

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC149197.10 - phase: 0
(330 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
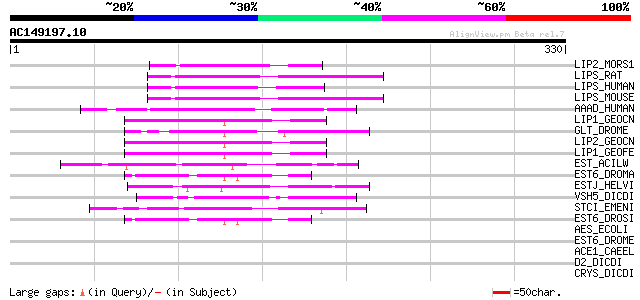
Score E
Sequences producing significant alignments: (bits) Value
LIP2_MORS1 (P24484) Lipase 2 (EC 3.1.1.3) (Triacylglycerol lipase) 66 1e-10
LIPS_RAT (P15304) Hormone sensitive lipase (EC 3.1.1.-) (HSL) 65 2e-10
LIPS_HUMAN (Q05469) Hormone sensitive lipase (EC 3.1.1.-) (HSL) 65 3e-10
LIPS_MOUSE (P54310) Hormone sensitive lipase (EC 3.1.1.-) (HSL) 64 4e-10
AAAD_HUMAN (P22760) Arylacetamide deacetylase (EC 3.1.1.-) (AADAC) 53 9e-07
LIP1_GEOCN (P17573) Lipase 1 precursor (EC 3.1.1.3) (Lipase I) (... 49 1e-05
GLT_DROME (P33438) Glutactin precursor 48 3e-05
LIP2_GEOCN (P22394) Lipase 2 precursor (EC 3.1.1.3) (Lipase II) ... 47 5e-05
LIP1_GEOFE (P79066) Lipase 1 precursor (EC 3.1.1.3) (Lipase I) (... 47 5e-05
EST_ACILW (P18773) Esterase (EC 3.1.1.-) 47 7e-05
EST6_DROMA (P47982) Esterase 6 precursor (EC 3.1.1.1) (Est-6) (C... 45 3e-04
ESTJ_HELVI (P12992) Juvenile hormone esterase precursor (EC 3.1.... 45 3e-04
VSH5_DICDI (P14326) Vegetative specific protein H5 44 4e-04
STCI_EMENI (Q00675) Putative sterigmatocystin biosynthesis lipas... 44 4e-04
EST6_DROSI (Q08662) Esterase 6 precursor (EC 3.1.1.1) (Est-6) (C... 44 6e-04
AES_ECOLI (P23872) Acetyl esterase (EC 3.1.1.-) 43 0.001
EST6_DROME (P08171) Esterase 6 precursor (EC 3.1.1.1) (Est-6) (C... 42 0.002
ACE1_CAEEL (P38433) Acetylcholinesterase 1 precursor (EC 3.1.1.7... 42 0.002
D2_DICDI (P18142) cAMP-regulated D2 protein precursor 42 0.002
CRYS_DICDI (P21837) Crystal protein precursor 40 0.006
>LIP2_MORS1 (P24484) Lipase 2 (EC 3.1.1.3) (Triacylglycerol lipase)
Length = 433
Score = 66.2 bits (160), Expect = 1e-10
Identities = 35/103 (33%), Positives = 55/103 (52%), Gaps = 12/103 (11%)
Query: 84 LVYFHGGVFMFESTFSKKYHEHLKTFASQANVIIVSIEYSLAPEYPLPTCYHDCWAALKW 143
+++FHGG F + +HE T +Q +VS++Y +APEYP PT DC AA W
Sbjct: 161 MLFFHGGGFCIGDIDT--HHEFCHTVCAQTGWAVVSVDYRMAPEYPAPTALKDCLAAYAW 218
Query: 144 ISSHSNNNINNPEPWLIEHGNFNKLFIGGDSAGANIAHNIAIQ 186
++ HS + +P +++ + GDSAG +A +A Q
Sbjct: 219 LAEHSQSLGASP----------SRIVLSGDSAGGCLAALVAQQ 251
>LIPS_RAT (P15304) Hormone sensitive lipase (EC 3.1.1.-) (HSL)
