

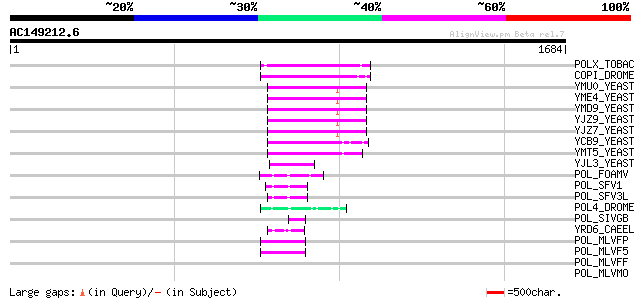
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC149212.6 - phase: 0 /pseudo
(1684 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
POLX_TOBAC (P10978) Retrovirus-related Pol polyprotein from tran... 163 3e-39
COPI_DROME (P04146) Copia protein (Gag-int-pol protein) [Contain... 147 2e-34
YMU0_YEAST (Q04670) Transposon Ty1 protein B 94 3e-18
YME4_YEAST (Q04711) Transposon Ty1 protein B 94 3e-18
YMD9_YEAST (Q03434) Transposon Ty1 protein B 94 3e-18
YJZ9_YEAST (P47100) Transposon Ty1 protein B 94 3e-18
YJZ7_YEAST (P47098) Transposon Ty1 protein B 93 5e-18
YCB9_YEAST (P25384) Transposon Ty2 protein B (Ty1-17 protein B) 93 7e-18
YMT5_YEAST (Q04214) Transposon Ty1 protein B 92 9e-18
YJL3_YEAST (P47024) Transposon Ty4 207.7 kDa hypothetical protein 69 1e-10
POL_FOAMV (P14350) Pol polyprotein [Contains: Reverse transcript... 62 2e-08
POL_SFV1 (P23074) Pol polyprotein [Contains: Protease (EC 3.4.23... 55 2e-06
POL_SFV3L (P27401) Pol polyprotein [Contains: Protease (EC 3.4.2... 52 1e-05
POL4_DROME (P10394) Retrovirus-related Pol polyprotein from tran... 48 3e-04
POL_SIVGB (P22382) Pol polyprotein [Contains: Protease (Retropep... 47 6e-04
YRD6_CAEEL (Q09575) Hypothetical protein K02A2.6 in chromosome II 46 8e-04
POL_MLVFP (P26808) Pol polyprotein [Contains: Protease (EC 3.4.2... 46 8e-04
POL_MLVF5 (P26810) Pol polyprotein [Contains: Protease (EC 3.4.2... 46 8e-04
POL_MLVFF (P26809) Pol polyprotein [Contains: Protease (EC 3.4.2... 46 0.001
POL_MLVMO (P03355) Pol polyprotein [Contains: Protease (EC 3.4.2... 45 0.001
>POLX_TOBAC (P10978) Retrovirus-related Pol polyprotein from
transposon TNT 1-94 [Contains: Protease (EC 3.4.23.-);
Reverse transcriptase (EC 2.7.7.49); Endonuclease]
Length = 1328
Score = 163 bits (413), Expect = 3e-39
Identities = 103/343 (30%), Positives = 169/343 (49%), Gaps = 16/343 (4%)
Query: 761 CQKGKIVKSSFKSKDIVSTSRPLELLHI---DLFGQVNTTSLYGSKYGLVIVDDYSRWTW 817
C GK + SF++ S+ R L +L + D+ G + S+ G+KY + +DD SR W
Sbjct: 460 CLFGKQHRVSFQT----SSERKLNILDLVYSDVCGPMEIESMGGNKYFVTFIDDASRKLW 515
Query: 818 VKFIKSKDYACEVFSSFCTQIQSEKELKILKVRSDHGGEFENETFESFCEKHGILHEFSS 877
V +K+KD +VF F ++ E K+ ++RSD+GGE+ + FE +C HGI HE +
Sbjct: 516 VYILKTKDQVFQVFQKFHALVERETGRKLKRLRSDNGGEYTSREFEEYCSSHGIRHEKTV 575
Query: 878 PRTPQQNGVVERKNRTLQEMARTMIHENNLAKHFWAEAINTSCYIQNRIYIRPMLEKTAY 937
P TPQ NGV ER NRT+ E R+M+ L K FW EA+ T+CY+ NR P+ +
Sbjct: 576 PGTPQHNGVAERMNRTIVEKVRSMLRMAKLPKSFWGEAVQTACYLINRSPSVPLAFEIPE 635
Query: 938 ELFKGTRPNISYFHQFGCTCYILNTKDYLKKFDAKAQRGIFLGYSERSKAYRVYNSETHC 997
++ + S+ FGC + K+ K D K+ IF+GY + YR+++
Sbjct: 636 RVWTNKEVSYSHLKVFGCRAFAHVPKEQRTKLDDKSIPCIFIGYGDEEFGYRLWDPVKKK 695
Query: 998 VEESMHVKFDDREPGSKTPEQSESNAG------TIDSEDASESDQLSDSEKHSEVESSPE 1051
