

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC149207.4 - phase: 0
(168 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
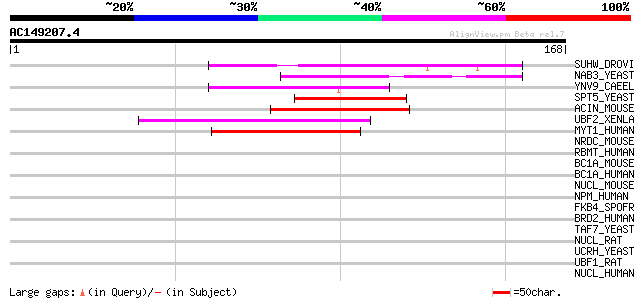
Score E
Sequences producing significant alignments: (bits) Value
SUHW_DROVI (Q08876) Suppressor of hairy wing protein 47 3e-05
NAB3_YEAST (P38996) Nuclear polyadenylated RNA-binding protein 3 45 1e-04
YNV9_CAEEL (P34572) Hypothetical protein T16H12.9 in chromosome III 44 2e-04
SPT5_YEAST (P27692) Transcription initiation protein SPT5 44 2e-04
ACIN_MOUSE (Q9JIX8) Apoptotic chromatin condensation inducer in ... 42 5e-04
UBF2_XENLA (P25980) Nucleolar transcription factor 2 (Upstream b... 42 9e-04
MYT1_HUMAN (Q01538) Myelin transcription factor 1 (MYT1) (MYTI) ... 42 9e-04
NRDC_MOUSE (Q8BHG1) Nardilysin precursor (EC 3.4.24.61) (N-argin... 41 0.001
RBMT_HUMAN (Q9NW13) RNA-binding protein 28 (RNA binding motif pr... 40 0.002
BC1A_MOUSE (Q9QYE3) B-cell lymphoma/leukemia 11A (B-cell CLL/lym... 40 0.002
BC1A_HUMAN (Q9H165) B-cell lymphoma/leukemia 11A (B-cell CLL/lym... 40 0.002
NUCL_MOUSE (P09405) Nucleolin (Protein C23) 40 0.003
NPM_HUMAN (P06748) Nucleophosmin (NPM) (Nucleolar phosphoprotein... 40 0.003
FKB4_SPOFR (Q26486) 46 kDa FK506-binding nuclear protein (EC 5.2... 40 0.003
BRD2_HUMAN (P25440) Bromodomain-containing protein 2 (RING3 prot... 40 0.003
TAF7_YEAST (Q05021) Transcription initiation factor TFIID subuni... 39 0.004
NUCL_RAT (P13383) Nucleolin (Protein C23) 39 0.006
UCRH_YEAST (P00127) Ubiquinol-cytochrome C reductase complex 17 ... 39 0.007
UBF1_RAT (P25977) Nucleolar transcription factor 1 (Upstream bin... 39 0.007
NUCL_HUMAN (P19338) Nucleolin (Protein C23) 38 0.010
>SUHW_DROVI (Q08876) Suppressor of hairy wing protein
Length = 899
Score = 46.6 bits (109), Expect = 3e-05
Identities = 35/106 (33%), Positives = 54/106 (50%), Gaps = 17/106 (16%)
Query: 61 DITDLLSTATEHRKSFVSHQIPNQLQEEEEEDDDDDDDDEEVDEEEAAKRARMNRLAELD 120
D +L + A E+ + FV + +E++DDDDDD+DE V E A +R+ N L E+
Sbjct: 163 DEVELGAGAKENGEEFVVSGV------DEDDDDDDDDEDEGVVEGGAKRRSGNNELKEMV 216
Query: 121 NLLVG--------VRSIKRELEIIKVDT---LAMKDRMKCLESFLK 155
+ G V+S+K+ LE + D+ L D +K LE K
Sbjct: 217 EHVCGKCYKTFRRVKSLKKHLEFCRYDSGYHLRKADMLKNLEKIEK 262
>NAB3_YEAST (P38996) Nuclear polyadenylated RNA-binding protein 3
Length = 802
Score = 44.7 bits (104), Expect = 1e-04
Identities = 30/73 (41%), Positives = 38/73 (51%), Gaps = 8/73 (10%)
Query: 83 NQLQEEEEEDDDDDDDDEEVDEEEAAKRARMNRLAELDNLLVGVRSIKRELEIIKVDTLA 142
N+ +EEEEEDDDDDDDD++ DEEE + DN VG S + E D
Sbjct: 107 NEEEEEEEEDDDDDDDDDDDDEEEEEEEEEEGN----DNSSVGSDSAAEDGE----DEED 158
Query: 143 MKDRMKCLESFLK 155
KD+ K E L+
Sbjct: 159 KKDKTKDKEVELR 171
Score = 37.7 bits (86), Expect = 0.012
Identities = 15/36 (41%), Positives = 26/36 (71%)
Query: 71 EHRKSFVSHQIPNQLQEEEEEDDDDDDDDEEVDEEE 106
EH++ + + +EEEEE++DDDDDD++ D++E
Sbjct: 93 EHQQKGGNDDDDDDNEEEEEEEEDDDDDDDDDDDDE 128
Score = 32.0 bits (71), Expect = 0.68
Identities = 16/32 (50%), Positives = 21/32 (65%), Gaps = 5/32 (15%)
Query: 80 QIPNQLQEEEEED-----DDDDDDDEEVDEEE 106
++ N +EEEEE +DDDDDD E +EEE
Sbjct: 82 EVINTEEEEEEEHQQKGGNDDDDDDNEEEEEE 113
>YNV9_CAEEL (P34572) Hypothetical protein T16H12.9 in chromosome III
Length = 289
Score = 43.5 bits (101), Expect = 2e-04
Identities = 21/58 (36%), Positives = 33/58 (56%), Gaps = 3/58 (5%)
Query: 61 DITDLLSTATEHRKSFVSHQIPNQLQEEEEEDDDDDDD---DEEVDEEEAAKRARMNR 115
DI D++++ + + + Q Q++ +DDDDDDD DE +EEE KR R +R
