

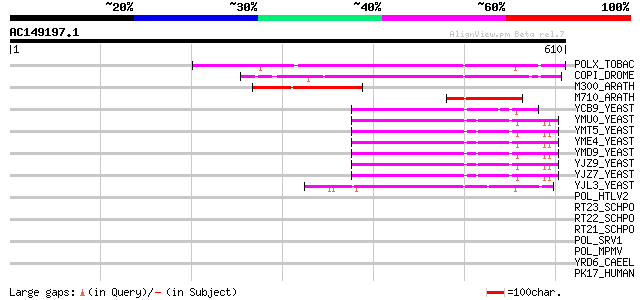
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC149197.1 + phase: 0 /pseudo
(610 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
POLX_TOBAC (P10978) Retrovirus-related Pol polyprotein from tran... 295 2e-79
COPI_DROME (P04146) Copia protein (Gag-int-pol protein) [Contain... 164 5e-40
M300_ARATH (P93293) Hypothetical mitochondrial protein AtMg00300... 115 4e-25
M710_ARATH (P92512) Hypothetical mitochondrial protein AtMg00710... 74 1e-12
YCB9_YEAST (P25384) Transposon Ty2 protein B (Ty1-17 protein B) 67 2e-10
YMU0_YEAST (Q04670) Transposon Ty1 protein B 63 3e-09
YMT5_YEAST (Q04214) Transposon Ty1 protein B 63 3e-09
YME4_YEAST (Q04711) Transposon Ty1 protein B 63 3e-09
YMD9_YEAST (Q03434) Transposon Ty1 protein B 63 3e-09
YJZ9_YEAST (P47100) Transposon Ty1 protein B 63 3e-09
YJZ7_YEAST (P47098) Transposon Ty1 protein B 63 3e-09
YJL3_YEAST (P47024) Transposon Ty4 207.7 kDa hypothetical protein 60 2e-08
POL_HTLV2 (P03363) Pol polyprotein [Contains: Reverse transcript... 40 0.014
RT23_SCHPO (Q9UR07) Retrotransposable element Tf2 155 kDa protei... 40 0.023
RT22_SCHPO (Q9C0R2) Retrotransposable element Tf2 155 kDa protei... 40 0.023
RT21_SCHPO (Q05654) Retrotransposable element Tf2 155 kDa protei... 40 0.023
POL_SRV1 (P04025) Pol polyprotein [Contains: Reverse transcripta... 39 0.030
POL_MPMV (P07572) Pol polyprotein [Contains: Reverse transcripta... 39 0.030
YRD6_CAEEL (Q09575) Hypothetical protein K02A2.6 in chromosome II 39 0.040
PK17_HUMAN (P63136) HERV-K_11q22.1 provirus ancestral Pol protei... 38 0.088
>POLX_TOBAC (P10978) Retrovirus-related Pol polyprotein from
transposon TNT 1-94 [Contains: Protease (EC 3.4.23.-);
Reverse transcriptase (EC 2.7.7.49); Endonuclease]
Length = 1328
Score = 295 bits (755), Expect = 2e-79
Identities = 165/429 (38%), Positives = 243/429 (56%), Gaps = 27/429 (6%)
Query: 202 LLDT*YNFHMCPYRDLFTTFEPVDCGVVLMGNDVQCKVA*IGIVHFKTNDGVVRTLTNES 261
++DT + H P RDLF + D G V MGN K+A IG + KTN G L +
Sbjct: 295 VVDTAASHHATPVRDLFCRYVAGDFGTVKMGNTSYSKIAGIGDICIKTNVGCTLVLKDVR 354
Query: 262 LV--CKYSAEGGVLM---------------VSKGYLVLLKASRIDSLYVLQGIVVTGSAA 304
V + + G+ + ++KG LV+ K +LY + G
Sbjct: 355 HVPDLRMNLISGIALDRDGYESYFANQKWRLTKGSLVIAKGVARGTLYRTNAEICQGELN 414
Query: 305 VSSSMPKKDVTKLWHMHLGHMTEKGMHLLSKQGRLGKQGIDKLEFCKHYVFWKQKKVSFS 364
+ D LWH +GHM+EKG+ +L+K+ + ++ C + +F KQ +VSF