V S V F + E + + G TI S + + S +++ SE P
Sbjct: 696 VIRSRDVVFRESEVRTAADMSEKVKNGIIPNFVTIPSTSNNPTSAESTTDEVSEQGEQP- 754
Query: 1052 AEITPEAESNSEAEPSPEIQNENASEDIQDNSQQAIQPKFKHK 1094
E+ + E E E+++ E+ +++ +P+ + +
Sbjct: 755 GEVIEQGEQLDEG--VEEVEHPTQGEEQHQPLRRSERPRVESR 795
>COPI_DROME (P04146) Copia protein (Gag-int-pol protein) [Contains:
Copia VLP protein; Copia protease (EC 3.4.23.-)]
Length = 1409
Score = 147 bits (371), Expect = 2e-34
Identities = 99/338 (29%), Positives = 170/338 (50%), Gaps = 14/338 (4%)
Query: 761 CQKGKIVKSSFKS-KDIVSTSRPLELLHIDLFGQVNTTSLYGSKYGLVIVDDYSRWTWVK 819
C GK + FK KD RPL ++H D+ G + +L Y ++ VD ++ +
Sbjct: 458 CLNGKQARLPFKQLKDKTHIKRPLFVVHSDVCGPITPVTLDDKNYFVIFVDQFTHYCVTY 517
Query: 820 FIKSKDYACEVFSSFCTQIQSEKELKILKVRSDHGGEFENETFESFCEKHGILHEFSSPR 879
IK K +F F + ++ LK++ + D+G E+ + FC K GI + + P
Sbjct: 518 LIKYKSDVFSMFQDFVAKSEAHFNLKVVYLYIDNGREYLSNEMRQFCVKKGISYHLTVPH 577
Query: 880 TPQQNGVVERKNRTLQEMARTMIHENNLAKHFWAEAINTSCYIQNRIYIRPMLE--KTAY 937
TPQ NGV ER RT+ E ARTM+ L K FW EA+ T+ Y+ NRI R +++ KT Y
Sbjct: 578 TPQLNGVSERMIRTITEKARTMVSGAKLDKSFWGEAVLTATYLINRIPSRALVDSSKTPY 637
Query: 938 ELFKGTRPNISYFHQFGCTCYILNTKDYLKKFDAKAQRGIFLGYSERSKAYRVYNSETHC 997
E++ +P + + FG T Y+ + K+ KFD K+ + IF+GY ++++++
Sbjct: 638 EMWHNKKPYLKHLRVFGATVYV-HIKNKQGKFDDKSFKSIFVGY--EPNGFKLWDAVNEK 694
Query: 998 VEESMHVKFDDREPGSKTPEQSESNAGTIDSEDASESDQLSDSEKHSEVESSPEAEITPE 1057
+ V D+ + + E+ DS+++ + +DS K + E E++ E
Sbjct: 695 FIVARDVVVDETNMVNSRAVKFET-VFLKDSKESENKNFPNDSRKIIQTEFPNESK---E 750
Query: 1058 AESNSEAEPSPEIQNENASEDIQDNSQQAIQPKFKHKS 1095
++ + S E +N+N D S++ IQ +F ++S
Sbjct: 751 CDNIQFLKDSKESENKNFPND----SRKIIQTEFPNES 784
>YMU0_YEAST (Q04670) Transposon Ty1 protein B
Length = 1328
Score = 94.0 bits (232), Expect = 3e-18
Identities = 79/321 (24%), Positives = 144/321 (44%), Gaps = 23/321 (7%)
Query: 782 PLELLHIDLFGQVNTTSLYGSKYGLVIVDDYSRWTWVKFI--KSKDYACEVFSSFCTQIQ 839
P + LH D+FG V+ Y + D+ +++ WV + + +D +VF++ I+
Sbjct: 237 PFQYLHTDIFGPVHNLPKSAPSYFISFTDETTKFRWVYPLHDRREDSILDVFTTILAFIK 296
Query: 840 SEKELKILKVRSDHGGEFENETFESFCEKHGILHEFSSPRTPQQNGVVERKNRTLQEMAR 899
++ + +L ++ D G E+ N T F EK+GI +++ + +GV ER NRTL + R
Sbjct: 297 NQFQASVLVIQMDRGSEYTNRTLHKFLEKNGITPCYTTTADSRAHGVAERLNRTLLDDCR 356
Query: 900 TMIHENNLAKHFWAEAINTSCYIQNRIYIRPMLEKTAYELFKGTRPNISYFHQFGCTCYI 959
T + + L H W AI S ++N + P +K+A + +IS FG I
Sbjct: 357 TQLQCSGLPNHLWFSAIEFSTIVRNSL-ASPKSKKSARQHAGLAGLDISTLLPFG-QPVI 414
Query: 960 LNTKDYLKKFDAKAQRGIFLGYSERSKAYRVY--------NSETHCVEESMHVKFDDREP 1011
+N + K + G L S S Y +Y ++ + + + + D
Sbjct: 415 VNDHNPNSKIHPRGIPGYALHPSRNSYGYIIYLPSLKKTVDTTNYVILQGKESRLDQFNY 474
Query: 1012 GSKTPEQ-----SESNAGTIDSEDASESDQL---SDSEKHSEVESSPE--AEITPEAESN 1061
+ T ++ + S I S + +SD L SD + S++E PE + +A S
Sbjct: 475 DALTFDEDLNRLTASYQSFIASNEIQQSDDLNIESDHDFQSDIELHPEQPRNVLSKAVSP 534