Length = 768
Score = 65.5 bits (158), Expect = 2e-10
Identities = 39/140 (27%), Positives = 69/140 (48%), Gaps = 12/140 (8%)
Query: 83 ILVYFHGGVFMFESTFSKKYHEHLKTFASQANVIIVSIEYSLAPEYPLPTCYHDCWAALK 142
++V+ HGG F+ ++ SK + +LK +A + V I+SI+YSLAPE P P +C+ A
Sbjct: 344 LVVHIHGGGFVAQT--SKSHEPYLKNWAQELGVPIISIDYSLAPEAPFPRALEECFFAYC 401
Query: 143 WISSHSNNNINNPEPWLIEHGNFNKLFIGGDSAGANIAHNIAIQAGLENLPCDVKILGAI 202
W H + ++ + GDSAG N+ ++++A + I+ A
Sbjct: 402 WAVKHCE----------LLGSTGERICLAGDSAGGNLCITVSLRAAAYGVRVPDGIMAAY 451
Query: 203 IIHPYFYSANPIGSEPIIEP 222
+ SA+P +++P
Sbjct: 452 PVTTLQSSASPSRLLSLMDP 471
>LIPS_HUMAN (Q05469) Hormone sensitive lipase (EC 3.1.1.-) (HSL)
Length = 775
Score = 64.7 bits (156), Expect = 3e-10
Identities = 33/105 (31%), Positives = 57/105 (53%), Gaps = 12/105 (11%)
Query: 83 ILVYFHGGVFMFESTFSKKYHEHLKTFASQANVIIVSIEYSLAPEYPLPTCYHDCWAALK 142
++V+FHGG F+ ++ S+ + +LK++A + I+SI+YSLAPE P P +C+ A
Sbjct: 345 LIVHFHGGGFVAQT--SRSHEPYLKSWAQELGAPIISIDYSLAPEAPFPRALEECFFAYC 402
Query: 143 WISSHSNNNINNPEPWLIEHGNFNKLFIGGDSAGANIAHNIAIQA 187
W H + ++ + GDSAG N+ +A++A
Sbjct: 403 WAIKHC----------ALLGSTGERICLAGDSAGGNLCFTVALRA 437
>LIPS_MOUSE (P54310) Hormone sensitive lipase (EC 3.1.1.-) (HSL)
Length = 759
Score = 64.3 bits (155), Expect = 4e-10
Identities = 39/140 (27%), Positives = 69/140 (48%), Gaps = 12/140 (8%)
Query: 83 ILVYFHGGVFMFESTFSKKYHEHLKTFASQANVIIVSIEYSLAPEYPLPTCYHDCWAALK 142
++V+ HGG F+ ++ SK + +LK +A + V I SI+YSLAPE P P +C+ A
Sbjct: 344 LVVHIHGGGFVAQT--SKSHEPYLKNWAQELGVPIFSIDYSLAPEAPFPRALEECFFAYC 401
Query: 143 WISSHSNNNINNPEPWLIEHGNFNKLFIGGDSAGANIAHNIAIQAGLENLPCDVKILGAI 202
W H + + ++ + GDSAG N+ ++++A + I+ A
Sbjct: 402 WAVKHCD----------LLGSTGERICLAGDSAGGNLCITVSLRAAAYGVRVPDGIMAAY 451
Query: 203 IIHPYFYSANPIGSEPIIEP 222
+ SA+P +++P
Sbjct: 452 PVTTLQSSASPSRLLSLMDP 471
>AAAD_HUMAN (P22760) Arylacetamide deacetylase (EC 3.1.1.-) (AADAC)
Length = 398
Score = 53.1 bits (126), Expect = 9e-07
Identities = 44/165 (26%), Positives = 74/165 (44%), Gaps = 17/165 (10%)
Query: 43 PSLNDPNTGISSKDIQIPHNPTISSRIYLPKITNPLSKFPILVYFHGGVFMFESTFSKKY 102
P +D N ++ I R+Y+PK + + L Y HGG + S Y
Sbjct: 71 PPTSDENVTVTETKFN-----NILVRVYVPKRKSEALRRG-LFYIHGGGWCVGSAALSGY 124