Sbjct: 415 AAQDEISVD---LWHKRMGHMSEKGLQILAKKSLISYAKGTTVKPCDYCLFGKQHRVSFQ 471
Query: 365 TATHRTKGILDYIHSDLWGPSKVTSYGGRRYMMTIIDDFPRKVWVYFLRYKNETFPTFKK 424
T++ R ILD ++SD+ GP ++ S GG +Y +T IDD RK+WVY L+ K++ F F+K
Sbjct: 472 TSSERKLNILDLVYSDVCGPMEIESMGGNKYFVTFIDDASRKLWVYILKTKDQVFQVFQK 531
Query: 425 WRILVETQTGKNVKKLITDN*LEFCSSDFNEFCTNHGIARHKTIPRNPQQNGVAERMIRT 484
+ LVE +TG+ +K+L +DN E+ S +F E+C++HGI KT+P PQ NGVAERM RT
Sbjct: 532 FHALVERETGRKLKRLRSDNGGEYTSREFEEYCSSHGIRHEKTVPGTPQHNGVAERMNRT 591
Query: 485 LLERARCMLSNAGL*N*RDLWVEAASTACHLVNRSPHSALDFKVPEDIWSGNLVDYSNLR 544
++E+ R ML A L + W EA TAC+L+NRSP L F++PE +W+ V YS+L+
Sbjct: 592 IVEKVRSMLRMAKL--PKSFWGEAVQTACYLINRSPSVPLAFEIPERVWTNKEVSYSHLK 649
Query: 545 IFGCPAYALV---NDGKLAPRAVECIFLSYASESKGYRLWCSDPKSQKLILSRDVTFNED 601
+FGC A+A V KL +++ CIF+ Y E GYRLW DP +K+I SRDV F E
Sbjct: 650 VFGCRAFAHVPKEQRTKLDDKSIPCIFIGYGDEEFGYRLW--DPVKKKVIRSRDVVFRES 707
Query: 602 ALLSSGKQS 610
+ ++ S
Sbjct: 708 EVRTAADMS 716
>COPI_DROME (P04146) Copia protein (Gag-int-pol protein) [Contains:
Copia VLP protein; Copia protease (EC 3.4.23.-)]
Length = 1409
Score = 164 bits (416), Expect = 5e-40
Identities = 111/365 (30%), Positives = 181/365 (49%), Gaps = 26/365 (7%)
Query: 254 VRTLTNESLVCKYSAEGGVLMVSKGYLVLLKASRIDSLYVLQGIVVTGSAAVSSSMPKKD 313
V+ L + ++ G + +SK L+++K S + L + V A S + K+
Sbjct: 361 VKRLQEAGMSIEFDKSG--VTISKNGLMVVKNSGM-----LNNVPVINFQAYSINAKHKN 413
Query: 314 VTKLWHMHLGHMTE------KGMHLLSKQGRLGKQGIDKLEFCKHYVFWKQKKVSFSTAT 367
+LWH GH+++ K ++ S Q L + E C+ + KQ ++ F
Sbjct: 414 NFRLWHERFGHISDGKLLEIKRKNMFSDQSLLNNLELS-CEICEPCLNGKQARLPFKQLK 472
Query: 368 HRT--KGILDYIHSDLWGPSKVTSYGGRRYMMTIIDDFPRKVWVYFLRYKNETFPTFKKW 425
+T K L +HSD+ GP + + Y + +D F Y ++YK++ F F+ +
Sbjct: 473 DKTHIKRPLFVVHSDVCGPITPVTLDDKNYFVIFVDQFTHYCVTYLIKYKSDVFSMFQDF 532
Query: 426 RILVETQTGKNVKKLITDN*LEFCSSDFNEFCTNHGIARHKTIPRNPQQNGVAERMIRTL 485
E V L DN E+ S++ +FC GI+ H T+P PQ NGV+ERMIRT+
Sbjct: 533 VAKSEAHFNLKVVYLYIDNGREYLSNEMRQFCVKKGISYHLTVPHTPQLNGVSERMIRTI 592
Query: 486 LERARCMLSNAGL*N*RDLWVEAASTACHLVNRSPHSAL--DFKVPEDIWSGNLVDYSNL 543
E+AR M+S A L + W EA TA +L+NR P AL K P ++W +L
Sbjct: 593 TEKARTMVSGAKL--DKSFWGEAVLTATYLINRIPSRALVDSSKTPYEMWHNKKPYLKHL 650
Query: 544 RIFGCPAYALVND--GKLAPRAVECIFLSYASESKGYRLWCSDPKSQKLILSRDVTFNED 601
R+FG Y + + GK ++ + IF+ Y E G++LW D ++K I++RDV +E
Sbjct: 651 RVFGATVYVHIKNKQGKFDDKSFKSIFVGY--EPNGFKLW--DAVNEKFIVARDVVVDET 706