Query: 1062 SEAEPSPEIQNENASEDIQDN 1082
+++ P P E++ + N
Sbjct: 535 TDSTP-PSTHTEDSKRVSKTN 554
>YME4_YEAST (Q04711) Transposon Ty1 protein B
Length = 1328
Score = 94.0 bits (232), Expect = 3e-18
Identities = 79/321 (24%), Positives = 144/321 (44%), Gaps = 23/321 (7%)
Query: 782 PLELLHIDLFGQVNTTSLYGSKYGLVIVDDYSRWTWVKFI--KSKDYACEVFSSFCTQIQ 839
P + LH D+FG V+ Y + D+ +++ WV + + +D +VF++ I+
Sbjct: 237 PFQYLHTDIFGPVHNLPKSAPSYFISFTDETTKFRWVYPLHDRREDSILDVFTTILAFIK 296
Query: 840 SEKELKILKVRSDHGGEFENETFESFCEKHGILHEFSSPRTPQQNGVVERKNRTLQEMAR 899
++ + +L ++ D G E+ N T F EK+GI +++ + +GV ER NRTL + R
Sbjct: 297 NQFQASVLVIQMDRGSEYTNRTLHKFLEKNGITPCYTTTADSRAHGVAERLNRTLLDDCR 356
Query: 900 TMIHENNLAKHFWAEAINTSCYIQNRIYIRPMLEKTAYELFKGTRPNISYFHQFGCTCYI 959
T + + L H W AI S ++N + P +K+A + +IS FG I
Sbjct: 357 TQLQCSGLPNHLWFSAIEFSTIVRNSL-ASPKSKKSARQHAGLAGLDISTLLPFG-QPVI 414
Query: 960 LNTKDYLKKFDAKAQRGIFLGYSERSKAYRVY--------NSETHCVEESMHVKFDDREP 1011
+N + K + G L S S Y +Y ++ + + + + D
Sbjct: 415 VNDHNPNSKIHPRGIPGYALHPSRNSYGYIIYLPSLKKTVDTTNYVILQGKESRLDQFNY 474
Query: 1012 GSKTPEQ-----SESNAGTIDSEDASESDQL---SDSEKHSEVESSPE--AEITPEAESN 1061
+ T ++ + S I S + +SD L SD + S++E PE + +A S
Sbjct: 475 DALTFDEDLNRLTASYQSFIASNEIQQSDDLNIESDHDFQSDIELHPEQPRNVLSKAVSP 534
Query: 1062 SEAEPSPEIQNENASEDIQDN 1082
+++ P P E++ + N
Sbjct: 535 TDSTP-PSTHTEDSKRVSKTN 554
>YMD9_YEAST (Q03434) Transposon Ty1 protein B
Length = 1328
Score = 94.0 bits (232), Expect = 3e-18
Identities = 79/321 (24%), Positives = 144/321 (44%), Gaps = 23/321 (7%)
Query: 782 PLELLHIDLFGQVNTTSLYGSKYGLVIVDDYSRWTWVKFI--KSKDYACEVFSSFCTQIQ 839
P + LH D+FG V+ Y + D+ +++ WV + + +D +VF++ I+
Sbjct: 237 PFQYLHTDIFGPVHNLPKSAPSYFISFTDETTKFRWVYPLHDRREDSILDVFTTILAFIK 296
Query: 840 SEKELKILKVRSDHGGEFENETFESFCEKHGILHEFSSPRTPQQNGVVERKNRTLQEMAR 899
++ + +L ++ D G E+ N T F EK+GI +++ + +GV ER NRTL + R
Sbjct: 297 NQFQASVLVIQMDRGSEYTNRTLHKFLEKNGITPCYTTTADSRAHGVAERLNRTLLDDCR 356
Query: 900 TMIHENNLAKHFWAEAINTSCYIQNRIYIRPMLEKTAYELFKGTRPNISYFHQFGCTCYI 959
T + + L H W AI S ++N + P +K+A + +IS FG I
Sbjct: 357 TQLQCSGLPNHLWFSAIEFSTIVRNSL-ASPKSKKSARQHAGLAGLDISTLLPFG-QPVI 414
Query: 960 LNTKDYLKKFDAKAQRGIFLGYSERSKAYRVY--------NSETHCVEESMHVKFDDREP 1011
+N + K + G L S S Y +Y ++ + + + + D
Sbjct: 415 VNDHNPNSKIHPRGIPGYALHPSRNSYGYIIYLPSLKKTVDTTNYVILQGKESRLDQFNY 474
Query: 1012 GSKTPEQ-----SESNAGTIDSEDASESDQL---SDSEKHSEVESSPE--AEITPEAESN 1061
+ T ++ + S I S + +SD L SD + S++E PE + +A S
Sbjct: 475 DALTFDEDLNRLTASYQSFIASNEIQQSDDLNIESDHDFQSDIELHPEQPRNVLSKAVSP 534
Query: 1062 SEAEPSPEIQNENASEDIQDN 1082
+++ P P E++ + N
Sbjct: 535 TDSTP-PSTHTEDSKRVSKTN 554
>YJZ9_YEAST (P47100) Transposon Ty1 protein B
Length = 1755
Score = 94.0 bits (232), Expect = 3e-18
Identities = 79/321 (24%), Positives = 144/321 (44%), Gaps = 23/321 (7%)
