

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC149130.9 + phase: 0 /pseudo
(1214 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
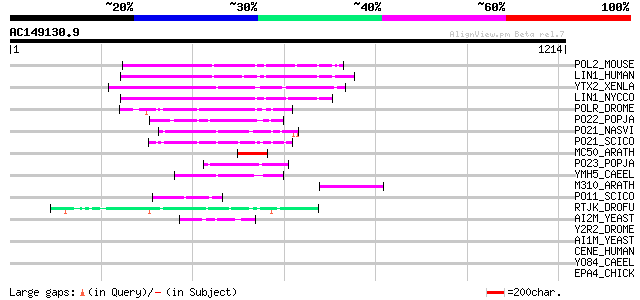
Score E
Sequences producing significant alignments: (bits) Value
POL2_MOUSE (P11369) Retrovirus-related Pol polyprotein [Contains... 159 3e-38
LIN1_HUMAN (P08547) LINE-1 reverse transcriptase homolog 159 3e-38
YTX2_XENLA (P14381) Transposon TX1 hypothetical 149 kDa protein ... 157 1e-37
LIN1_NYCCO (P08548) LINE-1 reverse transcriptase homolog 145 8e-34
POLR_DROME (P16423) Retrovirus-related Pol polyprotein from type... 104 1e-21
PO22_POPJA (Q03274) Retrovirus-related Pol polyprotein from type... 82 7e-15
PO21_NASVI (Q03278) Retrovirus-related Pol polyprotein from type... 76 6e-13
PO21_SCICO (Q03279) Retrovirus-related Pol polyprotein from type... 73 5e-12
MC50_ARATH (P92555) Hypothetical mitochondrial protein AtMg01250... 70 5e-11
PO23_POPJA (Q05118) Retrovirus-related Pol polyprotein from type... 67 4e-10
YMH5_CAEEL (P34472) Hypothetical protein F58A4.5 in chromosome III 65 1e-09
M310_ARATH (P93295) Hypothetical mitochondrial protein AtMg00310... 62 9e-09
PO11_SCICO (Q03277) Retrovirus-related Pol polyprotein from type... 60 5e-08
RTJK_DROFU (P21329) RNA-directed DNA polymerase from mobile elem... 54 2e-06
AI2M_YEAST (P03876) Putative COX1/OXI3 intron 2 protein 47 2e-04
Y2R2_DROME (P16425) Hypothetical 115 kDa protein in type I retro... 44 0.003
AI1M_YEAST (P03875) COX1/OXI3 intron 1 protein 44 0.003
CENE_HUMAN (Q02224) Centromeric protein E (CENP-E protein) 41 0.017
YO84_CAEEL (P34620) Hypothetical protein ZK1236.4 in chromosome III 35 1.6
EPA4_CHICK (Q07496) Ephrin type-A receptor 4 precursor (EC 2.7.1... 34 2.1
>POL2_MOUSE (P11369) Retrovirus-related Pol polyprotein [Contains:
Reverse transcriptase (EC 2.7.7.49); Endonuclease]
Length = 1300
Score = 159 bits (403), Expect = 3e-38
Identities = 134/495 (27%), Positives = 228/495 (45%), Gaps = 28/495 (5%)
Query: 248 IHKLQLPNGISTSDSNILQEEALKYFKKFFCG---SQIPYSRFFNEGRHPALDDTGKTSL 304
I+K++ G T+D +Q ++K+ + + +F + + P L+ L
Sbjct: 416 INKIRNEKGDITTDPEEIQNTIRSFYKRLYSTKLENLDEMDKFLDRYQVPKLNQDQVDHL 475