Sbjct: 104 DINDIVASQVDSNSTCTKYGFTRQQQKKSHDDDDDDDDSDSDESKEEEEKKKRDRKHR 161
>SPT5_YEAST (P27692) Transcription initiation protein SPT5
Length = 1063
Score = 43.5 bits (101), Expect = 2e-04
Identities = 19/34 (55%), Positives = 24/34 (69%)
Query: 87 EEEEEDDDDDDDDEEVDEEEAAKRARMNRLAELD 120
E+++EDDDDDDDDE+ D+E KR R R LD
Sbjct: 149 EDDDEDDDDDDDDEDDDDEAPTKRRRQERNRFLD 182
>ACIN_MOUSE (Q9JIX8) Apoptotic chromatin condensation inducer in the
nucleus (Acinus)
Length = 1338
Score = 42.4 bits (98), Expect = 5e-04
Identities = 17/42 (40%), Positives = 30/42 (70%)
Query: 80 QIPNQLQEEEEEDDDDDDDDEEVDEEEAAKRARMNRLAELDN 121
+I + +EEEEE+DDDD+++EEVDE + ++ A L + ++
Sbjct: 267 KIEEEEEEEEEEEDDDDEEEEEVDEAQKSREAEAPTLKQFED 308
Score = 33.9 bits (76), Expect = 0.18
Identities = 14/38 (36%), Positives = 27/38 (70%)
Query: 85 LQEEEEEDDDDDDDDEEVDEEEAAKRARMNRLAELDNL 122
L+ EEEE+++++++D++ +EEE A+ +R AE L
Sbjct: 266 LKIEEEEEEEEEEEDDDDEEEEEVDEAQKSREAEAPTL 303
>UBF2_XENLA (P25980) Nucleolar transcription factor 2 (Upstream
binding factor-2) (UBF-2) (xUBF-2)
Length = 701
Score = 41.6 bits (96), Expect = 9e-04
Identities = 17/70 (24%), Positives = 37/70 (52%)
Query: 40 WWKSLVVDDQFVQKHLHKSLADITDLLSTATEHRKSFVSHQIPNQLQEEEEEDDDDDDDD 99
W K L D+ K + + T + + K + + + +E++ED++DDDDD
Sbjct: 573 WMKGLSTQDRAAYKEQNTNKRKSTTKIQAPSSKSKLVIQSKSDDDEDDEDDEDEEDDDDD 632
Query: 100 EEVDEEEAAK 109
++ D+E++++
Sbjct: 633 DDEDKEDSSE 642
Score = 32.3 bits (72), Expect = 0.52
Identities = 11/33 (33%), Positives = 24/33 (72%)
Query: 86 QEEEEEDDDDDDDDEEVDEEEAAKRARMNRLAE 118
++EE+E+DDD+D++E+ D+ E+ + + A+
Sbjct: 662 EDEEDEEDDDEDNEEDDDDNESGSSSSSSSSAD 694
Score = 31.6 bits (70), Expect = 0.89
Identities = 10/21 (47%), Positives = 18/21 (85%)
Query: 86 QEEEEEDDDDDDDDEEVDEEE 106
+ E+EED++DDD+D E D+++
Sbjct: 660 ENEDEEDEEDDDEDNEEDDDD 680
Score = 29.3 bits (64), Expect = 4.4
Identities = 9/20 (45%), Positives = 18/20 (90%)
Query: 87 EEEEEDDDDDDDDEEVDEEE 106
EE E+++D++DDDE+ +E++
Sbjct: 659 EENEDEEDEEDDDEDNEEDD 678
Score = 28.9 bits (63), Expect = 5.8
Identities = 9/22 (40%), Positives = 20/22 (90%)
Query: 83 NQLQEEEEEDDDDDDDDEEVDE 104
N+ +E+EE+DD+D+++D++ +E
Sbjct: 661 NEDEEDEEDDDEDNEEDDDDNE 682
>MYT1_HUMAN (Q01538) Myelin transcription factor 1 (MYT1) (MYTI)
(Proteolipid protein binding protein) (PLPB1)
Length = 1121
Score = 41.6 bits (96), Expect = 9e-04
Identities = 16/45 (35%), Positives = 32/45 (70%)
Query: 62 ITDLLSTATEHRKSFVSHQIPNQLQEEEEEDDDDDDDDEEVDEEE 106
+ D S + +K +SH+ ++ +EEEEE++++D+++EE +EEE
Sbjct: 240 LEDAASEESSKQKGILSHEEEDEEEEEEEEEEEEDEEEEEEEEEE 284
Score = 33.9 bits (76), Expect = 0.18
Identities = 12/23 (52%), Positives = 22/23 (95%)
Query: 86 QEEEEEDDDDDDDDEEVDEEEAA 108
+EEEEE++++++++EE +EEEAA
Sbjct: 286 EEEEEEEEEEEEEEEEEEEEEAA 308
Score = 32.0 bits (71), Expect = 0.68
Identities = 11/26 (42%), Positives = 22/26 (84%)
Query: 86 QEEEEEDDDDDDDDEEVDEEEAAKRA 111
+EEEEE++++++++EE +EEE + A
Sbjct: 282 EEEEEEEEEEEEEEEEEEEEEEEEEA 307
Score = 31.6 bits (70), Expect = 0.89
Identities = 11/26 (42%), Positives = 22/26 (84%)
Query: 86 QEEEEEDDDDDDDDEEVDEEEAAKRA 111
+EEEEE++++++++EE +EEE + A
Sbjct: 283 EEEEEEEEEEEEEEEEEEEEEEEEAA 308
Score = 31.6 bits (70), Expect = 0.89
Identities = 13/42 (30%), Positives = 27/42 (63%)
Query: 65 LLSTATEHRKSFVSHQIPNQLQEEEEEDDDDDDDDEEVDEEE 106
+LS E + + + +EEEEE++++++++EE +EEE
Sbjct: 254 ILSHEEEDEEEEEEEEEEEEDEEEEEEEEEEEEEEEEEEEEE 295
Score = 30.8 bits (68), Expect = 1.5
Identities = 10/21 (47%), Positives = 20/21 (94%)
Query: 86 QEEEEEDDDDDDDDEEVDEEE 106