Query: 782 PLELLHIDLFGQVNTTSLYGSKYGLVIVDDYSRWTWVKFI--KSKDYACEVFSSFCTQIQ 839
P + LH D+FG V+ Y + D+ +++ WV + + +D +VF++ I+
Sbjct: 664 PFQYLHTDIFGPVHNLPKSAPSYFISFTDETTKFRWVYPLHDRREDSILDVFTTILAFIK 723
Query: 840 SEKELKILKVRSDHGGEFENETFESFCEKHGILHEFSSPRTPQQNGVVERKNRTLQEMAR 899
++ + +L ++ D G E+ N T F EK+GI +++ + +GV ER NRTL + R
Sbjct: 724 NQFQASVLVIQMDRGSEYTNRTLHKFLEKNGITPCYTTTADSRAHGVAERLNRTLLDDCR 783
Query: 900 TMIHENNLAKHFWAEAINTSCYIQNRIYIRPMLEKTAYELFKGTRPNISYFHQFGCTCYI 959
T + + L H W AI S ++N + P +K+A + +IS FG I
Sbjct: 784 TQLQCSGLPNHLWFSAIEFSTIVRNSL-ASPKSKKSARQHAGLAGLDISTLLPFG-QPVI 841
Query: 960 LNTKDYLKKFDAKAQRGIFLGYSERSKAYRVY--------NSETHCVEESMHVKFDDREP 1011
+N + K + G L S S Y +Y ++ + + + + D
Sbjct: 842 VNDHNPNSKIHPRGIPGYALHPSRNSYGYIIYLPSLKKTVDTTNYVILQGKESRLDQFNY 901
Query: 1012 GSKTPEQ-----SESNAGTIDSEDASESDQL---SDSEKHSEVESSPE--AEITPEAESN 1061
+ T ++ + S I S + +SD L SD + S++E PE + +A S
Sbjct: 902 DALTFDEDLNRLTASYQSFIASNEIQQSDDLNIESDHDFQSDIELHPEQPRNVLSKAVSP 961
Query: 1062 SEAEPSPEIQNENASEDIQDN 1082
+++ P P E++ + N
Sbjct: 962 TDSTP-PSTHTEDSKRVSKTN 981
>YJZ7_YEAST (P47098) Transposon Ty1 protein B
Length = 1755
Score = 93.2 bits (230), Expect = 5e-18
Identities = 79/321 (24%), Positives = 144/321 (44%), Gaps = 23/321 (7%)
Query: 782 PLELLHIDLFGQVNTTSLYGSKYGLVIVDDYSRWTWVKFI--KSKDYACEVFSSFCTQIQ 839
P + LH D+FG V+ Y + D+ +++ WV + + +D +VF++ I+
Sbjct: 664 PFQYLHTDIFGPVHNLPKSAPSYFISFTDETTKFRWVYPLHDRREDSILDVFTTILAFIK 723
Query: 840 SEKELKILKVRSDHGGEFENETFESFCEKHGILHEFSSPRTPQQNGVVERKNRTLQEMAR 899
++ + +L ++ D G E+ N T F EK+GI +++ + +GV ER NRTL + R
Sbjct: 724 NQFQASVLVIQMDRGSEYTNRTLHKFLEKNGITPCYTTTADSRAHGVAERLNRTLLDDCR 783
Query: 900 TMIHENNLAKHFWAEAINTSCYIQNRIYIRPMLEKTAYELFKGTRPNISYFHQFGCTCYI 959
T + + L H W AI S ++N + P +K+A + +IS FG I
Sbjct: 784 TQLQCSGLPNHLWFSAIEFSTIVRNSL-ASPKSKKSARQHAGLAGLDISTLLPFG-QPVI 841
Query: 960 LNTKDYLKKFDAKAQRGIFLGYSERSKAYRVY--------NSETHCVEESMHVKFDDREP 1011
+N + K + G L S S Y +Y ++ + + + + D
Sbjct: 842 VNDHNPNSKIHPRGIPGYALHPSRNSYGYIIYLPSLKKTVDTTNYVILQGKESRLDQFNY 901
Query: 1012 GSKTPEQ-----SESNAGTIDSEDASESDQL---SDSEKHSEVESSPE--AEITPEAESN 1061
+ T ++ + S I S + ES+ L SD + S++E PE + +A S
Sbjct: 902 DALTFDEDLNRLTASYQSFIASNEIQESNDLNIESDHDFQSDIELHPEQPRNVLSKAVSP 961
Query: 1062 SEAEPSPEIQNENASEDIQDN 1082
+++ P P E++ + N
Sbjct: 962 TDSTP-PSTHTEDSKRVSKTN 981
>YCB9_YEAST (P25384) Transposon Ty2 protein B (Ty1-17 protein B)
Length = 1770
Score = 92.8 bits (229), Expect = 7e-18
Identities = 78/311 (25%), Positives = 140/311 (44%), Gaps = 15/311 (4%)
Query: 782 PLELLHIDLFGQVNTTSLYGSKYGLVIVDDYSRWTWVKFI--KSKDYACEVFSSFCTQIQ 839
P + LH D+FG V+ Y + D+ +R+ WV + + ++ VF+S I+
Sbjct: 660 PFQYLHTDIFGPVHHLPKSAPSYFISFTDEKTRFQWVYPLHDRREESILNVFTSILAFIK 719
Query: 840 SEKELKILKVRSDHGGEFENETFESFCEKHGILHEFSSPRTPQQNGVVERKNRTLQEMAR 899