Query: 602 ALLSS 606
+++S
Sbjct: 707 NMVNS 711
>M300_ARATH (P93293) Hypothetical mitochondrial protein AtMg00300
(ORF145a) (ORF1451)
Length = 145
Score = 115 bits (287), Expect = 4e-25
Identities = 57/121 (47%), Positives = 77/121 (63%), Gaps = 2/121 (1%)
Query: 267 SAEGGVLMVSKGYLVLLKASRIDSLYVLQGIVVTGSAAVSSSMPKKDVTKLWHMHLGHMT 326
S GVL V KG +LK +R DSLY+LQG V TG + ++ + KD T+LWH L HM+
Sbjct: 23 SCSEGVLKVLKGCRTILKGNRHDSLYILQGSVETGESNLAETA--KDETRLWHSRLAHMS 80
Query: 327 EKGMHLLSKQGRLGKQGIDKLEFCKHYVFWKQKKVSFSTATHRTKGILDYIHSDLWGPSK 386
++GM LL K+G L + L+FC+ ++ K +V+FST H TK LDY+HSDLWG
Sbjct: 81 QRGMELLVKKGFLDSSKVSSLKFCEDCIYGKTHRVNFSTGQHTTKNPLDYVHSDLWGAPS 140
Query: 387 V 387
V
Sbjct: 141 V 141
>M710_ARATH (P92512) Hypothetical mitochondrial protein AtMg00710
(ORF120)
Length = 120
Score = 73.9 bits (180), Expect = 1e-12
Identities = 36/83 (43%), Positives = 55/83 (65%), Gaps = 2/83 (2%)
Query: 481 MIRTLLERARCMLSNAGL*N*RDLWVEAASTACHLVNRSPHSALDFKVPEDIWSGNLVDY 540
M RT++E+ R ML GL + +AA+TA H++N+ P +A++F VP+++W ++ Y
Sbjct: 1 MNRTIIEKVRSMLCECGLP--KTFRADAANTAVHIINKYPSTAINFHVPDEVWFQSVPTY 58
Query: 541 SNLRIFGCPAYALVNDGKLAPRA 563
S LR FGC AY ++GKL PRA
Sbjct: 59 SYLRRFGCVAYIHCDEGKLKPRA 81
>YCB9_YEAST (P25384) Transposon Ty2 protein B (Ty1-17 protein B)
Length = 1770
Score = 66.6 bits (161), Expect = 2e-10
Identities = 58/212 (27%), Positives = 90/212 (42%), Gaps = 11/212 (5%)
Query: 376 YIHSDLWGPSKVTSYGGRRYMMTIIDDFPRKVWVYFLRYKNET--FPTFKKWRILVETQT 433
Y+H+D++GP Y ++ D+ R WVY L + E F ++ Q
Sbjct: 663 YLHTDIFGPVHHLPKSAPSYFISFTDEKTRFQWVYPLHDRREESILNVFTSILAFIKNQF 722
Query: 434 GKNVKKLITDN*LEFCSSDFNEFCTNHGIARHKTIPRNPQQNGVAERMIRTLLERARCML 493
V + D E+ + ++F TN GI T + + +GVAER+ RTLL R +L
Sbjct: 723 NARVLVIQMDRGSEYTNKTLHKFFTNRGITACYTTTADSRAHGVAERLNRTLLNDCRTLL 782
Query: 494 SNAGL*N*RDLWVEAASTACHLVNRSPHSALDFKVPEDIWSGNLVDYSNLRIFGCPAYAL 553
+GL N LW A + + N D + L D + + FG P +
Sbjct: 783 HCSGLPN--HLWFSAVEFSTIIRNSLVSPKNDKSARQHAGLAGL-DITTILPFGQP--VI 837
Query: 554 VN----DGKLAPRAVECIFLSYASESKGYRLW 581
VN D K+ PR + L + S GY ++
Sbjct: 838 VNNHNPDSKIHPRGIPGYALHPSRNSYGYIIY 869
>YMU0_YEAST (Q04670) Transposon Ty1 protein B
Length = 1328
Score = 62.8 bits (151), Expect = 3e-09
Identities = 68/243 (27%), Positives = 104/243 (41%), Gaps = 20/243 (8%)
Query: 376 YIHSDLWGPSKVTSYGGRRYMMTIIDDFPRKVWVYFL--RYKNETFPTFKKWRILVETQT 433
Y+H+D++GP Y ++ D+ + WVY L R ++ F ++ Q