Query: 103 HEHLKTFASQANVIIVSIEYSLAPEYPLPTCYHDCWAALKWISSHSNNNINNPEPWLIEH 162
+ A + + ++VS Y LAP+Y P + D + AL+W + L ++
Sbjct: 125 DLLSRWTADRLDAVVVSTNYRLAPKYHFPIQFEDVYNALRWFLR---------KKVLAKY 175
Query: 163 G-NFNKLFIGGDSAGANIAHNIAIQAGLENLPCDVKILGAIIIHP 206
G N ++ I GDSAG N+A + Q L++ +K+ +I+P
Sbjct: 176 GVNPERIGISGDSAGGNLAAAVTQQL-LDDPDVKIKLKIQSLIYP 219
>LIP1_GEOCN (P17573) Lipase 1 precursor (EC 3.1.1.3) (Lipase I) (GCL
I)
Length = 563
Score = 49.3 bits (116), Expect = 1e-05
Identities = 37/133 (27%), Positives = 60/133 (44%), Gaps = 23/133 (17%)
Query: 69 IYLPKITNPLSKFPILVYFHGGVFMFESTFSKKYHEHLKTFASQAN-VIIVSIEYSLAP- 126
++ P T P +K P++V+ +GG F+F S+ S + ++K V+ VSI Y P
Sbjct: 129 VFRPAGTKPDAKLPVMVWIYGGAFVFGSSASYPGNGYVKESVEMGQPVVFVSINYRTGPY 188
Query: 127 ----------EYPLPTCYHDCWAALKWISSHSNNNINNPEPWLIEHGNFNKLFIGGDSAG 176
E HD L+W+S + N +P+ K+ I G+SAG
Sbjct: 189 GFLGGDAITAEGNTNAGLHDQRKGLEWVSDNIANFGGDPD----------KVMIFGESAG 238
Query: 177 A-NIAHNIAIQAG 188
A ++AH + G
Sbjct: 239 AMSVAHQLVAYGG 251
>GLT_DROME (P33438) Glutactin precursor
Length = 1026
Score = 48.1 bits (113), Expect = 3e-05
Identities = 40/157 (25%), Positives = 65/157 (40%), Gaps = 31/157 (19%)
Query: 69 IYLPKITNPLSKFPILVYFHGGVFMFESTFSKKYHEHLKTFASQANVIIVSIEYSLAP-- 126
IY P+ N L P+LV+ HG E F E + + +V++VSI Y LAP
Sbjct: 150 IYAPEGANQL---PVLVFVHG-----EMLFDGGSEEAQPDYVLEKDVLLVSINYRLAPFG 201
Query: 127 -------EYPLPTCYHDCWAALKWISSHSNNNINNPEPWLIEH--GNFNKLFIGGDSAGA 177
E P D AL+W+ + + H GN ++ + G + GA
Sbjct: 202 FLSALTDELPGNVALSDLQLALEWLQRN------------VVHFGGNAGQVTLVGQAGGA 249
Query: 178 NIAHNIAIQAGLENLPCDVKILGAIIIHPYFYSANPI 214
+AH +++ NL + + ++PY P+
Sbjct: 250 TLAHALSLSGRAGNLFQQLILQSGTALNPYLIDNQPL 286
>LIP2_GEOCN (P22394) Lipase 2 precursor (EC 3.1.1.3) (Lipase II)
(GCL II)
Length = 563
Score = 47.4 bits (111), Expect = 5e-05
Identities = 35/133 (26%), Positives = 61/133 (45%), Gaps = 23/133 (17%)
Query: 69 IYLPKITNPLSKFPILVYFHGGVFMFESTFSKKYHEHLKTFASQAN-VIIVSIEYSLAP- 126
++ P T P +K P++V+ +GG F++ S+ + + ++K + V+ VSI Y P
Sbjct: 129 VFRPAGTKPDAKLPVMVWIYGGAFVYGSSAAYPGNSYVKESINMGQPVVFVSINYRTGPF 188
Query: 127 ----------EYPLPTCYHDCWAALKWISSHSNNNINNPEPWLIEHGNFNKLFIGGDSAG 176
E HD L+W+S + N +P+ K+ I G+SAG
Sbjct: 189 GFLGGDAITAEGNTNAGLHDQRKGLEWVSDNIANFGGDPD----------KVMIFGESAG 238