++ ++L ++ D G E+ N+T F GI +++ + +GV ER NRTL R
Sbjct: 720 NQFNARVLVIQMDRGSEYTNKTLHKFFTNRGITACYTTTADSRAHGVAERLNRTLLNDCR 779
Query: 900 TMIHENNLAKHFWAEAINTSCYIQNRIYIRPMLEKTAYELFKGTRPNISYFHQFGCTCYI 959
T++H + L H W A+ S I+N + + P +K+A + +I+ FG I
Sbjct: 780 TLLHCSGLPNHLWFSAVEFSTIIRNSL-VSPKNDKSARQHAGLAGLDITTILPFG-QPVI 837
Query: 960 LNTKDYLKKFDAKAQRGIFLGYSERSKAYRVY-NSETHCVEESMHVKFDDREPGSKTPEQ 1018
+N + K + G L S S Y +Y S V+ + +V D K +
Sbjct: 838 VNNHNPDSKIHPRGIPGYALHPSRNSYGYIIYLPSLKKTVDTTNYVILQD-----KQSKL 892
Query: 1019 SESNAGTIDSEDASESDQLSDSEKHSEVESSPEAEITPEAESNSEAEPSPEIQNENASED 1078
+ N T+ +D + ++L+ + ++ E ES+ + + EI N+
Sbjct: 893 DQFNYDTLTFDD--DLNRLTAHNQSFIEQNETEQSYDQNTESDHDYQSEIEI---NSDPL 947
Query: 1079 IQDNSQQAIQP 1089
+ D S Q+I P
Sbjct: 948 VNDFSSQSINP 958
>YMT5_YEAST (Q04214) Transposon Ty1 protein B
Length = 1328
Score = 92.4 bits (228), Expect = 9e-18
Identities = 74/291 (25%), Positives = 131/291 (44%), Gaps = 8/291 (2%)
Query: 782 PLELLHIDLFGQVNTTSLYGSKYGLVIVDDYSRWTWVKFI--KSKDYACEVFSSFCTQIQ 839
P + LH D+FG V+ Y + D+ +++ WV + + +D +VF++ I+
Sbjct: 237 PFQYLHTDIFGPVHNLPKSAPSYFISFTDETTKFRWVYPLHDRREDSILDVFTTILAFIK 296
Query: 840 SEKELKILKVRSDHGGEFENETFESFCEKHGILHEFSSPRTPQQNGVVERKNRTLQEMAR 899
++ + +L ++ D G E+ N T F EK+GI +++ + +GV ER NRTL + R
Sbjct: 297 NQFQASVLVIQMDRGSEYTNRTLHKFLEKNGITPCYTTTADSRAHGVAERLNRTLLDDCR 356
Query: 900 TMIHENNLAKHFWAEAINTSCYIQNRIYIRPMLEKTAYELFKGTRPNISYFHQFGCTCYI 959
T + + L H W AI S ++N + P +K+A + +IS FG I
Sbjct: 357 TQLQCSGLPNHLWFSAIEFSTIVRNSL-ASPKSKKSARQHAGLAGLDISTLLPFG-QPVI 414
Query: 960 LNTKDYLKKFDAKAQRGIFLGYSERSKAYRVY-NSETHCVEESMHVKFDDREPGSKTPEQ 1018
+N + K + G L S S Y +Y S V+ + +V +E +Q
Sbjct: 415 VNDHNPNSKIHPRGIPGYALHPSRNSYGYIIYLPSLKKTVDTTNYVILQGKE---SRLDQ 471
Query: 1019 SESNAGTIDSEDASESDQLSDSEKHSEVESSPEAEITPEAESNSEAEPSPE 1069
+A T D + + +E++ S + I + + S+ E PE
Sbjct: 472 FNYDALTFDEDLNRLTASYHSFIASNEIQQSNDLNIESDHDFQSDIELHPE 522
>YJL3_YEAST (P47024) Transposon Ty4 207.7 kDa hypothetical protein
Length = 1803
Score = 68.9 bits (167), Expect = 1e-10
Identities = 39/139 (28%), Positives = 74/139 (53%), Gaps = 2/139 (1%)
Query: 788 IDLFGQVNTTSLYGSKYGLVIVDDYSRW--TWVKFIKSKDYACEVFSSFCTQIQSEKELK 845
+D+FG V++++ +Y L++VD+ +R+ T F K+ + ++++ + K
Sbjct: 630 MDIFGPVSSSNADTKRYMLIMVDNNTRYCMTSTHFNKNAETILAQVRKNIQYVETQFDRK 689
Query: 846 ILKVRSDHGGEFENETFESFCEKHGILHEFSSPRTPQQNGVVERKNRTLQEMARTMIHEN 905
+ ++ SD G EF N+ E + GI H +S + NG ER RT+ A T++ ++
Sbjct: 690 VREINSDRGTEFTNDQIEEYFISKGIHHILTSTQDHAANGRAERYIRTIITDATTLLRQS 749
Query: 906 NLAKHFWAEAINTSCYIQN 924
NL FW A+ ++ I+N
Sbjct: 750 NLRVKFWEYAVTSATNIRN 768
>POL_FOAMV (P14350) Pol polyprotein [Contains: Reverse
transcriptase/ribonuclease H (EC 2.7.7.49) (EC 3.1.26.4)
(RT); Integrase (IN)]
Length = 886
Score = 61.6 bits (148), Expect = 2e-08