Sbjct: 240 YLHTDIFGPVHNLPKSAPSYFISFTDETTKFRWVYPLHDRREDSILDVFTTILAFIKNQF 299
Query: 434 GKNVKKLITDN*LEFCSSDFNEFCTNHGIARHKTIPRNPQQNGVAERMIRTLLERARCML 493
+V + D E+ + ++F +GI T + + +GVAER+ RTLL+ R L
Sbjct: 300 QASVLVIQMDRGSEYTNRTLHKFLEKNGITPCYTTTADSRAHGVAERLNRTLLDDCRTQL 359
Query: 494 SNAGL*N*RDLWVEAASTACHLVNRSPHSALDFKVPEDIWSGNLVDYSNLRIFGCPAYAL 553
+GL N LW A + +V S S K +D S L FG P +
Sbjct: 360 QCSGLPN--HLWFSAIEFST-IVRNSLASPKSKKSARQHAGLAGLDISTLLPFGQP--VI 414
Query: 554 VND----GKLAPRAVECIFLSYASESKGYRLWCSDPK-----SQKLIL----SRDVTFNE 600
VND K+ PR + L + S GY ++ K + +IL SR FN
Sbjct: 415 VNDHNPNSKIHPRGIPGYALHPSRNSYGYIIYLPSLKKTVDTTNYVILQGKESRLDQFNY 474
Query: 601 DAL 603
DAL
Sbjct: 475 DAL 477
>YMT5_YEAST (Q04214) Transposon Ty1 protein B
Length = 1328
Score = 62.8 bits (151), Expect = 3e-09
Identities = 68/243 (27%), Positives = 104/243 (41%), Gaps = 20/243 (8%)
Query: 376 YIHSDLWGPSKVTSYGGRRYMMTIIDDFPRKVWVYFL--RYKNETFPTFKKWRILVETQT 433
Y+H+D++GP Y ++ D+ + WVY L R ++ F ++ Q
Sbjct: 240 YLHTDIFGPVHNLPKSAPSYFISFTDETTKFRWVYPLHDRREDSILDVFTTILAFIKNQF 299
Query: 434 GKNVKKLITDN*LEFCSSDFNEFCTNHGIARHKTIPRNPQQNGVAERMIRTLLERARCML 493
+V + D E+ + ++F +GI T + + +GVAER+ RTLL+ R L
Sbjct: 300 QASVLVIQMDRGSEYTNRTLHKFLEKNGITPCYTTTADSRAHGVAERLNRTLLDDCRTQL 359
Query: 494 SNAGL*N*RDLWVEAASTACHLVNRSPHSALDFKVPEDIWSGNLVDYSNLRIFGCPAYAL 553
+GL N LW A + +V S S K +D S L FG P +
Sbjct: 360 QCSGLPN--HLWFSAIEFST-IVRNSLASPKSKKSARQHAGLAGLDISTLLPFGQP--VI 414
Query: 554 VND----GKLAPRAVECIFLSYASESKGYRLWCSDPK-----SQKLIL----SRDVTFNE 600
VND K+ PR + L + S GY ++ K + +IL SR FN
Sbjct: 415 VNDHNPNSKIHPRGIPGYALHPSRNSYGYIIYLPSLKKTVDTTNYVILQGKESRLDQFNY 474
Query: 601 DAL 603
DAL
Sbjct: 475 DAL 477
>YME4_YEAST (Q04711) Transposon Ty1 protein B
Length = 1328
Score = 62.8 bits (151), Expect = 3e-09
Identities = 68/243 (27%), Positives = 104/243 (41%), Gaps = 20/243 (8%)
Query: 376 YIHSDLWGPSKVTSYGGRRYMMTIIDDFPRKVWVYFL--RYKNETFPTFKKWRILVETQT 433
Y+H+D++GP Y ++ D+ + WVY L R ++ F ++ Q
Sbjct: 240 YLHTDIFGPVHNLPKSAPSYFISFTDETTKFRWVYPLHDRREDSILDVFTTILAFIKNQF 299
Query: 434 GKNVKKLITDN*LEFCSSDFNEFCTNHGIARHKTIPRNPQQNGVAERMIRTLLERARCML 493
+V + D E+ + ++F +GI T + + +GVAER+ RTLL+ R L
Sbjct: 300 QASVLVIQMDRGSEYTNRTLHKFLEKNGITPCYTTTADSRAHGVAERLNRTLLDDCRTQL 359
Query: 494 SNAGL*N*RDLWVEAASTACHLVNRSPHSALDFKVPEDIWSGNLVDYSNLRIFGCPAYAL 553
+GL N LW A + +V S S K +D S L FG P +