Query: 177 A-NIAHNIAIQAG 188
A ++AH + G
Sbjct: 239 AMSVAHQLIAYGG 251
>LIP1_GEOFE (P79066) Lipase 1 precursor (EC 3.1.1.3) (Lipase I) (TFL
I)
Length = 563
Score = 47.4 bits (111), Expect = 5e-05
Identities = 35/133 (26%), Positives = 61/133 (45%), Gaps = 23/133 (17%)
Query: 69 IYLPKITNPLSKFPILVYFHGGVFMFESTFSKKYHEHLKTFASQAN-VIIVSIEYSLAP- 126
++ P T P +K P++V+ +GG F++ S+ + + ++K + V+ VSI Y P
Sbjct: 129 VFRPAGTKPDAKLPVMVWIYGGAFVYGSSAAYPGNSYVKESINMGQPVVFVSINYRTGPF 188
Query: 127 ----------EYPLPTCYHDCWAALKWISSHSNNNINNPEPWLIEHGNFNKLFIGGDSAG 176
E HD L+W+S + N +P+ K+ I G+SAG
Sbjct: 189 GFLGGDAITAEGNTNAGLHDQRKGLEWVSDNIANFGGDPD----------KVMIFGESAG 238
Query: 177 A-NIAHNIAIQAG 188
A ++AH + G
Sbjct: 239 AMSVAHQLIAYGG 251
>EST_ACILW (P18773) Esterase (EC 3.1.1.-)
Length = 303
Score = 47.0 bits (110), Expect = 7e-05
Identities = 49/187 (26%), Positives = 78/187 (41%), Gaps = 36/187 (19%)
Query: 31 SIERPKQSPFAPPSLNDPNTGISSKDIQIPHNPTISSR------IYLPKITNPLSKFPIL 84
+I P Q AP +L + P NPT+ R + +I S ++
Sbjct: 19 TIRTPSQLNLAPNALRPVLDQLCRL---FPQNPTVQIRPIRLAGVRGEEIKAQASATQLI 75
Query: 85 VYFHGGVFMFESTFSKKYHEHLKT-FASQANVIIVSIEYSLAPEYPLP---TCYHDCWAA 140
+ HGG F S + H L T AS+ + ++ ++Y LAPE+P P D + A
Sbjct: 76 FHIHGGAFFLGSLNT---HRALMTDLASRTQMQVIHVDYPLAPEHPYPEAIDAIFDVYQA 132
Query: 141 LKWISSHSNNNINNPEPWLIEHGNFNKLFIGGDSAGANIAHNIAIQAGLENLPCDVKILG 200
L L++ + I GDS GAN+A +A+ L+ P ++ G
Sbjct: 133 L-----------------LVQGIKPKDIIISGDSCGANLA--LALSLRLKQQP-ELMPSG 172
Query: 201 AIIIHPY 207
I++ PY
Sbjct: 173 LILMSPY 179
>EST6_DROMA (P47982) Esterase 6 precursor (EC 3.1.1.1) (Est-6)
(Carboxylic-ester hydrolase 6) (Carboxylesterase-6)
Length = 542
Score = 45.1 bits (105), Expect = 3e-04
Identities = 36/120 (30%), Positives = 53/120 (44%), Gaps = 23/120 (19%)
Query: 69 IYLPKITNPLSKFPILVYFHGGVFMFESTFSKKYHEHLKTFASQANVIIVSIEYSLAP-- 126
IY PK + S FP++ + HGG FMF + + + ++ + I+V I Y L P
Sbjct: 108 IYKPK-NSKRSSFPVVAHIHGGAFMFGAAWQNGHENVMR----EGKFILVKISYRLGPLG 162
Query: 127 -----EYPLPTCY--HDCWAALKWISSHSNNNINNPEPWLIEHGNFNKLFIGGDSAGANI 179
+ LP Y D ALKWI + + PE N L IG + GA++
Sbjct: 163 FASTGDRDLPGNYGLKDQRLALKWIKQNIASFGGEPE---------NILLIGHSAGGASV 213
>ESTJ_HELVI (P12992) Juvenile hormone esterase precursor (EC