+EEEEE++++++++EE +EEE
Sbjct: 278 EEEEEEEEEEEEEEEEEEEEE 298
Score = 30.8 bits (68), Expect = 1.5
Identities = 10/21 (47%), Positives = 20/21 (94%)
Query: 86 QEEEEEDDDDDDDDEEVDEEE 106
+EEEEE++++++++EE +EEE
Sbjct: 280 EEEEEEEEEEEEEEEEEEEEE 300
Score = 30.8 bits (68), Expect = 1.5
Identities = 10/21 (47%), Positives = 20/21 (94%)
Query: 86 QEEEEEDDDDDDDDEEVDEEE 106
+EEEEE++++++++EE +EEE
Sbjct: 281 EEEEEEEEEEEEEEEEEEEEE 301
Score = 30.8 bits (68), Expect = 1.5
Identities = 10/21 (47%), Positives = 20/21 (94%)
Query: 86 QEEEEEDDDDDDDDEEVDEEE 106
+EEEEE++++++++EE +EEE
Sbjct: 277 EEEEEEEEEEEEEEEEEEEEE 297
Score = 30.8 bits (68), Expect = 1.5
Identities = 10/21 (47%), Positives = 20/21 (94%)
Query: 86 QEEEEEDDDDDDDDEEVDEEE 106
+EEEEE++++++++EE +EEE
Sbjct: 279 EEEEEEEEEEEEEEEEEEEEE 299
Score = 30.4 bits (67), Expect = 2.0
Identities = 16/62 (25%), Positives = 34/62 (54%), Gaps = 5/62 (8%)
Query: 50 FVQKHLHKSLADIT-----DLLSTATEHRKSFVSHQIPNQLQEEEEEDDDDDDDDEEVDE 104
F+Q + + ++T DL + E S S + L EEE+++++++++EE ++
Sbjct: 215 FIQPEDAEEVVEVTTERSQDLCPQSLEDAASEESSKQKGILSHEEEDEEEEEEEEEEEED 274
Query: 105 EE 106
EE
Sbjct: 275 EE 276
>NRDC_MOUSE (Q8BHG1) Nardilysin precursor (EC 3.4.24.61) (N-arginine
dibasic convertase) (NRD convertase) (NRD-C)
Length = 1161
Score = 40.8 bits (94), Expect = 0.001
Identities = 16/24 (66%), Positives = 21/24 (86%)
Query: 86 QEEEEEDDDDDDDDEEVDEEEAAK 109
+EEEEEDDDDDDDD++ DE+ A+
Sbjct: 146 EEEEEEDDDDDDDDDDDDEDSGAE 169
Score = 38.5 bits (88), Expect = 0.007
Identities = 14/22 (63%), Positives = 21/22 (94%)
Query: 86 QEEEEEDDDDDDDDEEVDEEEA 107
+EEEEE+DDDDDDD++ D+E++
Sbjct: 145 EEEEEEEDDDDDDDDDDDDEDS 166
Score = 36.6 bits (83), Expect = 0.028
Identities = 13/21 (61%), Positives = 20/21 (94%)
Query: 86 QEEEEEDDDDDDDDEEVDEEE 106
+EEEEE++DDDDDD++ D++E
Sbjct: 144 EEEEEEEEDDDDDDDDDDDDE 164
Score = 34.7 bits (78), Expect = 0.11
Identities = 15/42 (35%), Positives = 26/42 (61%)
Query: 62 ITDLLSTATEHRKSFVSHQIPNQLQEEEEEDDDDDDDDEEVD 103
I+DL + + + + + +EEE++DDDDDDDD++ D
Sbjct: 124 ISDLSNVEGKTGNATDEEEEEEEEEEEEDDDDDDDDDDDDED 165
Score = 33.9 bits (76), Expect = 0.18
Identities = 11/21 (52%), Positives = 20/21 (94%)
Query: 86 QEEEEEDDDDDDDDEEVDEEE 106
+EEEEE+++DDDDD++ D+++
Sbjct: 143 EEEEEEEEEDDDDDDDDDDDD 163
Score = 32.3 bits (72), Expect = 0.52
Identities = 10/21 (47%), Positives = 20/21 (94%)
Query: 86 QEEEEEDDDDDDDDEEVDEEE 106
+EEEEE++++DDDD++ D+++
Sbjct: 142 EEEEEEEEEEDDDDDDDDDDD 162
Score = 31.2 bits (69), Expect = 1.2
Identities = 14/35 (40%), Positives = 23/35 (65%), Gaps = 7/35 (20%)
Query: 86 QEEEEEDDDDDDDDEEVDEEEAAKRARMNRLAELD 120
+EE ++DDDD+ DD++++ EE N L EL+
Sbjct: 181 EEEFDDDDDDEHDDDDLENEE-------NELEELE 208
Score = 30.8 bits (68), Expect = 1.5
Identities = 9/21 (42%), Positives = 20/21 (94%)
Query: 86 QEEEEEDDDDDDDDEEVDEEE 106
+EEEEE+++++DDD++ D+++
Sbjct: 141 EEEEEEEEEEEDDDDDDDDDD 161
Score = 30.4 bits (67), Expect = 2.0
Identities = 24/76 (31%), Positives = 36/76 (46%), Gaps = 14/76 (18%)
Query: 87 EEEEEDDDDDDDDEEVDEEEAAKRARMNRLAELDNLLVGVRSIKRELE-------IIKVD 139
++EEE DDDDDD+ + D+ E N EL+ L V + K+ E + V
Sbjct: 179 DDEEEFDDDDDDEHDDDDLE-------NEENELEELEERVEARKKTTEKQSAAALCVGVG 231
Query: 140 TLAMKDRMKCLESFLK 155
+ A D + L FL+
Sbjct: 232 SFADPDDLPGLAHFLE 247
Score = 28.9 bits (63), Expect = 5.8
Identities = 16/64 (25%), Positives = 30/64 (46%), Gaps = 8/64 (12%)
Query: 51 VQKHLHKSLADITDLLSTATEHRKSFVSHQIPNQLQEEEEEDDDD--------DDDDEEV 102
+Q L L+++ AT+ + + +++++DDDD DDD+E