Sbjct: 360 QCSGLPN--HLWFSAIEFST-IVRNSLASPKSKKSARQHAGLAGLDISTLLPFGQP--VI 414
Query: 554 VND----GKLAPRAVECIFLSYASESKGYRLWCSDPK-----SQKLIL----SRDVTFNE 600
VND K+ PR + L + S GY ++ K + +IL SR FN
Sbjct: 415 VNDHNPNSKIHPRGIPGYALHPSRNSYGYIIYLPSLKKTVDTTNYVILQGKESRLDQFNY 474
Query: 601 DAL 603
DAL
Sbjct: 475 DAL 477
>YMD9_YEAST (Q03434) Transposon Ty1 protein B
Length = 1328
Score = 62.8 bits (151), Expect = 3e-09
Identities = 68/243 (27%), Positives = 104/243 (41%), Gaps = 20/243 (8%)
Query: 376 YIHSDLWGPSKVTSYGGRRYMMTIIDDFPRKVWVYFL--RYKNETFPTFKKWRILVETQT 433
Y+H+D++GP Y ++ D+ + WVY L R ++ F ++ Q
Sbjct: 240 YLHTDIFGPVHNLPKSAPSYFISFTDETTKFRWVYPLHDRREDSILDVFTTILAFIKNQF 299
Query: 434 GKNVKKLITDN*LEFCSSDFNEFCTNHGIARHKTIPRNPQQNGVAERMIRTLLERARCML 493
+V + D E+ + ++F +GI T + + +GVAER+ RTLL+ R L
Sbjct: 300 QASVLVIQMDRGSEYTNRTLHKFLEKNGITPCYTTTADSRAHGVAERLNRTLLDDCRTQL 359
Query: 494 SNAGL*N*RDLWVEAASTACHLVNRSPHSALDFKVPEDIWSGNLVDYSNLRIFGCPAYAL 553
+GL N LW A + +V S S K +D S L FG P +
Sbjct: 360 QCSGLPN--HLWFSAIEFST-IVRNSLASPKSKKSARQHAGLAGLDISTLLPFGQP--VI 414
Query: 554 VND----GKLAPRAVECIFLSYASESKGYRLWCSDPK-----SQKLIL----SRDVTFNE 600
VND K+ PR + L + S GY ++ K + +IL SR FN
Sbjct: 415 VNDHNPNSKIHPRGIPGYALHPSRNSYGYIIYLPSLKKTVDTTNYVILQGKESRLDQFNY 474
Query: 601 DAL 603
DAL
Sbjct: 475 DAL 477
>YJZ9_YEAST (P47100) Transposon Ty1 protein B
Length = 1755
Score = 62.8 bits (151), Expect = 3e-09
Identities = 68/243 (27%), Positives = 104/243 (41%), Gaps = 20/243 (8%)
Query: 376 YIHSDLWGPSKVTSYGGRRYMMTIIDDFPRKVWVYFL--RYKNETFPTFKKWRILVETQT 433
Y+H+D++GP Y ++ D+ + WVY L R ++ F ++ Q
Sbjct: 667 YLHTDIFGPVHNLPKSAPSYFISFTDETTKFRWVYPLHDRREDSILDVFTTILAFIKNQF 726
Query: 434 GKNVKKLITDN*LEFCSSDFNEFCTNHGIARHKTIPRNPQQNGVAERMIRTLLERARCML 493
+V + D E+ + ++F +GI T + + +GVAER+ RTLL+ R L
Sbjct: 727 QASVLVIQMDRGSEYTNRTLHKFLEKNGITPCYTTTADSRAHGVAERLNRTLLDDCRTQL 786
Query: 494 SNAGL*N*RDLWVEAASTACHLVNRSPHSALDFKVPEDIWSGNLVDYSNLRIFGCPAYAL 553
+GL N LW A + +V S S K +D S L FG P +
Sbjct: 787 QCSGLPN--HLWFSAIEFST-IVRNSLASPKSKKSARQHAGLAGLDISTLLPFGQP--VI 841
Query: 554 VND----GKLAPRAVECIFLSYASESKGYRLWCSDPK-----SQKLIL----SRDVTFNE 600
VND K+ PR + L + S GY ++ K + +IL SR FN
Sbjct: 842 VNDHNPNSKIHPRGIPGYALHPSRNSYGYIIYLPSLKKTVDTTNYVILQGKESRLDQFNY 901
Query: 601 DAL 603
DAL
Sbjct: 902 DAL 904
>YJZ7_YEAST (P47098) Transposon Ty1 protein B
Length = 1755
Score = 62.8 bits (151), Expect = 3e-09