Query: 305 TSPITKKEVFAALNSMKPYKAPGPDGFHCIFFKQYWHIVGDDVFHLVRSAFLTGHFDPAI 364
SPI+ KE+ A +NS+ K+PGPDGF F++ + + + L + G +
Sbjct: 476 NSPISPKEIEAVINSLPTKKSPGPDGFSAEFYQTFKEDLIPILHKLFHKIEVEGTLPNSF 535
Query: 365 SNTLIALIPKIDS-PNTYKDFRPISLCNTLYKIITKVLVHRLRPFLNNLIGPYQSSFLPG 423
I LIPK P ++FRPISL N KI+ K+L +R++ + +I P Q F+PG
Sbjct: 536 YEATITLIPKPQKDPTKIENFRPISLMNIDAKILNKILANRIQEHIKAIIHPDQVGFIPG 595
Query: 424 RGTADNSIILQEILHFMKRSKRKKGYVAFKLDLEKAFDNVNWDFLNSCLLDFGFPDIIVK 483
N ++H++ + K K ++ LD EKAFD + F+ L G +
Sbjct: 596 MQGWFNIRKSINVIHYINKLK-DKNHMIISLDAEKAFDKIQHPFMIKVLERSGIQGPYLN 654
Query: 484 LIMHCVSSANYSLLWNGNKMPPFKPTHGLRQGDPLSPYLFILCMEKLSVAIQDAVLQGSW 543
+I S ++ NG K+ G RQG PLSPYLF + +E L+ AI+ Q
Sbjct: 655 MIKAIYSKPVANIKVNGEKLEAIPLKSGTRQGCPLSPYLFNIVLEVLARAIRQ---QKEI 711
Query: 544 EPIHIINDGPQISHLLFADDVLLFTKAKSSQLQFITNLFDRFSRASGLKINISKSRA--Y 601
+ I I + +IS L ADD++++ + + + NL + F G KIN +KS A Y
Sbjct: 712 KGIQIGKEEVKIS--LLADDMIVYISDPKNSTRELLNLINSFGEVVGYKINSNKSMAFLY 769
Query: 602 YSSGTPNGKINNLTAISGIQSTTTLGKYLGFPMLQGRPKRSDFNF--ILEKMQTRLASWK 659
+ +I T S + + KYLG + + D NF + ++++ L WK
Sbjct: 770 TKNKQAEKEIRETTPFSIVTNNI---KYLGVTLTKEVKDLYDKNFKSLKKEIKEDLRRWK 826
Query: 660 NRLLNRTGRLTLATSVLSTIPTYYMQINWLPQNI----CDSIDQTARNFLWKGSNNKGIH 715
+ + GR+ + ++ +P + N +P I + ++ F+W NNK
Sbjct: 827 DLPCSWIGRINIVK--MAILPKAIYRFNAIPIKIPTQFFNELEGAICKFVW---NNKKPR 881
Query: 716 LVNWKTITRPKSIGG 730
+ K++ + K G
Sbjct: 882 IA--KSLLKDKRTSG 894
>LIN1_HUMAN (P08547) LINE-1 reverse transcriptase homolog
Length = 1259
Score = 159 bits (403), Expect = 3e-38
Identities = 136/526 (25%), Positives = 242/526 (45%), Gaps = 29/526 (5%)
Query: 242 RRKWNKIHKLQLPNGISTSDSNILQEEALKYFKKFFCG---SQIPYSRFFNEGRHPALDD 298
+R+ N+I ++ G T+D +Q +Y+K + + +F + P L+
Sbjct: 383 KREKNQIDTIKNDRGDITTDPTEIQTTIREYYKHLYANKLENLEEMDKFLDTYTLPRLNQ 442
Query: 299 TGKTSLTSPITKKEVFAALNSMKPYKAPGPDGFHCIFFKQYWHIVGDDVFHLVRSAFLTG 358
SL PIT E+ A +NS+ K+PGP+GF F+++Y + + L +S G
Sbjct: 443 EEVESLNRPITSSEIEAIINSLPNKKSPGPEGFTAEFYQRYKEELVPFLLKLFQSIEKEG 502
Query: 359 HFDPAISNTLIALIPKIDSPNTYKD-FRPISLCNTLYKIITKVLVHRLRPFLNNLIGPYQ 417
+ I LIPK T K+ FRPISL N KI+ K+L ++++ + LI Q
Sbjct: 503 ILPNSFYEASIILIPKPGRDTTKKENFRPISLMNIDAKILNKILANQIQQHIKKLIHHDQ 562