Sbjct: 119 LQALLISDLSNVEGKTGNATDEEEEEEEEEEEEDDDDDDDDDDDDEDSGAEIQDDDEEGF 178
Query: 103 DEEE 106
D+EE
Sbjct: 179 DDEE 182
>RBMT_HUMAN (Q9NW13) RNA-binding protein 28 (RNA binding motif
protein 28)
Length = 758
Score = 40.4 bits (93), Expect = 0.002
Identities = 26/81 (32%), Positives = 43/81 (52%), Gaps = 11/81 (13%)
Query: 57 KSLADITDLLSTATEHRKSFVSHQIPNQLQEEEEEDDDDDDDDEE------VDEEEAAKR 110
+S++ I + S ++H++S + EEEE DDDDDDDDEE DEEE
Sbjct: 198 QSVSAIGEEKSHESKHQESVKKKGREEEDMEEEENDDDDDDDDEEDGVFDDEDEEEENIE 257
Query: 111 ARMNRLAELDNLLVGVRSIKR 131
+++ + ++ R++KR
Sbjct: 258 SKVTKPVQIQK-----RAVKR 273
Score = 30.8 bits (68), Expect = 1.5
Identities = 16/61 (26%), Positives = 32/61 (52%)
Query: 45 VVDDQFVQKHLHKSLADITDLLSTATEHRKSFVSHQIPNQLQEEEEEDDDDDDDDEEVDE 104
V D V K +K ++ + + K S + + +E+ EE+++DDDDD++ +E
Sbjct: 183 VAVDWAVAKDKYKDTQSVSAIGEEKSHESKHQESVKKKGREEEDMEEEENDDDDDDDDEE 242
Query: 105 E 105
+
Sbjct: 243 D 243
Score = 28.1 bits (61), Expect = 9.9
Identities = 16/47 (34%), Positives = 25/47 (53%), Gaps = 3/47 (6%)
Query: 63 TDLLSTATEHRKSFVSHQIPNQLQEEEEEDDDD---DDDDEEVDEEE 106
T +S E + HQ + + EEED ++ DDDD++ DEE+
Sbjct: 197 TQSVSAIGEEKSHESKHQESVKKKGREEEDMEEEENDDDDDDDDEED 243
>BC1A_MOUSE (Q9QYE3) B-cell lymphoma/leukemia 11A (B-cell
CLL/lymphoma 11A) (COUP-TF interacting protein 1)
(Ecotropic viral integration site 9 protein) (EVI-9)
Length = 773
Score = 40.4 bits (93), Expect = 0.002
Identities = 21/60 (35%), Positives = 37/60 (61%), Gaps = 8/60 (13%)
Query: 63 TDLLSTATEHRKSFVS--------HQIPNQLQEEEEEDDDDDDDDEEVDEEEAAKRARMN 114
+DL+ +A+ KS V+ + IP EEEEEDD++++++EE +EEE + R++
Sbjct: 453 SDLVGSASSALKSVVAKFKSENDPNLIPENGDEEEEEDDEEEEEEEEEEEEELTESERVD 512
>BC1A_HUMAN (Q9H165) B-cell lymphoma/leukemia 11A (B-cell
CLL/lymphoma 11A) (COUP-TF interacting protein 1)
(Ecotropic viral integration site 9 protein) (EVI-9)
Length = 835
Score = 40.4 bits (93), Expect = 0.002
Identities = 21/60 (35%), Positives = 37/60 (61%), Gaps = 8/60 (13%)
Query: 63 TDLLSTATEHRKSFVS--------HQIPNQLQEEEEEDDDDDDDDEEVDEEEAAKRARMN 114
+DL+ +A+ KS V+ + IP EEEEEDD++++++EE +EEE + R++
Sbjct: 453 SDLVGSASSALKSVVAKFKSENDPNLIPENGDEEEEEDDEEEEEEEEEEEEELTESERVD 512
>NUCL_MOUSE (P09405) Nucleolin (Protein C23)
Length = 706
Score = 40.0 bits (92), Expect = 0.003
Identities = 18/49 (36%), Positives = 31/49 (62%)
Query: 86 QEEEEEDDDDDDDDEEVDEEEAAKRARMNRLAELDNLLVGVRSIKRELE 134
++E++E++DD+DDDEE +EEE K A R E+ + K+++E
Sbjct: 252 EDEDDEEEDDEDDDEEEEEEEPVKAAPGKRKKEMTKQKEAPEAKKQKVE 300
Score = 35.0 bits (79), Expect = 0.081
Identities = 12/26 (46%), Positives = 21/26 (80%)
Query: 81 IPNQLQEEEEEDDDDDDDDEEVDEEE 106
+ + +EEE++DD+D+DDEE D+E+
Sbjct: 238 VAEEEDDEEEDEDDEDEDDEEEDDED 263
Score = 34.7 bits (78), Expect = 0.11
Identities = 13/19 (68%), Positives = 18/19 (94%)
Query: 88 EEEEDDDDDDDDEEVDEEE 106
E+EEDD+D+DD+E+ DEEE
Sbjct: 189 EDEEDDEDEDDEEDDDEEE 207
Score = 33.5 bits (75), Expect = 0.24
Identities = 11/21 (52%), Positives = 19/21 (90%)
Query: 86 QEEEEEDDDDDDDDEEVDEEE 106
+++E+EDD++DDD+EE D+ E
Sbjct: 192 EDDEDEDDEEDDDEEEEDDSE 212
Score = 33.1 bits (74), Expect = 0.31
Identities = 12/20 (60%), Positives = 18/20 (90%)
Query: 87 EEEEEDDDDDDDDEEVDEEE 106
E+E+E+D+DD D++E DEEE
Sbjct: 146 EDEDEEDEDDSDEDEDDEEE 165
Score = 33.1 bits (74), Expect = 0.31
Identities = 13/20 (65%), Positives = 17/20 (85%)
Query: 87 EEEEEDDDDDDDDEEVDEEE 106
EE+E+D D+D+DDEE DE E
Sbjct: 150 EEDEDDSDEDEDDEEEDEFE 169
Score = 33.1 bits (74), Expect = 0.31
Identities = 11/26 (42%), Positives = 21/26 (80%)
