VAKKTEQIVEMLKRLPNNQLLVTVIKRCLTLDFLPQSAKASGVNPVSLVTVRKRCELMCK 448
V +K ++ L L L + + SG++ V V++ + +
Sbjct: 1468 VTTMCSVAQHCIKDAMSDYL------EQLILHIVKNGQEGSGMS----VAVKQIIDSLVD 1517
Query: 449 CLL--------EKILQVDEMNSNELEKHALPYVLVLHAFCLVDPTLCAPASNPSQFVLTL 500
+L E + +V+ M E E+ + Y+ L F + P L + V L
Sbjct: 1518 HILNLEQHKSSENVSEVELMRRKEQEEKYMAYLSTLAVFSKIRPLLL------TSHVEVL 1571
Query: 501 QPYL-----KTQVKTNLKTYILGSLQ 521
PYL KT + + ++G L+
Sbjct: 1572 LPYLTFSGAKTNAENQVTKEMIGMLE 1597
>NPBL_COPCI (Q00333) Rad9 protein (SCC2 homolog)
Length = 2157
Score = 88.6 bits (218), Expect = 4e-17
Identities = 44/121 (36%), Positives = 75/121 (61%)
Query: 197 IKKITLALGQNSSFCRGFDKIFHTLLVSLKENSPVIRAKALRAVSIIVEADPEVLGDKFV 256
I +++ +G S F I + +L +L +R KALRA+ IV +D +LG V
Sbjct: 1190 IDRLSEEIGTIQSLRNSFQPILNVILSALDAPVIFMRTKALRALGQIVTSDATILGTASV 1249
Query: 257 QSSVEGRFCDTAISVREAALELVGRHIASHPDVGFKYFEKITERIKDTGVSVRKRAIKII 316
+ +E D++ +VR+AA+EL+G+++ P+V Y++KI ER+ DTG++VRKR IK++
Sbjct: 1250 RQGIENHLLDSSPAVRDAAVELIGKYMIDSPEVAGNYYQKIAERMADTGLAVRKRVIKLL 1309
Query: 317 R 317
+
Sbjct: 1310 K 1310
>CHD3_CAEEL (Q22516) Chromodomain helicase-DNA-binding protein 3
homolog (CHD-3)
Length = 1787
Score = 46.6 bits (109), Expect = 2e-04
Identities = 31/136 (22%), Positives = 52/136 (37%), Gaps = 20/136 (14%)
Query: 30 DLLSEDAAPQHYPKDTCCVCLGGRVENLFKCSGCDRLFHADCLDVKENEVPNRNWYCLMC 89
D+L + P D C +C N+ C C +HA C+D E+P W C C
Sbjct: 315 DVLIVEEEPAKANMDYCRICK--ETSNILLCDTCPSSYHAYCIDPPLTEIPEGEWSCPRC 372
Query: 90 ICSKQLLVLQSYCNSQRKD-------DAKKNRKVSKDDSTFSNHEIVQQLLLNYFQDVTS 142
I + ++ + + K+ + K+ + SKDD + L + +
Sbjct: 373 IIPEPAQRIEKILSWRWKEISYPEPLECKEGEEASKDD-----------VFLKPPRKMEP 421
Query: 143 ADDLHHFICWFYLCSW 158
+ F+ W YL W
Sbjct: 422 RREREFFVKWKYLAYW 437
Score = 33.9 bits (76), Expect = 1.1
Identities = 15/56 (26%), Positives = 23/56 (40%), Gaps = 2/56 (3%)
Query: 34 EDAAPQHYPKDTCCVCLGGRVENLFKCSGCDRLFHADCLDVKENEVPNRNWYCLMC 89
E + ++ C VC + L C C R +H C+D + P +W C C
Sbjct: 256 EQGVVEENHQENCEVC--NQDGELMLCDTCTRAYHVACIDENMEQPPEGDWSCPHC 309
>BA2B_HUMAN (Q9UIF8) Bromodomain adjacent to zinc finger domain 2B
(hWALp4)
Length = 1972
Score = 46.2 bits (108), Expect = 2e-04
Identities = 27/99 (27%), Positives = 46/99 (46%), Gaps = 4/99 (4%)
Query: 28 LQDLLSEDAAPQHYPKDTCCVCL-GGRVENLFKCSGCDRLFHADCLDVKENEVPNRNWYC 86
+Q L A + K C +C G E L C GCD+ H C K +P+ +W+C
Sbjct: 1720 IQQLQKSIAWEKSIMKVYCQICRKGDNEELLLLCDGCDKGCHTYCHRPKITTIPDGDWFC 1779
Query: 87 LMCIC---SKQLLVLQSYCNSQRKDDAKKNRKVSKDDST 122
CI + L + + + ++ +++KK +KV+ T
Sbjct: 1780 PACIAKASGQTLKIKKLHVKGKKTNESKKGKKVTLTGDT 1818
>AIRE_HUMAN (O43918) Autoimmune regulator (Autoimmune
polyendocrinopathy candidiasis ectodermal dystrophy
protein) (APECED protein)
Length = 545
Score = 44.3 bits (103), Expect = 8e-04