3.1.1.59) (JH esterase)
Length = 564
Score = 44.7 bits (104), Expect = 3e-04
Identities = 41/155 (26%), Positives = 64/155 (40%), Gaps = 27/155 (17%)
Query: 71 LPKITNPLSKFPILVYFHGGVFMFESTFSKKYHE--HLKTFASQANVIIVSIEYSL---- 124
LP++ PILV+ HGG F F S HE H + NVI+++ Y L
Sbjct: 121 LPRVRGTTPLRPILVFIHGGGFAFGSG-----HEDLHGPEYLVTKNVIVITFNYRLNVFG 175
Query: 125 -----APEYPLPTCYHDCWAALKWISSHSNNNINNPEPWLIEHGNFNKLFIGGDSAGANI 179
+ P D L+W+ ++ N +P + + I G SAGA+
Sbjct: 176 FLSMNTTKIPGNAGLRDQVTLLRWVQRNAKNFGGDP----------SDITIAGQSAGASA 225
Query: 180 AHNIAIQAGLENLPCDVKILGAIIIHPYFYSANPI 214
AH + + E L IL + YF++ +P+
Sbjct: 226 AHLLTLSKATEGL-FKRAILMSGTGMSYFFTTSPL 259
>VSH5_DICDI (P14326) Vegetative specific protein H5
Length = 316
Score = 44.3 bits (103), Expect = 4e-04
Identities = 35/132 (26%), Positives = 60/132 (44%), Gaps = 14/132 (10%)
Query: 76 NPLS-KFPILVYFHGGVFMFESTFSKKYHEHLKTFASQANVIIVSIEYSLAPEYPLPTCY 134
NP+ K+ + Y HGG FM + KK + + + N I++ +Y L PE+ P
Sbjct: 56 NPVDHKYKAIFYIHGGGFMVDGI--KKLPREI---SDRTNSILIYPDYGLTPEFKYPLGL 110
Query: 135 HDCWAALKWISSHSNNNINNPEPWLIEHGNFNKLFIGGDSAGANIAHNIAIQAGLENLPC 194
C+ I + + N N+ LI + + I G+S+G N A ++ + L N
Sbjct: 111 KQCYQLFTDIMNGNFNPFND----LIN----DSISIVGESSGGNFALSLPLMLKLNNSTF 162
Query: 195 DVKILGAIIIHP 206
KI ++ +P
Sbjct: 163 FKKISKVLVYYP 174
>STCI_EMENI (Q00675) Putative sterigmatocystin biosynthesis
lipase/esterase stcI
Length = 286
Score = 44.3 bits (103), Expect = 4e-04
Identities = 43/169 (25%), Positives = 70/169 (40%), Gaps = 28/169 (16%)
Query: 48 PNTGISSKDIQIPHNPTISSRIYLPK-ITNPLSKFPILVYFHGGVFMFESTFSKKYHEHL 106
P+ + ++D + PT RIY P + +P P+ +YFH G ++ S + +
Sbjct: 20 PDLSVQAEDKILGGVPT---RIYTPPDVADP----PLALYFHAGGWVMGSIDEED--GFV 70
Query: 107 KTFASQANVIIVSIEYSLAPEYPLPTCYHDCWAALKWISSHSNNNINNPEPWLIEHGNFN 166
+T A I S+ Y LAPE+ P DC + + +E
Sbjct: 71 RTLCKLARTRIFSVGYRLAPEFRFPMALDDCLTVARSV---------------LETYPVQ 115
Query: 167 KLFIGGDSAGANIAHNIA---IQAGLENLPCDVKILGAIIIHPYFYSAN 212
+ G SAG N+A + A + GL + V L + +HP SA+
Sbjct: 116 SICFIGASAGGNMAFSTALTLVSDGLGDRVQGVVALAPVTVHPDSVSAD 164
>EST6_DROSI (Q08662) Esterase 6 precursor (EC 3.1.1.1) (Est-6)
(Carboxylic-ester hydrolase 6) (Carboxylesterase-6)
Length = 542
Score = 43.9 bits (102), Expect = 6e-04