Query: 82 PNQLQEEEEEDDDDDDDDEEVDEEEA 107
P EE++ED+DD++DD+E +E+++
Sbjct: 186 PASEDEEDDEDEDDEEDDDEEEEDDS 211
Score = 32.7 bits (73), Expect = 0.40
Identities = 11/21 (52%), Positives = 19/21 (90%)
Query: 86 QEEEEEDDDDDDDDEEVDEEE 106
++E+EED+DD D+DE+ +EE+
Sbjct: 146 EDEDEEDEDDSDEDEDDEEED 166
Score = 32.0 bits (71), Expect = 0.68
Identities = 11/20 (55%), Positives = 18/20 (90%)
Query: 87 EEEEEDDDDDDDDEEVDEEE 106
+E+E+D++DDD++EE D EE
Sbjct: 194 DEDEDDEEDDDEEEEDDSEE 213
Score = 31.2 bits (69), Expect = 1.2
Identities = 10/20 (50%), Positives = 18/20 (90%)
Query: 86 QEEEEEDDDDDDDDEEVDEE 105
++EE+EDD D+D+D+E ++E
Sbjct: 148 EDEEDEDDSDEDEDDEEEDE 167
Score = 31.2 bits (69), Expect = 1.2
Identities = 10/20 (50%), Positives = 18/20 (90%)
Query: 87 EEEEEDDDDDDDDEEVDEEE 106
+E+E+++D+DD DE+ D+EE
Sbjct: 145 DEDEDEEDEDDSDEDEDDEE 164
Score = 30.4 bits (67), Expect = 2.0
Identities = 9/28 (32%), Positives = 23/28 (82%)
Query: 86 QEEEEEDDDDDDDDEEVDEEEAAKRARM 113
++EE+++D+DD++D++ +EE+ ++ M
Sbjct: 189 EDEEDDEDEDDEEDDDEEEEDDSEEEVM 216
Score = 29.6 bits (65), Expect = 3.4
Identities = 9/20 (45%), Positives = 18/20 (90%)
Query: 87 EEEEEDDDDDDDDEEVDEEE 106
E+EE++DD D+D+++ +E+E
Sbjct: 148 EDEEDEDDSDEDEDDEEEDE 167
>NPM_HUMAN (P06748) Nucleophosmin (NPM) (Nucleolar phosphoprotein
B23) (Numatrin) (Nucleolar protein NO38)
Length = 294
Score = 40.0 bits (92), Expect = 0.003
Identities = 14/29 (48%), Positives = 25/29 (85%)
Query: 87 EEEEEDDDDDDDDEEVDEEEAAKRARMNR 115
+++EEDDD+DDDD++ D+EEA ++A + +
Sbjct: 166 DDDEEDDDEDDDDDDFDDEEAEEKAPVKK 194
Score = 34.7 bits (78), Expect = 0.11
Identities = 14/27 (51%), Positives = 22/27 (80%), Gaps = 1/27 (3%)
Query: 86 QEEEEEDDDDDD-DDEEVDEEEAAKRA 111
+E+++EDDDDDD DDEE +E+ K++
Sbjct: 169 EEDDDEDDDDDDFDDEEAEEKAPVKKS 195
Score = 30.0 bits (66), Expect = 2.6
Identities = 13/42 (30%), Positives = 25/42 (58%)
Query: 65 LLSTATEHRKSFVSHQIPNQLQEEEEEDDDDDDDDEEVDEEE 106
LLS + + ++P + + ++DDDDDD+E+ DE++
Sbjct: 135 LLSISGKRSAPGGGSKVPQKKVKLAADEDDDDDDEEDDDEDD 176
>FKB4_SPOFR (Q26486) 46 kDa FK506-binding nuclear protein (EC
5.2.1.8) (Peptidyl-prolyl cis-trans isomerase) (PPIase)
(Rotamase)
Length = 412
Score = 39.7 bits (91), Expect = 0.003
Identities = 15/37 (40%), Positives = 28/37 (75%)
Query: 87 EEEEEDDDDDDDDEEVDEEEAAKRARMNRLAELDNLL 123
++E+E+D++DDD+++ +EEEA K+ + AE D+ L
Sbjct: 197 DDEDEEDEEDDDEDDEEEEEAPKKKKKQPAAEQDSTL 233
Score = 29.6 bits (65), Expect = 3.4
Identities = 18/61 (29%), Positives = 29/61 (47%), Gaps = 8/61 (13%)
Query: 69 ATEHRKSFVSHQIPNQLQEEEEEDDDDDDDD--------EEVDEEEAAKRARMNRLAELD 120
AT ++K+ + E D DDDD+D E++D +E + +MN AE D
Sbjct: 131 ATANKKAKPDKKAGKNSAPAAESDSDDDDEDQLQKFLDGEDIDTDENDESFKMNTSAEGD 190
Query: 121 N 121
+
Sbjct: 191 D 191
>BRD2_HUMAN (P25440) Bromodomain-containing protein 2 (RING3
protein) (O27.1.1)
Length = 801
Score = 39.7 bits (91), Expect = 0.003
Identities = 20/71 (28%), Positives = 39/71 (54%), Gaps = 3/71 (4%)
Query: 78 SHQIPNQLQEEEEEDDDDDDDDEEVDEEEAAKRARMNRLAELDNLLVGVRSIKRELEIIK 137
S + ++ EEEE++D++D++EE E ++ R +RLAEL L R++ +L +
Sbjct: 482 SEESSSESSSEEEEEEDEEDEEEEESESSDSEEERAHRLAELQEQL---RAVHEQLAALS 538
Query: 138 VDTLAMKDRMK 148
++ R +
Sbjct: 539 QGPISKPKRKR 549
>TAF7_YEAST (Q05021) Transcription initiation factor TFIID subunit 7
(TBP-associated factor 7) (TBP-associated factor 67 kDa)
(TAFII-67)
Length = 590
Score = 39.3 bits (90), Expect = 0.004
Identities = 23/81 (28%), Positives = 41/81 (50%), Gaps = 10/81 (12%)