Identities = 68/243 (27%), Positives = 104/243 (41%), Gaps = 20/243 (8%)
Query: 376 YIHSDLWGPSKVTSYGGRRYMMTIIDDFPRKVWVYFL--RYKNETFPTFKKWRILVETQT 433
Y+H+D++GP Y ++ D+ + WVY L R ++ F ++ Q
Sbjct: 667 YLHTDIFGPVHNLPKSAPSYFISFTDETTKFRWVYPLHDRREDSILDVFTTILAFIKNQF 726
Query: 434 GKNVKKLITDN*LEFCSSDFNEFCTNHGIARHKTIPRNPQQNGVAERMIRTLLERARCML 493
+V + D E+ + ++F +GI T + + +GVAER+ RTLL+ R L
Sbjct: 727 QASVLVIQMDRGSEYTNRTLHKFLEKNGITPCYTTTADSRAHGVAERLNRTLLDDCRTQL 786
Query: 494 SNAGL*N*RDLWVEAASTACHLVNRSPHSALDFKVPEDIWSGNLVDYSNLRIFGCPAYAL 553
+GL N LW A + +V S S K +D S L FG P +
Sbjct: 787 QCSGLPN--HLWFSAIEFST-IVRNSLASPKSKKSARQHAGLAGLDISTLLPFGQP--VI 841
Query: 554 VND----GKLAPRAVECIFLSYASESKGYRLWCSDPK-----SQKLIL----SRDVTFNE 600
VND K+ PR + L + S GY ++ K + +IL SR FN
Sbjct: 842 VNDHNPNSKIHPRGIPGYALHPSRNSYGYIIYLPSLKKTVDTTNYVILQGKESRLDQFNY 901
Query: 601 DAL 603
DAL
Sbjct: 902 DAL 904
>YJL3_YEAST (P47024) Transposon Ty4 207.7 kDa hypothetical protein
Length = 1803
Score = 60.1 bits (144), Expect = 2e-08
Identities = 72/304 (23%), Positives = 125/304 (40%), Gaps = 37/304 (12%)
Query: 325 MTEKGMHLLSKQGRLGKQGIDKLEFC---KHYV-----------FWKQK-KVSFSTATHR 369
+ ++ + L R+G GI ++E HY FW Q K+S +T +
Sbjct: 551 LNKRSITLEDAHKRMGHTGIQQIENSIKHNHYEESLDLIKEPNEFWCQTCKISKATKRNH 610
Query: 370 TKGILDYIHS-----------DLWGPSKVTSYGGRRYMMTIIDDFPRKVWV--YFLRYKN 416
G ++ HS D++GP ++ +RYM+ ++D+ R +F +
Sbjct: 611 YTGSMNN-HSTDHEPGSSWCMDIFGPVSSSNADTKRYMLIMVDNNTRYCMTSTHFNKNAE 669
Query: 417 ETFPTFKKWRILVETQTGKNVKKLITDN*LEFCSSDFNEFCTNHGIARHKTIPRNPQQNG 476
+K VETQ + V+++ +D EF + E+ + GI T ++ NG
Sbjct: 670 TILAQVRKNIQYVETQFDRKVREINSDRGTEFTNDQIEEYFISKGIHHILTSTQDHAANG 729
Query: 477 VAERMIRTLLERARCMLSNAGL*N*RDLWVEAASTACHLVNRSPHSALDFKVPEDIWSGN 536
AER IRT++ A +L + L W A ++A ++ N H + K+P S
Sbjct: 730 RAERYIRTIITDATTLLRQSNL--RVKFWEYAVTSATNIRNYLEHKSTG-KLPLKAISRQ 786
Query: 537 LVDYSNLRIFGCPAYALV---NDGKLAPRAVECIFLSYASESKGYRLWCSDPKSQKLILS 593
V + ++ N KL P + I L S GY+ + P K++ S
Sbjct: 787 PVTVRLMSFLPFGEKGIIWNHNHKKLKPSGLPSIILCKDPNSYGYKFFI--PSKNKIVTS 844
Query: 594 RDVT 597
+ T
Sbjct: 845 DNYT 848
>POL_HTLV2 (P03363) Pol polyprotein [Contains: Reverse
transcriptase/ribonuclease H (EC 2.7.7.49) (EC 3.1.26.4)
(RT); Integrase (IN)]
Length = 982
Score = 40.4 bits (93), Expect = 0.014
Identities = 21/50 (42%), Positives = 29/50 (58%), Gaps = 3/50 (6%)
Query: 442 TDN*LEFCSSDFNEFCTNHGIARHKTIPRNPQQNGVAER---MIRTLLER 488
TDN F S +F EFCT++ I IP NP +G+ ER +I+ LL +