Query: 418 SSFLPGRGTADNSIILQEILHFMKRSKRKKGYVAFKLDLEKAFDNVNWDFLNSCLLDFGF 477
F+P N I+ + R+K ++ +D EKAFD + F+ L G
Sbjct: 563 VGFIPAMQGWFNIRKSINIIQHINRTK-DTNHMIISIDAEKAFDKIQQPFMLKPLNKLGI 621
Query: 478 PDIIVKLIMHCVSSANYSLLWNGNKM--PPFKPTHGLRQGDPLSPYLFILCMEKLSVAI- 534
+K+I +++ NG K+ PP K G RQG PLSP L + +E L+ AI
Sbjct: 622 DGTYLKIIRAIYDKPTANIILNGQKLEAPPLKT--GTRQGCPLSPLLPNIVLEVLARAIR 679
Query: 535 QDAVLQGSWEPIHIINDGPQISHLLFADDVLLFTKAKSSQLQFITNLFDRFSRASGLKIN 594
Q+ ++G I + + ++S LFADD++++ + Q + L FS+ SG KIN
Sbjct: 680 QEKEIKG----IQLGKEEVKLS--LFADDMIVYLENPIVSAQNLLKLISNFSKVSGYKIN 733
Query: 595 ISKSRAYYSSGTPNGKINNLTAISGIQSTTTLGKYLGFPMLQGRPK--RSDFNFILEKMQ 652
+ KS+A+ + + ++ + ++ + KYLG + + + ++ +L +++
Sbjct: 734 VQKSQAFLYTNNRQTESQIMSELPFTIASKRI-KYLGIQLTRDVKDLFKENYKPLLNEIK 792
Query: 653 TRLASWKNRLLNRTGRLTLATSVLSTIPTYYMQIN----WLPQNICDSIDQTARNFLWKG 708
WKN + GR+ + ++ +P + N LP +++T F+W
Sbjct: 793 EDTNKWKNIPCSWVGRINIVK--MAILPKVIYRFNAIPIKLPMTFFTELEKTTLKFIW-- 848
Query: 709 SNNKGIHLVNWKTITRPKSIGGLGIRSARDANICLLGKLVWDMVQS 754
N K H+ T+++ GG+ + + + K W Q+
Sbjct: 849 -NQKRAHIAK-STLSQKNKAGGITLPDFKLYYKATVTKTAWYWYQN 892
>YTX2_XENLA (P14381) Transposon TX1 hypothetical 149 kDa protein
(ORF 2)
Length = 1308
Score = 157 bits (398), Expect = 1e-37
Identities = 143/526 (27%), Positives = 241/526 (45%), Gaps = 29/526 (5%)
Query: 217 YQKSREQWVKLGDKNTAFFHAQTVIRRKWNKIHKLQLPNGISTSDSNILQEEALKYFKKF 276
+ +SR Q + D+ + FF+A + +I L +G D +++ A +++
Sbjct: 357 FVRSRMQLLCDMDRGSRFFYALEKKKGNRKQITCLFAEDGTPLEDPEAIRDRARSFYQNL 416
Query: 277 FCGSQI-PYSRFFNEGRHPALDDTGKTSLTSPITKKEVFAALNSMKPYKAPGPDGFHCIF 335
F I P + P + + K L +PIT E+ AL M K+PG DG F
Sbjct: 417 FSPDPISPDACEELWDGLPVVSERRKERLETPITLDELSQALRLMPHNKSPGLDGLTIEF 476
Query: 336 FKQYWHIVGDDVFHLVRSAFLTGHFDPAISNTLIALIPKIDSPNTYKDFRPISLCNTLYK 395
F+ +W +G D ++ AF G + +++L+PK K++RP+SL +T YK
Sbjct: 477 FQFFWDTLGPDFHRVLTEAFKKGELPLSCRRAVLSLLPKKGDLRLIKNWRPVSLLSTDYK 536
Query: 396 IITKVLVHRLRPFLNNLIGPYQSSFLPGRGTADNSIILQEILHFMKRSKRKKGYVAFKLD 455
I+ K + RL+ L +I P QS +PGR DN +++++LHF +R+ +++ LD
Sbjct: 537 IVAKAISLRLKSVLAEVIHPDQSYTVPGRTIFDNVFLIRDLLHFARRTGLSLAFLS--LD 594
Query: 456 LEKAFDNVNWDFLNSCLLDFGFPDIIVKLIMHCVSSANYSLLWNGNKMPPFKPTHGLRQG 515
EKAFD V+ +L L + F V + +SA + N + P G+RQG