Query: 26 VDSNNPLQLRSVCKWWKSLVVDDQFVQKHLHKSLADITDLLSTATEHRKSFVSHQIPNQL 85
+DS L+ + K + +D ++H H+S A+ + + E +
Sbjct: 476 IDSIKKLEKEAELKRKQLQQTEDSVQKQHQHRSDAETANNVEEEEEEEEE---------- 525
Query: 86 QEEEEEDDDDDDDDEEVDEEE 106
+EEE+E D+D++DDEE DE+E
Sbjct: 526 EEEEDEVDEDEEDDEENDEDE 546
>NUCL_RAT (P13383) Nucleolin (Protein C23)
Length = 712
Score = 38.9 bits (89), Expect = 0.006
Identities = 17/49 (34%), Positives = 31/49 (62%)
Query: 86 QEEEEEDDDDDDDDEEVDEEEAAKRARMNRLAELDNLLVGVRSIKRELE 134
++E+EED++DD+D++E +EEE K A R E+ + K+++E
Sbjct: 254 EDEDEEDEEDDEDEDEEEEEEPVKAAPGKRKKEMTKQKEAPEAKKQKIE 302
Score = 37.0 bits (84), Expect = 0.021
Identities = 13/21 (61%), Positives = 20/21 (94%)
Query: 86 QEEEEEDDDDDDDDEEVDEEE 106
++EE++DD+DDDDD+E +EEE
Sbjct: 189 EDEEDDDDEDDDDDDEEEEEE 209
Score = 37.0 bits (84), Expect = 0.021
Identities = 14/25 (56%), Positives = 20/25 (80%)
Query: 82 PNQLQEEEEEDDDDDDDDEEVDEEE 106
P E+EE+DDD+DDDD++ +EEE
Sbjct: 184 PASEDEDEEDDDDEDDDDDDEEEEE 208
Score = 36.6 bits (83), Expect = 0.028
Identities = 14/27 (51%), Positives = 22/27 (80%)
Query: 87 EEEEEDDDDDDDDEEVDEEEAAKRARM 113
EE+++D+DDDDDDEE +EE+ ++ M
Sbjct: 191 EEDDDDEDDDDDDEEEEEEDDSEEEVM 217
Score = 35.4 bits (80), Expect = 0.062
Identities = 13/20 (65%), Positives = 19/20 (95%)
Query: 87 EEEEEDDDDDDDDEEVDEEE 106
EEE+ED++D++DDE+ DEEE
Sbjct: 252 EEEDEDEEDEEDDEDEDEEE 271
Score = 34.3 bits (77), Expect = 0.14
Identities = 12/20 (60%), Positives = 19/20 (95%)
Query: 87 EEEEEDDDDDDDDEEVDEEE 106
EEEE+D+DD+D++E+ DEE+
Sbjct: 241 EEEEDDEDDEDEEEDEDEED 260
Score = 34.3 bits (77), Expect = 0.14
Identities = 13/19 (68%), Positives = 18/19 (94%)
Query: 88 EEEEDDDDDDDDEEVDEEE 106
EEEEDD+DD+D+EE ++EE
Sbjct: 241 EEEEDDEDDEDEEEDEDEE 259
Score = 33.5 bits (75), Expect = 0.24
Identities = 10/22 (45%), Positives = 21/22 (95%)
Query: 86 QEEEEEDDDDDDDDEEVDEEEA 107
+E+++++DDDDDD+EE +E+++
Sbjct: 191 EEDDDDEDDDDDDEEEEEEDDS 212
Score = 31.2 bits (69), Expect = 1.2
Identities = 10/20 (50%), Positives = 18/20 (90%)
Query: 87 EEEEEDDDDDDDDEEVDEEE 106
+E+E+++D+DD DE+ DEE+
Sbjct: 145 DEDEDEEDEDDSDEDEDEED 164
Score = 31.2 bits (69), Expect = 1.2
Identities = 10/20 (50%), Positives = 18/20 (90%)
Query: 86 QEEEEEDDDDDDDDEEVDEE 105
++E+EED+DD D+DE+ ++E
Sbjct: 146 EDEDEEDEDDSDEDEDEEDE 165
Score = 30.4 bits (67), Expect = 2.0
Identities = 10/20 (50%), Positives = 18/20 (90%)
Query: 87 EEEEEDDDDDDDDEEVDEEE 106
E+E+E+D+DD D++E +E+E
Sbjct: 146 EDEDEEDEDDSDEDEDEEDE 165
Score = 30.4 bits (67), Expect = 2.0
Identities = 11/20 (55%), Positives = 17/20 (85%)
Query: 87 EEEEEDDDDDDDDEEVDEEE 106
E+EE++DD D+D++E DE E
Sbjct: 148 EDEEDEDDSDEDEDEEDEFE 167
Score = 30.0 bits (66), Expect = 2.6
Identities = 11/20 (55%), Positives = 17/20 (85%)
Query: 86 QEEEEEDDDDDDDDEEVDEE 105
++EE+EDD D+D+DEE + E
Sbjct: 148 EDEEDEDDSDEDEDEEDEFE 167
Score = 30.0 bits (66), Expect = 2.6
Identities = 19/58 (32%), Positives = 31/58 (52%), Gaps = 1/58 (1%)
Query: 81 IPNQLQEEEEEDDDD-DDDDEEVDEEEAAKRARMNRLAELDNLLVGVRSIKRELEIIK 137
+P + + EE++DD DD+DEE DE+E + + E + V KR+ E+ K
Sbjct: 232 VPVKAKSVAEEEEDDEDDEDEEEDEDEEDEEDDEDEDEEEEEEPVKAAPGKRKKEMTK 289
Score = 28.9 bits (63), Expect = 5.8
Identities = 8/21 (38%), Positives = 18/21 (85%)
Query: 86 QEEEEEDDDDDDDDEEVDEEE 106
++ +E++D++D+DD + DE+E
Sbjct: 142 EDSDEDEDEEDEDDSDEDEDE 162
>UCRH_YEAST (P00127) Ubiquinol-cytochrome C reductase complex 17 kDa
protein (EC 1.10.2.2) (Mitochondrial hinge protein)
(Complex III polypeptide VI)
Length = 147
Score = 38.5 bits (88), Expect = 0.007