Identities = 35/120 (29%), Positives = 53/120 (44%), Gaps = 23/120 (19%)
Query: 69 IYLPKITNPLSKFPILVYFHGGVFMFESTFSKKYHEHLKTFASQANVIIVSIEYSLAP-- 126
IY PK + S FP++ + HGG FMF + + + ++ + I+V I Y L P
Sbjct: 108 IYKPK-NSKRSTFPVVAHIHGGAFMFGAAWQNGHENVMR----EGKFILVKISYRLGPLG 162
Query: 127 -----EYPLPTCY--HDCWAALKWISSHSNNNINNPEPWLIEHGNFNKLFIGGDSAGANI 179
+ LP Y D ALKWI + + P+ N L IG + GA++
Sbjct: 163 FASTGDRDLPGNYGLKDQRLALKWIKQNIASFGGEPQ---------NVLLIGHSAGGASV 213
>AES_ECOLI (P23872) Acetyl esterase (EC 3.1.1.-)
Length = 319
Score = 43.1 bits (100), Expect = 0.001
Identities = 33/139 (23%), Positives = 66/139 (46%), Gaps = 17/139 (12%)
Query: 65 ISSRIYLPKITNPLSKFPILVYFHGGVFMFESTFSKKYHEHLKTFASQANVIIVSIEYSL 124
+ +R++ P+ +P + F Y HGG F+ + + + ++ AS + ++ I+Y+L
Sbjct: 72 VETRLFCPQPDSPATLF----YLHGGGFILGNLDT--HDRIMRLLASYSQCTVIGIDYTL 125
Query: 125 APEYPLPTCYHDCWAALKWISSHSNNNINNPEPWLIEHGNFNKLFIGGDSAGANIAHNIA 184
+PE P + AA + + E + I N +++ GDSAGA +A A
Sbjct: 126 SPEARFPQAIEEIVAACCYFHQQA-------EDYQI---NMSRIGFAGDSAGAMLALASA 175
Query: 185 IQAGLENLPCDVKILGAII 203
+ + + C K+ G ++
Sbjct: 176 LWLRDKQIDCG-KVAGVLL 193
>EST6_DROME (P08171) Esterase 6 precursor (EC 3.1.1.1) (Est-6)
(Carboxylic-ester hydrolase 6) (Carboxylesterase-6)
Length = 544
Score = 42.4 bits (98), Expect = 0.002
Identities = 32/120 (26%), Positives = 53/120 (43%), Gaps = 23/120 (19%)
Query: 69 IYLPKITNPLSKFPILVYFHGGVFMFESTFSKKYHEHLKTFASQANVIIVSIEYSLAP-- 126
+Y PK + + FP++ + HGG FMF + + + ++ + I+V I Y L P
Sbjct: 110 VYKPK-NSKRNSFPVVAHIHGGAFMFGAAWQNGHENVMR----EGKFILVKISYRLGPLG 164
Query: 127 -----EYPLPTCY--HDCWAALKWISSHSNNNINNPEPWLIEHGNFNKLFIGGDSAGANI 179
+ LP Y D ALKWI + + P+ N L +G + GA++
Sbjct: 165 FVSTGDRDLPGNYGLKDQRLALKWIKQNIASFGGEPQ---------NVLLVGHSAGGASV 215
>ACE1_CAEEL (P38433) Acetylcholinesterase 1 precursor (EC 3.1.1.7)
(AChE 1)
Length = 620
Score = 42.4 bits (98), Expect = 0.002
Identities = 44/182 (24%), Positives = 77/182 (42%), Gaps = 43/182 (23%)
Query: 69 IYLPKITNPLSKFPILVYFHGGVFMFESTFSKKYHEHLKTFASQANVIIVSIEYSLA--- 125
+Y+P +P K ++V+ +GG F + Y + T + NVI+V++ Y ++
Sbjct: 114 VYVPGKVDPNKKLAVMVWVYGGGFWSGTATLDVYDGRILTV--EENVILVAMNYRVSIFG 171
Query: 126 ------PEYPLPTCYHDCWAALKWISSHSNNNINNPEPWLIEHGNFNKLFIGGDSAGANI 179
PE P D A+KW+ H N ++ G+ +++ + G+SAGA