Sbjct: 806 TDNGPAFLSQEFQEFCTSYRIKHSTHIPYNPTSSGLVERTNGVIKNLLNK 855
>RT23_SCHPO (Q9UR07) Retrotransposable element Tf2 155 kDa protein
type 3
Length = 1333
Score = 39.7 bits (91), Expect = 0.023
Identities = 25/88 (28%), Positives = 40/88 (45%), Gaps = 5/88 (5%)
Query: 436 NVKKLITDN*LEFCSSDFNEFCTNHGIARHKTIPRNPQQNGVAERMIRTLLERARCMLSN 495
N K++I DN F S + +F + ++P PQ +G ER +T+ + RC+ S
Sbjct: 1043 NPKEIIADNDHIFTSQTWKDFAHKYNFVMKFSLPYRPQTDGQTERTNQTVEKLLRCVCST 1102
Query: 496 AGL*N*RDLWVEAASTACHLVNRSPHSA 523
+ WV+ S N + HSA
Sbjct: 1103 H-----PNTWVDHISLVQQSYNNAIHSA 1125
>RT22_SCHPO (Q9C0R2) Retrotransposable element Tf2 155 kDa protein
type 2
Length = 1333
Score = 39.7 bits (91), Expect = 0.023
Identities = 25/88 (28%), Positives = 40/88 (45%), Gaps = 5/88 (5%)
Query: 436 NVKKLITDN*LEFCSSDFNEFCTNHGIARHKTIPRNPQQNGVAERMIRTLLERARCMLSN 495
N K++I DN F S + +F + ++P PQ +G ER +T+ + RC+ S
Sbjct: 1043 NPKEIIADNDHIFTSQTWKDFAHKYNFVMKFSLPYRPQTDGQTERTNQTVEKLLRCVCST 1102
Query: 496 AGL*N*RDLWVEAASTACHLVNRSPHSA 523
+ WV+ S N + HSA
Sbjct: 1103 H-----PNTWVDHISLVQQSYNNAIHSA 1125
>RT21_SCHPO (Q05654) Retrotransposable element Tf2 155 kDa protein
type 1
Length = 1333
Score = 39.7 bits (91), Expect = 0.023
Identities = 25/88 (28%), Positives = 40/88 (45%), Gaps = 5/88 (5%)
Query: 436 NVKKLITDN*LEFCSSDFNEFCTNHGIARHKTIPRNPQQNGVAERMIRTLLERARCMLSN 495
N K++I DN F S + +F + ++P PQ +G ER +T+ + RC+ S
Sbjct: 1043 NPKEIIADNDHIFTSQTWKDFAHKYNFVMKFSLPYRPQTDGQTERTNQTVEKLLRCVCST 1102
Query: 496 AGL*N*RDLWVEAASTACHLVNRSPHSA 523
+ WV+ S N + HSA
Sbjct: 1103 H-----PNTWVDHISLVQQSYNNAIHSA 1125
>POL_SRV1 (P04025) Pol polyprotein [Contains: Reverse
transcriptase/ribonuclease H (EC 2.7.7.49) (EC 3.1.26.4)
(RT); Integrase (IN)]
Length = 867
Score = 39.3 bits (90), Expect = 0.030
Identities = 29/99 (29%), Positives = 45/99 (45%), Gaps = 21/99 (21%)
Query: 412 LRYKNETFPTFKKWRILVETQTGKNVKKLIT-----------------DN*LEFCSSDFN 454
L+Y + + TF + +L QTG+ K +IT DN + S +F
Sbjct: 667 LKYIHVSIDTFSGF-LLATLQTGETTKHVITHLLHCFSIIGLPKQIKTDNGPGYTSKNFQ 725
Query: 455 EFCTNHGIARHKTIPRNPQQNGVAER---MIRTLLERAR 490
EFC+ I IP NPQ G+ ER ++T +E+ +
Sbjct: 726 EFCSTLQIKHVTGIPYNPQGQGIVERAHLSLKTTIEKIK 764
>POL_MPMV (P07572) Pol polyprotein [Contains: Reverse
transcriptase/ribonuclease H (EC 2.7.7.49) (EC 3.1.26.4)
(RT); Integrase (IN)]
Length = 867
Score = 39.3 bits (90), Expect = 0.030