Identities = 29/88 (32%), Positives = 46/88 (51%), Gaps = 11/88 (12%)
Query: 87 EEEEEDDDDDDDDEEVDEEEAAKRARMNRLAE-LDNLLVG---VRSIKRELEIIKVDT-- 140
E+E+ED+DDDDDD+E +EEE ++ L E N G V + E +K+
Sbjct: 52 EDEDEDEDDDDDDDEDEEEEEEVTDQLEDLREHFKNTEEGKALVHHYEECAERVKIQQQQ 111
Query: 141 ---LAMKDRMKCLESF--LKIYLKSATS 163
++ + C+E F L+ YL +AT+
Sbjct: 112 PGYADLEHKEDCVEEFFHLQHYLDTATA 139
Score = 35.8 bits (81), Expect = 0.047
Identities = 12/21 (57%), Positives = 20/21 (95%)
Query: 86 QEEEEEDDDDDDDDEEVDEEE 106
++E+E++D+DDDDD++ DEEE
Sbjct: 50 EDEDEDEDEDDDDDDDEDEEE 70
Score = 33.9 bits (76), Expect = 0.18
Identities = 12/33 (36%), Positives = 25/33 (75%)
Query: 87 EEEEEDDDDDDDDEEVDEEEAAKRARMNRLAEL 119
+E+E++D+D+DDD++ DE+E + ++L +L
Sbjct: 49 DEDEDEDEDEDDDDDDDEDEEEEEEVTDQLEDL 81
>UBF1_RAT (P25977) Nucleolar transcription factor 1 (Upstream
binding factor 1) (UBF-1)
Length = 764
Score = 38.5 bits (88), Expect = 0.007
Identities = 21/68 (30%), Positives = 39/68 (56%), Gaps = 3/68 (4%)
Query: 40 WWKSLVVDDQFVQK-HLHKSLADITDLLSTATEHRKSFVSHQIPNQLQEEEEEDDDDDDD 98
W KSL D+ K ++ ++T L + S + Q ++ +E+++E+DDDDDD
Sbjct: 633 WVKSLSPQDRAAYKEYISNKRKNMTKL--RGPNPKSSRTTLQSKSESEEDDDEEDDDDDD 690
Query: 99 DEEVDEEE 106
+EE +++E
Sbjct: 691 EEEEEDDE 698
Score = 35.4 bits (80), Expect = 0.062
Identities = 13/20 (65%), Positives = 18/20 (90%)
Query: 87 EEEEEDDDDDDDDEEVDEEE 106
+E E+DDDD+DDDE+ DE+E
Sbjct: 722 DENEDDDDDEDDDEDDDEDE 741
Score = 31.2 bits (69), Expect = 1.2
Identities = 9/24 (37%), Positives = 20/24 (82%)
Query: 86 QEEEEEDDDDDDDDEEVDEEEAAK 109
+ E+++DD+DDD+D++ DE+ ++
Sbjct: 723 ENEDDDDDEDDDEDDDEDEDNESE 746
Score = 31.2 bits (69), Expect = 1.2
Identities = 10/22 (45%), Positives = 19/22 (85%)
Query: 87 EEEEEDDDDDDDDEEVDEEEAA 108
++++EDDD+DDD++E +E E +
Sbjct: 727 DDDDEDDDEDDDEDEDNESEGS 748
Score = 30.8 bits (68), Expect = 1.5
Identities = 10/23 (43%), Positives = 19/23 (82%)
Query: 83 NQLQEEEEEDDDDDDDDEEVDEE 105
N+ +++E+DD+DDD+DE+ + E
Sbjct: 724 NEDDDDDEDDDEDDDEDEDNESE 746
Score = 30.4 bits (67), Expect = 2.0
Identities = 9/24 (37%), Positives = 20/24 (82%)
Query: 83 NQLQEEEEEDDDDDDDDEEVDEEE 106
++ ++ +E +DDDDD+D++ D++E
Sbjct: 716 DESEDGDENEDDDDDEDDDEDDDE 739
Score = 29.3 bits (64), Expect = 4.4
Identities = 10/29 (34%), Positives = 22/29 (75%)
Query: 78 SHQIPNQLQEEEEEDDDDDDDDEEVDEEE 106
S + ++ + E+ ++++DDDDDE+ DE++
Sbjct: 709 SSESSSEDESEDGDENEDDDDDEDDDEDD 737
>NUCL_HUMAN (P19338) Nucleolin (Protein C23)
Length = 706
Score = 38.1 bits (87), Expect = 0.010
Identities = 16/49 (32%), Positives = 31/49 (62%)
Query: 86 QEEEEEDDDDDDDDEEVDEEEAAKRARMNRLAELDNLLVGVRSIKRELE 134
+++E++DD+DD+++EE +EEE K A R E+ + K+++E
Sbjct: 250 EDDEDDDDEDDEEEEEEEEEEPVKEAPGKRKKEMAKQKAAPEAKKQKVE 298
Score = 37.7 bits (86), Expect = 0.012
Identities = 15/22 (68%), Positives = 19/22 (86%)
Query: 87 EEEEEDDDDDDDDEEVDEEEAA 108
+E++EDD+DDDDDEE D EE A
Sbjct: 188 DEDDEDDEDDDDDEEDDSEEEA 209
Score = 35.8 bits (81), Expect = 0.047
Identities = 12/21 (57%), Positives = 20/21 (95%)
Query: 86 QEEEEEDDDDDDDDEEVDEEE 106
+++E+EDDDDD+DDE+ D+E+
Sbjct: 239 EDDEDEDDDDDEDDEDDDDED 259
Score = 35.4 bits (80), Expect = 0.062
Identities = 12/21 (57%), Positives = 20/21 (95%)
Query: 86 QEEEEEDDDDDDDDEEVDEEE 106
++EEE+D+D+DDDD+E DE++
Sbjct: 235 EDEEEDDEDEDDDDDEDDEDD 255
Score = 34.7 bits (78), Expect = 0.11
Identities = 12/20 (60%), Positives = 19/20 (95%)
Query: 87 EEEEEDDDDDDDDEEVDEEE 106
E++++D+DD+DDD+E DEEE
Sbjct: 244 EDDDDDEDDEDDDDEDDEEE 263
Score = 34.3 bits (77), Expect = 0.14