Identities = 53/199 (26%), Positives = 87/199 (43%), Gaps = 16/199 (8%)
Query: 759 GACQKGKIVKSSFKSKDIV----STSRPLELLHIDLFGQVNTTSLYGSKYGLVIVDDYSR 814
G CQ+ I +S K+ + +P + ID G + + G Y LV+VD +
Sbjct: 648 GRCQQCLITNASNKASGPILRPDRPQKPFDKFFIDYIGPLPPSQ--GYLYVLVVVDGMTG 705
Query: 815 WTWVKFIKSKDYACEVFS-SFCTQIQSEKELKILKVRSDHGGEFENETFESFCEKHGILH 873
+TW+ K+ + V S + T I K + SD G F + TF + ++ GI
Sbjct: 706 FTWLYPTKAPSTSATVKSLNVLTSIAIPKV-----IHSDQGAAFTSSTFAEWAKERGIHL 760
Query: 874 EFSSPRTPQQNGVVERKNRTLQEMARTMIHENNLAKHFWAEAINTSCYIQNRIYIRPMLE 933
EFS+P PQ VERKN ++ + ++ W + + N Y P+L+
Sbjct: 761 EFSTPYHPQSGSKVERKNSDIKRLLTKLLVGRPTK---WYDLLPVVQLALNNTY-SPVLK 816
Query: 934 KTAYELFKGTRPNISYFHQ 952
T ++L G N + +Q
Sbjct: 817 YTPHQLLFGIDSNTPFANQ 835
>POL_SFV1 (P23074) Pol polyprotein [Contains: Protease (EC
3.4.23.-); Reverse transcriptase/ribonuclease H (EC
2.7.7.49) (EC 3.1.26.4) (RT); Integrase (IN)]
Length = 1161
Score = 55.1 bits (131), Expect = 2e-06
Identities = 37/127 (29%), Positives = 63/127 (49%), Gaps = 8/127 (6%)
Query: 777 VSTSRPLELLHIDLFGQVNTTSLYGSKYGLVIVDDYSRWTWVKFIKSKDYACEVFS-SFC 835
V +P + +ID G + ++ G + LV+VD + + W+ K+ + V + +
Sbjct: 878 VKPLKPFDKFYIDYIGPLPPSN--GYLHVLVVVDSMTGFVWLYPTKAPSTSATVKALNML 935
Query: 836 TQIQSEKELKILKVRSDHGGEFENETFESFCEKHGILHEFSSPRTPQQNGVVERKNRTLQ 895
T I K L SD G F + TF + ++ GI EFS+P PQ +G VERKN ++
Sbjct: 936 TSIAIPKVL-----HSDQGAAFTSSTFADWAKEKGIQLEFSTPYHPQSSGKVERKNSDIK 990
Query: 896 EMARTMI 902
+ ++
Sbjct: 991 RLLTKLL 997
>POL_SFV3L (P27401) Pol polyprotein [Contains: Protease (EC
3.4.23.-); Reverse transcriptase/ribonuclease H (EC
2.7.7.49) (EC 3.1.26.4) (RT); Integrase (IN)]
Length = 1157
Score = 52.4 bits (124), Expect = 1e-05
Identities = 35/123 (28%), Positives = 60/123 (48%), Gaps = 8/123 (6%)
Query: 781 RPLELLHIDLFGQVNTTSLYGSKYGLVIVDDYSRWTWVKFIKSKDYACEVFS-SFCTQIQ 839
+P + ID G + ++ G + LV+VD + + W+ K+ + V + + T I
Sbjct: 884 KPFDKFFIDYIGPLPPSN--GYLHVLVVVDSMTGFVWLYPTKAPSTSATVKALNMLTSIA 941
Query: 840 SEKELKILKVRSDHGGEFENETFESFCEKHGILHEFSSPRTPQQNGVVERKNRTLQEMAR 899
K + SD G F + TF + + GI EFS+P PQ +G VERKN ++ +
Sbjct: 942 VPKV-----IHSDQGAAFTSATFADWAKNKGIQLEFSTPYHPQSSGKVERKNSDIKRLLT 996
Query: 900 TMI 902
++
Sbjct: 997 KLL 999
>POL4_DROME (P10394) Retrovirus-related Pol polyprotein from
transposon 412 [Contains: Protease (EC 3.4.23.-); Reverse
transcriptase (EC 2.7.7.49); Endonuclease]
Length = 1237
Score = 47.8 bits (112), Expect = 3e-04
Identities = 63/268 (23%), Positives = 108/268 (39%), Gaps = 26/268 (9%)
Query: 761 CQKGKIVKSSFKSKDIVST-SRPLELLHIDLFGQVNTTSLYGSKYGLVIVDDYSRWTWVK 819
CQK K K + I T + + +D G + S G++Y + ++ D +++
Sbjct: 942 CQKAKTTKHTKTPMTITETPEHAFDRVVVDTIGPL-PKSENGNEYAVTLICDLTKYLVAI 1000
Query: 820 FIKSKD---YACEVFSSFCTQIQSEKELKILKVRSDHGGEFENETFESFCEKHGILHEFS 876