Identities = 22/55 (40%), Positives = 26/55 (47%), Gaps = 4/55 (7%)
Query: 38 PQHYPK--DTCCVCLGGRVENLFKCSGCDRLFHADCLDVKENEVPNRNWYCLMCI 90
PQ + K D C VC G L C GC R FH CL E+P+ W C C+
Sbjct: 289 PQLHQKNEDECAVCRDGG--ELICCDGCPRAFHLACLSPPLREIPSGTWRCSSCL 341
>BA2B_CHICK (Q9DE13) Bromodomain adjacent to zinc finger domain 2B
(Extracellular matrix protein F22)
Length = 2130
Score = 43.9 bits (102), Expect = 0.001
Identities = 29/96 (30%), Positives = 44/96 (45%), Gaps = 4/96 (4%)
Query: 28 LQDLLSEDAAPQHYPKDTCCVCL-GGRVENLFKCSGCDRLFHADCLDVKENEVPNRNWYC 86
+Q L A + K C +C G E L C GCD+ H C K +P+ +W+C
Sbjct: 1880 IQQLQKSIAWEKSIMKVYCQICRKGDNEELLLLCDGCDKGCHTYCHRPKITTIPDGDWFC 1939
Query: 87 LMCI--CSKQLLVLQS-YCNSQRKDDAKKNRKVSKD 119
CI S Q L L+ ++ ++ K+ RK+ D
Sbjct: 1940 PACIAKASGQTLKLKKLQIKGKKSNEQKRGRKLPGD 1975
>BA2A_MOUSE (Q91YE5) Bromodomain adjacent to zinc finger domain 2A
(Transcription termination factor-I interacting protein
5) (TTF-I interacting protein 5) (Tip5)
Length = 1850
Score = 43.9 bits (102), Expect = 0.001
Identities = 27/74 (36%), Positives = 36/74 (48%), Gaps = 6/74 (8%)
Query: 43 KDTCCVCL-GGRVENLFKCSGCDRLFHADCLDVKENEVPNRNWYCLMCICSKQLLVLQSY 101
K TC VC G E L C GCDR H C K VP +W+C +C+ + V + Y
Sbjct: 1623 KVTCLVCRKGDNDEFLLLCDGCDRGCHIYCHRPKMEAVPEGDWFCAVCLSQQ---VEEEY 1679
Query: 102 CNSQRKDDAKKNRK 115
+QR K+ +K
Sbjct: 1680 --TQRPGFPKRGQK 1691
>BA2A_HUMAN (Q9UIF9) Bromodomain adjacent to zinc finger domain 2A
(Transcription termination factor-I interacting protein
5) (TTF-I interacting protein 5) (Tip5) (hWALp3)
Length = 1878
Score = 43.9 bits (102), Expect = 0.001
Identities = 21/52 (40%), Positives = 26/52 (49%), Gaps = 1/52 (1%)
Query: 43 KDTCCVCL-GGRVENLFKCSGCDRLFHADCLDVKENEVPNRNWYCLMCICSK 93
K TC VC G E L C GCDR H C K VP +W+C +C+ +
Sbjct: 1649 KVTCLVCRKGDNDEFLLLCDGCDRGCHIYCHRPKMEAVPEGDWFCTVCLAQQ 1700
>MYS3_HUMAN (Q92794) MYST histone acetyltransferase 3 (Runt-related
transcription factor binding protein 2) (Monocytic
leukemia zinc finger protein) (Zinc finger protein 220)
Length = 2004
Score = 42.7 bits (99), Expect = 0.002
Identities = 26/87 (29%), Positives = 36/87 (40%), Gaps = 13/87 (14%)
Query: 45 TCCVCL--GGRVENLFKCSGCDRLFHADCLDVKENEVPNRNWYCLMC---------ICSK 93
TC C G +N+ C CDR FH +C D +P W C +C + K
Sbjct: 264 TCSSCRDQGKNADNMLFCDSCDRGFHMECCDPPLTRMPKGMWICQICRPRKKGRKLLQKK 323
Query: 94 QLLVLQSYCN--SQRKDDAKKNRKVSK 118
+ + Y N + K+ KK VSK
Sbjct: 324 AAQIKRRYTNPIGRPKNRLKKQNTVSK 350
>AIRE_MOUSE (Q9Z0E3) Autoimmune regulator (Autoimmune
polyendocrinopathy candidiasis ectodermal dystrophy
protein homolog) (APECED protein homolog)
Length = 552
Score = 42.4 bits (98), Expect = 0.003
Identities = 19/48 (39%), Positives = 23/48 (47%), Gaps = 2/48 (4%)
Query: 43 KDTCCVCLGGRVENLFKCSGCDRLFHADCLDVKENEVPNRNWYCLMCI 90
+D C VC G L C GC R FH CL E+P+ W C C+