Identities = 11/21 (52%), Positives = 20/21 (94%)
Query: 86 QEEEEEDDDDDDDDEEVDEEE 106
+E++E++DDDDD+D+E D++E
Sbjct: 238 EEDDEDEDDDDDEDDEDDDDE 258
Score = 33.5 bits (75), Expect = 0.24
Identities = 10/26 (38%), Positives = 23/26 (88%)
Query: 86 QEEEEEDDDDDDDDEEVDEEEAAKRA 111
++E++EDD+DD+DD++ +E+++ + A
Sbjct: 184 EDEDDEDDEDDEDDDDDEEDDSEEEA 209
Score = 33.1 bits (74), Expect = 0.31
Identities = 14/25 (56%), Positives = 19/25 (76%)
Query: 87 EEEEEDDDDDDDDEEVDEEEAAKRA 111
EE+EEDD+D+D+DE+ E A K A
Sbjct: 153 EEDEEDDEDEDEDEDEIEPAAMKAA 177
Score = 33.1 bits (74), Expect = 0.31
Identities = 11/27 (40%), Positives = 22/27 (80%)
Query: 87 EEEEEDDDDDDDDEEVDEEEAAKRARM 113
E+E+++DD+DD+D++ DEE+ ++ M
Sbjct: 184 EDEDDEDDEDDEDDDDDEEDDSEEEAM 210
Score = 32.3 bits (72), Expect = 0.52
Identities = 10/24 (41%), Positives = 21/24 (86%)
Query: 83 NQLQEEEEEDDDDDDDDEEVDEEE 106
N ++E+EE+DD+D+DD++ +++E
Sbjct: 230 NVAEDEDEEEDDEDEDDDDDEDDE 253
Score = 32.0 bits (71), Expect = 0.68
Identities = 11/21 (52%), Positives = 18/21 (85%)
Query: 86 QEEEEEDDDDDDDDEEVDEEE 106
++ +EE+DDD ++DEE DE+E
Sbjct: 142 EDSDEEEDDDSEEDEEDDEDE 162
Score = 30.8 bits (68), Expect = 1.5
Identities = 9/21 (42%), Positives = 19/21 (89%)
Query: 86 QEEEEEDDDDDDDDEEVDEEE 106
++++ E+D++DD+DE+ DE+E
Sbjct: 148 EDDDSEEDEEDDEDEDEDEDE 168
Score = 30.4 bits (67), Expect = 2.0
Identities = 12/26 (46%), Positives = 22/26 (84%), Gaps = 2/26 (7%)
Query: 86 QEEEEEDDDDDDDDEEVDEEEAAKRA 111
+E+EE+D+D+D+D++E+ E AA +A
Sbjct: 153 EEDEEDDEDEDEDEDEI--EPAAMKA 176
Score = 30.4 bits (67), Expect = 2.0
Identities = 11/22 (50%), Positives = 18/22 (81%)
Query: 87 EEEEEDDDDDDDDEEVDEEEAA 108
+ EE+++DD+D+DE+ DE E A
Sbjct: 151 DSEEDEEDDEDEDEDEDEIEPA 172
Score = 30.4 bits (67), Expect = 2.0
Identities = 9/21 (42%), Positives = 19/21 (89%)
Query: 86 QEEEEEDDDDDDDDEEVDEEE 106
+EE+++ ++D++DDE+ DE+E
Sbjct: 146 EEEDDDSEEDEEDDEDEDEDE 166
Score = 30.0 bits (66), Expect = 2.6
Identities = 11/33 (33%), Positives = 22/33 (66%)
Query: 87 EEEEEDDDDDDDDEEVDEEEAAKRARMNRLAEL 119
+E++EDDDDD++D+ +E A+ + A++
Sbjct: 191 DEDDEDDDDDEEDDSEEEAMETTPAKGKKAAKV 223
Score = 29.6 bits (65), Expect = 3.4
Identities = 9/20 (45%), Positives = 18/20 (90%)
Query: 87 EEEEEDDDDDDDDEEVDEEE 106
+EEE+DD ++D++++ DE+E
Sbjct: 145 DEEEDDDSEEDEEDDEDEDE 164
Score = 29.3 bits (64), Expect = 4.4
Identities = 9/24 (37%), Positives = 19/24 (78%)
Query: 83 NQLQEEEEEDDDDDDDDEEVDEEE 106
N +E+ +E++DDD +++E D+E+
Sbjct: 138 NAKKEDSDEEEDDDSEEDEEDDED 161
Score = 28.1 bits (61), Expect = 9.9
Identities = 10/24 (41%), Positives = 17/24 (70%)
Query: 83 NQLQEEEEEDDDDDDDDEEVDEEE 106
N ++E+ D+++DDD E DEE+
Sbjct: 135 NGKNAKKEDSDEEEDDDSEEDEED 158
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.313 0.129 0.348
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 17,875,766
Number of Sequences: 164201
Number of extensions: 743456
Number of successful extensions: 22805
Number of sequences better than 10.0: 900
Number of HSP's better than 10.0 without gapping: 685
Number of HSP's successfully gapped in prelim test: 225
Number of HSP's that attempted gapping in prelim test: 10911
Number of HSP's gapped (non-prelim): 5044
length of query: 168
length of database: 59,974,054
effective HSP length: 102
effective length of query: 66
effective length of database: 43,225,552
effective search space: 2852886432
effective search space used: 2852886432
T: 11
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 61 (28.1 bits)
Medicago: description of AC149207.4