I +K A +F SF + K +D G E++N C+ I + S
Sbjct: 1001 PIANKSAKTVAKAIFESFILKYGPMKTFI-----TDMGTEYKNSIITDLCKYLKIKNITS 1055
Query: 877 SPRTPQQNGVVERKNRTLQEMARTMIHENNLAKHFWAEAINTSCYIQNRIYIRPMLEKTA 936
+ Q GVVER +RTL E R+ I + W + C+ + +
Sbjct: 1056 TAHHHQTVGVVERSHRTLNEYIRSYISTDKTDWDVWLQYF-VYCFNTTQSMVH---NYCP 1111
Query: 937 YELFKGTRPNI-SYFHQFGCTCYILNTKDYLKKFDAKAQRGIFLGYSERSK---AYRVYN 992
YEL G N+ +F++ I N DY K+ + + + Y+ K A++ N
Sbjct: 1112 YELVFGRTSNLPKHFNKLHSIEPIYNIDDYAKESKYRLE----VAYARARKLLEAHKEKN 1167
Query: 993 SETHCVEESMHVKFDDREPGSKTPEQSE 1020
E + + VK + E G K ++E
Sbjct: 1168 KENY----DLKVKDIELEVGDKVLLRNE 1191
>POL_SIVGB (P22382) Pol polyprotein [Contains: Protease
(Retropepsin) (EC 3.4.23.-); Reverse
transcriptase/ribonuclease H (EC 2.7.7.49) (EC 3.1.26.4)
(RT); Integrase (IN)]
Length = 1009
Score = 46.6 bits (109), Expect = 6e-04
Identities = 22/52 (42%), Positives = 31/52 (59%)
Query: 846 ILKVRSDHGGEFENETFESFCEKHGILHEFSSPRTPQQNGVVERKNRTLQEM 897
I K+ +D+G F ++ E+ C GI H F P PQ GVVE KN+ L+E+
Sbjct: 834 ISKLHTDNGPNFTSQEVETMCWWLGIEHTFGIPYNPQSQGVVENKNKYLKEL 885
>YRD6_CAEEL (Q09575) Hypothetical protein K02A2.6 in chromosome II
Length = 1268
Score = 46.2 bits (108), Expect = 8e-04
Identities = 38/120 (31%), Positives = 55/120 (45%), Gaps = 21/120 (17%)
Query: 782 PLELLHIDLFGQVNTTSLYGSKYGLVIVDDYSRWTWVKFIKSK------DYACEVFSSFC 835
P + +HID G +N Y LV+VD +++ VK +S D E+FS
Sbjct: 848 PWKRIHIDFAGPLNGC------YLLVVVDAKTKYAEVKLTRSISAVTTIDLLEEIFS--- 898
Query: 836 TQIQSEKELKILKVRSDHGGEFENETFESFCEKHGILHEFSSPRTPQQNGVVERKNRTLQ 895
I E I SD+G + + F C+ HGI H+ S+ P+ NG ER TL+
Sbjct: 899 --IHGYPETII----SDNGTQLTSHLFAQMCQSHGIEHKTSAVYYPRSNGAAERFVDTLK 952
>POL_MLVFP (P26808) Pol polyprotein [Contains: Protease (EC 3.4.23.-);
Reverse transcriptase/ribonuclease H (EC 2.7.7.49) (EC
3.1.26.4) (RT); Integrase (IN)]
Length = 1204
Score = 46.2 bits (108), Expect = 8e-04
Identities = 42/137 (30%), Positives = 63/137 (45%), Gaps = 4/137 (2%)
Query: 760 ACQKGKIVKSSFKSKDIVSTSRPLELLHIDLFGQVNTTSLYGSKYGLVIVDDYSRWTWVK 819
AC + KS+ K V RP ID F +V LYG KY LV VD +S W
Sbjct: 892 ACAQVNASKSAVKQGTRVRGHRPGTHWEID-FTEVKP-GLYGYKYLLVFVDTFSGWVEA- 948
Query: 820 FIKSKDYACEVFSSFCTQIQSEKELKILKVRSDHGGEFENETFESFCEKHGILHEFSSPR 879
F K+ A V +I + + + +D+G F ++ ++ + G+ +
Sbjct: 949 FPTKKETAKVVTKKLLEEIFPRFGMPQV-LGTDNGPAFVSKVSQTVADLLGVDWKLHCAY 1007
Query: 880 TPQQNGVVERKNRTLQE 896
PQ +G VER NRT++E
Sbjct: 1008 RPQSSGQVERMNRTIKE 1024
>POL_MLVF5 (P26810) Pol polyprotein [Contains: Protease (EC 3.4.23.-);
Reverse transcriptase/ribonuclease H (EC 2.7.7.49) (EC
3.1.26.4) (RT); Integrase (IN)]
Length = 1204
Score = 46.2 bits (108), Expect = 8e-04
Identities = 42/137 (30%), Positives = 63/137 (45%), Gaps = 4/137 (2%)
Query: 760 ACQKGKIVKSSFKSKDIVSTSRPLELLHIDLFGQVNTTSLYGSKYGLVIVDDYSRWTWVK 819