Sbjct: 298 EDECAVCHDGG--ELICCDGCPRAFHLACLSPPLQEIPSGLWRCSCCL 343
>MIS4_SCHPO (Q09725) Sister chromatid cohesion protein mis4 (SCC2
homolog)
Length = 1583
Score = 41.6 bits (96), Expect = 0.005
Identities = 67/308 (21%), Positives = 125/308 (39%), Gaps = 50/308 (16%)
Query: 216 KIFHTLLVSLKENSPV-IRAKALRAVS------IIVEADPEVLGDKFVQSSVEGRFCDTA 268
K F +L++ ++ +R K LR ++ I+ PEVL +S+ D +
Sbjct: 774 KFFVSLIIGFLDSPQASLRTKCLRIINQMKTIPSILRTHPEVLAQIISKSN------DQS 827
Query: 269 ISVREAALELVGRHIASHPDVGFKYFEKITERIKDTGVSVRKRAIKIIRDMC-CSDGNFL 327
VR+ L+L+G +I ++ + + + I I D VRKRAIK + ++ ++ +
Sbjct: 828 AIVRDTVLDLLGTYIMAYRETIPQIYGCIISGISDPSTIVRKRAIKQLCEVYEATEDLNI 887
Query: 328 RV---YKSLHRDNFTCY***IEYPAYSASDLQDLVCKTFYEFWFEEPSTPQTQVFKDGST 384
RV K L R N + +L + + WF P++ + K
Sbjct: 888 RVDIASKLLTRSN------------DEEETISELSLEVLEKLWF-SPASNELDCQKGYEQ 934
Query: 385 VP-LEVAKKTEQIVEMLK-----RLPNNQLLVTVIKRCLTLDFLPQSAKASGVNPVSLVT 438
+ LE K Q +LK + LLVT +K LT ++L T
Sbjct: 935 LTFLEKQKLRVQYFPILKLCAEPSTERHVLLVTSLKTMLT-----------SKEEINLST 983
Query: 439 VRKRCELMCKCLLEKILQVDEMNSNELEKHALPYVLVLHAFCLVDPTLCAPASNPSQFVL 498
+ + L+ CL ++++V + + + Y ++ F + P ++
Sbjct: 984 LHTQIRLLLSCLFNQLIEVVTEDQVDESTKGILYEIMSTLFVF---SRAFPFLFDLSYLH 1040
Query: 499 TLQPYLKT 506
L+PYL++
Sbjct: 1041 LLKPYLRS 1048
>BRD1_HUMAN (O95696) Bromodomain-containing protein 1 (BR140-like
protein)
Length = 1058
Score = 41.6 bits (96), Expect = 0.005
Identities = 22/77 (28%), Positives = 33/77 (42%), Gaps = 13/77 (16%)
Query: 20 CSQEKFWVLQDLLSEDAAPQHYPKDTCCVCLGGRVEN---LFKCSGCDRLFHADCLDVKE 76
C +K Q L+ EDA CC+C+ G +N + C C+ H +C V
Sbjct: 199 CENQKQGEQQSLIDEDAV--------CCICMDGECQNSNVILFCDMCNLAVHQECYGVP- 249
Query: 77 NEVPNRNWYCLMCICSK 93
+P W C C+ S+
Sbjct: 250 -YIPEGQWLCRHCLQSR 265
>BA1A_HUMAN (Q9NRL2) Bromodomain adjacent to zinc finger domain
protein 1A (ATP-utilizing chromatin assembly and
remodeling factor 1) (hACF1) (ATP-dependent chromatin
remodelling protein) (Williams syndrome transcription
factor-related chromatin remodelin
Length = 1556
Score = 41.2 bits (95), Expect = 0.007
Identities = 18/45 (40%), Positives = 23/45 (51%), Gaps = 1/45 (2%)
Query: 46 CCVCLG-GRVENLFKCSGCDRLFHADCLDVKENEVPNRNWYCLMC 89
C +C G EN+ C GCDR H C+ K VP +W+C C
Sbjct: 1151 CKICRKKGDAENMVLCDGCDRGHHTYCVRPKLKTVPEGDWFCPEC 1195
>YA27_SCHPO (Q09698) Hypothetical protein C2F7.07c in chromosome I
Length = 607
Score = 40.8 bits (94), Expect = 0.009
Identities = 19/54 (35%), Positives = 24/54 (44%), Gaps = 4/54 (7%)
Query: 38 PQHYPKDTCCVCLGGRVENLFKCSGCDRLFHADCLD--VKENEVPNRNWYCLMC 89
P Y D C C G N C C FH C+D ++E +P+ WYC C
Sbjct: 258 PYRYNNDYCSACHGPG--NFLCCETCPNSFHFTCIDPPIEEKNLPDDAWYCNEC 309
>NSD1_HUMAN (Q96L73) Nuclear receptor binding SET domain containing
protein 1 (NR-binding SET domain containing protein)