Identities = 29/99 (29%), Positives = 45/99 (45%), Gaps = 21/99 (21%)
Query: 412 LRYKNETFPTFKKWRILVETQTGKNVKKLIT-----------------DN*LEFCSSDFN 454
L+Y + + TF + +L QTG+ K +IT DN + S +F
Sbjct: 667 LKYIHVSIDTFSGF-LLATLQTGETTKHVITHLLHCFSIIGLPKQIKTDNGPGYTSKNFQ 725
Query: 455 EFCTNHGIARHKTIPRNPQQNGVAER---MIRTLLERAR 490
EFC+ I IP NPQ G+ ER ++T +E+ +
Sbjct: 726 EFCSTLQIKHITGIPYNPQGQGIVERAHLSLKTTIEKIK 764
>YRD6_CAEEL (Q09575) Hypothetical protein K02A2.6 in chromosome II
Length = 1268
Score = 38.9 bits (89), Expect = 0.040
Identities = 29/99 (29%), Positives = 44/99 (44%), Gaps = 6/99 (6%)
Query: 440 LITDN*LEFCSSDFNEFCTNHGIARHKTIPRNPQQNGVAERMIRTLLERARCMLSNAGL* 499
+I+DN + S F + C +HGI + P+ NG AER + T L+R + G
Sbjct: 906 IISDNGTQLTSHLFAQMCQSHGIEHKTSAVYYPRSNGAAERFVDT-LKRGIAKIKGEGSV 964
Query: 500 N*RDLWVEAASTACHLVNRSPHSALDFKVPEDIWSGNLV 538
N + L S +PHSAL+ P + G +
Sbjct: 965 NQQILNKFLIS-----YRNTPHSALNGSTPAECHFGRKI 998
>PK17_HUMAN (P63136) HERV-K_11q22.1 provirus ancestral Pol protein
[Includes: Reverse transcriptase (RT) (EC 2.7.7.49);
Ribonuclease H (EC 3.1.26.4) (RNase H); Integrase (IN)]
Length = 954
Score = 37.7 bits (86), Expect = 0.088
Identities = 28/99 (28%), Positives = 43/99 (43%), Gaps = 3/99 (3%)
Query: 387 VTSYGGRRYMMTIIDDFPRKVWVYFLRYKNETFPTFKKWRILVETQTGKNVKKLITDN*L 446
V S+G Y+ +D + +W + E+ KK + G +K+ TDN
Sbjct: 655 VPSFGRLSYVHVTVDTYSHFIWATC--HTGESTSHVKKHLLSCFAVMGVP-EKIKTDNGP 711
Query: 447 EFCSSDFNEFCTNHGIARHKTIPRNPQQNGVAERMIRTL 485
+CS F +F + I+ IP N Q + ER RTL
Sbjct: 712 GYCSKAFQKFLSQWKISHTTGIPYNSQGQAIVERTNRTL 750
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.331 0.144 0.458
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 66,419,120
Number of Sequences: 164201
Number of extensions: 2662202
Number of successful extensions: 6469
Number of sequences better than 10.0: 74
Number of HSP's better than 10.0 without gapping: 65
Number of HSP's successfully gapped in prelim test: 9
Number of HSP's that attempted gapping in prelim test: 6381
Number of HSP's gapped (non-prelim): 78
length of query: 610
length of database: 59,974,054
effective HSP length: 116
effective length of query: 494
effective length of database: 40,926,738
effective search space: 20217808572
effective search space used: 20217808572
T: 11
A: 40
X1: 15 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.9 bits)
S2: 69 (31.2 bits)
Medicago: description of AC149197.1