AC + KS+ K V RP ID F +V LYG KY LV VD +S W
Sbjct: 892 ACAQVNASKSAVKQGTRVRGHRPGTHWEID-FTEVKP-GLYGYKYLLVFVDTFSGWVEA- 948
Query: 820 FIKSKDYACEVFSSFCTQIQSEKELKILKVRSDHGGEFENETFESFCEKHGILHEFSSPR 879
F K+ A V +I + + + +D+G F ++ ++ + G+ +
Sbjct: 949 FPTKKETAKVVTKKLLEEIFPRFGMPQV-LGTDNGPAFVSKVSQTVADLLGVDWKLHCAY 1007
Query: 880 TPQQNGVVERKNRTLQE 896
PQ +G VER NRT++E
Sbjct: 1008 RPQSSGQVERMNRTIKE 1024
>POL_MLVFF (P26809) Pol polyprotein [Contains: Protease (EC 3.4.23.-);
Reverse transcriptase/ribonuclease H (EC 2.7.7.49) (EC
3.1.26.4) (RT); Integrase (IN)]
Length = 1204
Score = 45.8 bits (107), Expect = 0.001
Identities = 41/137 (29%), Positives = 63/137 (45%), Gaps = 4/137 (2%)
Query: 760 ACQKGKIVKSSFKSKDIVSTSRPLELLHIDLFGQVNTTSLYGSKYGLVIVDDYSRWTWVK 819
AC + KS+ K V RP ID F +V LYG KY LV +D +S W
Sbjct: 892 ACAQVNASKSAVKQGTRVRGHRPGTHWEID-FTEVKP-GLYGYKYLLVFIDTFSGWVEA- 948
Query: 820 FIKSKDYACEVFSSFCTQIQSEKELKILKVRSDHGGEFENETFESFCEKHGILHEFSSPR 879
F K+ A V +I + + + +D+G F ++ ++ + G+ +
Sbjct: 949 FPTKKETAKVVTKKLLEEIFPRFGMPQV-LGTDNGPAFVSKVSQTVADLLGVDWKLHCAY 1007
Query: 880 TPQQNGVVERKNRTLQE 896
PQ +G VER NRT++E
Sbjct: 1008 RPQSSGQVERMNRTIKE 1024
>POL_MLVMO (P03355) Pol polyprotein [Contains: Protease (EC 3.4.23.-);
Reverse transcriptase/ribonuclease H (EC 2.7.7.49) (EC
3.1.26.4) (RT); Integrase (IN)]
Length = 1199
Score = 45.4 bits (106), Expect = 0.001
Identities = 41/137 (29%), Positives = 63/137 (45%), Gaps = 4/137 (2%)
Query: 760 ACQKGKIVKSSFKSKDIVSTSRPLELLHIDLFGQVNTTSLYGSKYGLVIVDDYSRWTWVK 819
AC + KS+ K V RP ID F ++ LYG KY LV +D +S W
Sbjct: 887 ACAQVNASKSAVKQGTRVRGHRPGTHWEID-FTEIKP-GLYGYKYLLVFIDTFSGWIEA- 943
Query: 820 FIKSKDYACEVFSSFCTQIQSEKELKILKVRSDHGGEFENETFESFCEKHGILHEFSSPR 879
F K+ A V +I + + + +D+G F ++ ++ + GI +
Sbjct: 944 FPTKKETAKVVTKKLLEEIFPRFGMPQV-LGTDNGPAFVSKVSQTVADLLGIDWKLHCAY 1002
Query: 880 TPQQNGVVERKNRTLQE 896
PQ +G VER NRT++E
Sbjct: 1003 RPQSSGQVERMNRTIKE 1019
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.359 0.159 0.563
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 151,887,264
Number of Sequences: 164201
Number of extensions: 5452926
Number of successful extensions: 33853
Number of sequences better than 10.0: 255
Number of HSP's better than 10.0 without gapping: 122
Number of HSP's successfully gapped in prelim test: 140
Number of HSP's that attempted gapping in prelim test: 32033
Number of HSP's gapped (non-prelim): 1165
length of query: 1684
length of database: 59,974,054
effective HSP length: 124
effective length of query: 1560
effective length of database: 39,613,130
effective search space: 61796482800
effective search space used: 61796482800
T: 11
A: 40
X1: 14 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 37 (21.8 bits)
S2: 73 (32.7 bits)
Medicago: description of AC149212.6