Sbjct: 172 FLYMNRPEAPGNMGMWDQLLAMKWV--HKNIDLFG--------GDLSRITLFGESAGAAS 221
Query: 180 A-------------HNIAIQAGLENLPC-----DVKILGAIIIHPYFYSANPIGSEPIIE 221
H IQ+G P DV + A+I+ Y+A G+ +I
Sbjct: 222 VSIHMLSPKSAPYFHRAIIQSGSATSPWAIEPRDVALARAVIL----YNAMKCGNMSLIN 277
Query: 222 PE 223
P+
Sbjct: 278 PD 279
>D2_DICDI (P18142) cAMP-regulated D2 protein precursor
Length = 535
Score = 42.0 bits (97), Expect = 0.002
Identities = 37/116 (31%), Positives = 54/116 (45%), Gaps = 20/116 (17%)
Query: 69 IYLPKITNPLSKFPILVYFHGGVFMFESTFSKKYHEHLKTFASQANVIIVSIEYSLAPEY 128
++ PK P SK+P++VY GG F S Y T +Q++VI+V+I Y L
Sbjct: 114 VFTPKDATPNSKYPVIVYIPGGAFSVGSGSVPLYD---ATKFAQSSVIVVNINYRLGVLG 170
Query: 129 PLPT-------CYHDCWAALKWISSHSNNNINNPEPWLIEHGNFNKLFIGGDSAGA 177
+ T + D AL+W+ NNI + GN + I G+SAGA
Sbjct: 171 FMGTDLMHGNYGFLDQIKALEWV----YNNIGS------FGGNKEMITIWGESAGA 216
>CRYS_DICDI (P21837) Crystal protein precursor
Length = 550
Score = 40.4 bits (93), Expect = 0.006
Identities = 35/116 (30%), Positives = 59/116 (50%), Gaps = 20/116 (17%)
Query: 69 IYLPKITNPLSKFPILVYFHGGVFMFESTFSKKYHEHLKTFASQANVIIVSIEYSLAPEY 128
+++P+ NP SK P++V+ GG F + T S ++ LK FA+ ++VI+V++ Y L
Sbjct: 116 VFIPRTVNPGSKVPVMVFIPGGAFT-QGTGSCPLYDGLK-FAN-SSVIVVNVNYRLGVLG 172
Query: 129 PLPT-------CYHDCWAALKWISSHSNNNINNPEPWLIEHGNFNKLFIGGDSAGA 177
L T + D AL W+ + + G+ N++ I G+SAGA
Sbjct: 173 FLCTGLLSGNFGFLDQVMALDWVQENIE----------VFGGDKNQVTIYGESAGA 218
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.318 0.138 0.434
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 43,961,547
Number of Sequences: 164201
Number of extensions: 2029581
Number of successful extensions: 4460
Number of sequences better than 10.0: 72
Number of HSP's better than 10.0 without gapping: 10
Number of HSP's successfully gapped in prelim test: 62
Number of HSP's that attempted gapping in prelim test: 4428
Number of HSP's gapped (non-prelim): 74
length of query: 330
length of database: 59,974,054
effective HSP length: 111
effective length of query: 219
effective length of database: 41,747,743
effective search space: 9142755717
effective search space used: 9142755717
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 66 (30.0 bits)
Medicago: description of AC149197.10