(Androgen receptor-associated coregulator 267)
Length = 2696
Score = 40.8 bits (94), Expect = 0.009
Identities = 19/56 (33%), Positives = 27/56 (47%), Gaps = 5/56 (8%)
Query: 39 QHYPKDTCCVCLGGRVENLFKCSGCDRLFHADCLDVKENEVPNRNWYCLMCICSKQ 94
+H C VC G +L C C FH +CL++ ++P NWYC C K+
Sbjct: 1703 EHVNVSWCFVCSEGG--SLLCCDSCPAAFHRECLNI---DIPEGNWYCNDCKAGKK 1753
Score = 33.9 bits (76), Expect = 1.1
Identities = 28/91 (30%), Positives = 41/91 (44%), Gaps = 16/91 (17%)
Query: 43 KDTCCVCLGGRVENLFKCS--GCDRLFHADCLDVKENEVPNRNWYC--LMC-ICSKQLLV 97
+D C C G L C GC +++HADCL++ + P W C C IC K+
Sbjct: 2118 EDECFSC--GDAGQLVSCKKPGCPKVYHADCLNLTKR--PAGKWECPWHQCDICGKE--- 2170
Query: 98 LQSYCNSQRKDDAKKNRK----VSKDDSTFS 124
S+C K++R+ +SK D S
Sbjct: 2171 AASFCEMCPSSFCKQHREGMLFISKLDGRLS 2201
>FALZ_HUMAN (Q12830) Fetal Alzheimer antigen (Fetal Alz-50-reactive
clone 1)
Length = 810
Score = 39.3 bits (90), Expect = 0.026
Identities = 20/78 (25%), Positives = 33/78 (41%), Gaps = 2/78 (2%)
Query: 44 DTCCVCLGGRVENLFKCSGCDRLFHADCLDVKENEVPNRNWYCLMCICSKQLLVLQSYCN 103
D C VC ++ +L C C ++H +C+ EVP W C +C+ K V
Sbjct: 252 DHCRVC--HKLGDLLCCETCSAVYHLECVKPPLEEVPEDEWQCEVCVAHKVPGVTDCVAE 309
Query: 104 SQRKDDAKKNRKVSKDDS 121
Q+ ++ + D S
Sbjct: 310 IQKNKPYIRHEPIGYDRS 327
>CHD4_MOUSE (Q6PDQ2) Chromodomain helicase-DNA-binding protein 4
(CHD-4)
Length = 1915
Score = 39.3 bits (90), Expect = 0.026
Identities = 18/65 (27%), Positives = 28/65 (42%), Gaps = 2/65 (3%)
Query: 27 VLQDLLSEDAAPQHYPKDTCCVCLGGRVENLFKCSGCDRLFHADCLDVKENEVPNRNWYC 86
+L+++ + + + C VC G L C C +H CL+ E+PN W C
Sbjct: 426 ILEEVGGDPEEEDDHHMEFCRVCKDGG--ELLCCDTCPSSYHIHCLNPPLPEIPNGEWLC 483
Query: 87 LMCIC 91
C C
Sbjct: 484 PRCTC 488
Score = 34.7 bits (78), Expect = 0.65
Identities = 16/47 (34%), Positives = 20/47 (42%), Gaps = 2/47 (4%)
Query: 43 KDTCCVCLGGRVENLFKCSGCDRLFHADCLDVKENEVPNRNWYCLMC 89
+D C VC G + C C R +H CLD + P W C C
Sbjct: 363 QDYCEVCQQGG--EIILCDTCPRAYHMVCLDPDMEKAPEGKWSCPHC 407
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.327 0.139 0.433
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 61,203,867
Number of Sequences: 164201
Number of extensions: 2485902
Number of successful extensions: 7436
Number of sequences better than 10.0: 92
Number of HSP's better than 10.0 without gapping: 47
Number of HSP's successfully gapped in prelim test: 45
Number of HSP's that attempted gapping in prelim test: 7323
Number of HSP's gapped (non-prelim): 153
length of query: 543
length of database: 59,974,054
effective HSP length: 115
effective length of query: 428
effective length of database: 41,090,939
effective search space: 17586921892
effective search space used: 17586921892
T: 11
A: 40
X1: 15 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.7 bits)
S2: 68 (30.8 bits)
Medicago: description of AC149305.6