Sbjct: 595 QEKAFDRVDHQYLIGTLQAYSFGPQFVGYLKTMYASAECLVKINWSLTAPLAFGRGVRQG 654
Query: 516 DPLSPYLFILCMEKLSVAIQDAVLQGSWEPIHIINDGPQISHLL--FADDVLLFTKAKSS 573
PLS L+ L +E ++ + ++ P + +L +ADDV+L +
Sbjct: 655 CPLSGQLYSLAIEPFLCLLRKRL-------TGLVLKEPDMRVVLSAYADDVILVAQ-DLV 706
Query: 574 QLQFITNLFDRFSRASGLKINISKSRAYYSSGTPNG--KINNL-TAISGIQSTTTLGKYL 630
L+ + ++ AS +IN SK SSG G K++ L A I + + KYL
Sbjct: 707 DLERAQECQEVYAAASSARINWSK-----SSGLLEGSLKVDFLPPAFRDISWESKIIKYL 761
Query: 631 G-FPMLQGRPKRSDFNFILEKMQTRLASWKN--RLLNRTGRLTLATSVLSTIPTYYMQIN 687
G + + P +F + E + TRL WK ++L+ GR + ++++ Y +
Sbjct: 762 GVYLSAEEYPVSQNFIELEECVLTRLGKWKGFAKVLSMRGRALVINQLVASQIWYRLICL 821
Query: 688 WLPQNICDSIDQTARNFLWKGSNNKGIHLVNWKTITRPKSIGGLGI 733
Q I + +FLW G H V+ + P GG G+
Sbjct: 822 SPTQEFIAKIQRRLLDFLWIGK-----HWVSAGVSSLPLKEGGQGV 862
>LIN1_NYCCO (P08548) LINE-1 reverse transcriptase homolog
Length = 1260
Score = 145 bits (365), Expect = 8e-34
Identities = 125/478 (26%), Positives = 220/478 (45%), Gaps = 25/478 (5%)
Query: 242 RRKWNKIHKLQLPNGISTSDSNILQEEALKYFKKFFCGSQIPYSRF--FNEGRH-PALDD 298
+R + I ++ N T+D + +Q+ +Y+KK + + E H P L
Sbjct: 383 KRVKSLISSIRNGNDEITTDPSEIQKILNEYYKKLYSHKYENLKEIDQYLEACHLPRLSQ 442
Query: 299 TGKTSLTSPITKKEVFAALNSMKPYKAPGPDGFHCIFFKQYWHIVGDDVFHLVRSAFLTG 358
L PI+ E+ + + ++ K+PGPDGF F++ + + + +L ++ G
Sbjct: 443 KEVEMLNRPISSSEIASTIQNLPKKKSPGPDGFTSEFYQTFKEELVPILLNLFQNIEKEG 502
Query: 359 HFDPAISNTLIALIPKIDSPNTYKD-FRPISLCNTLYKIITKVLVHRLRPFLNNLIGPYQ 417
I LIPK T K+ +RPISL N KI+ K+L +R++ + +I Q
Sbjct: 503 ILPNTFYEANITLIPKPGKDPTRKENYRPISLMNIDAKILNKILTNRIQQHIKKIIHHDQ 562
Query: 418 SSFLPG-RGTADNSIILQEILHFMKRSKRKKGYVAFKLDLEKAFDNVNWDFLNSCLLDFG 476
F+PG +G + + I H K + K ++ +D EKAFDN+ F+ L G
Sbjct: 563 VGFIPGSQGWFNIRKSINVIQHINKL--KNKDHMILSIDAEKAFDNIQHPFMIRTLKKIG 620
Query: 477 FPDIIVKLIMHCVSSANYSLLWNGNKMPPFKPTHGLRQGDPLSPYLFILCMEKLSVAIQD 536
+KLI S +++ NG K+ F G RQG PLSP LF + ME L++AI++
Sbjct: 621 IEGTFLKLIEAIYSKPTANIILNGVKLKSFPLRSGTRQGCPLSPLLFNIVMEVLAIAIRE 680
Query: 537 AVLQGSWEPIHIINDGPQISHLLFADDVLLFTKAKSSQLQFITNLFDRFSRASGLKINIS 596
+ + + IHI ++ ++S LFADD++++ + + + +S SG KIN
Sbjct: 681 ---EKAIKGIHIGSEEIKLS--LFADDMIVYLENTRDSTTKLLEVIKEYSNVSGYKINTH 735
Query: 597 KSRAYYSSGTPNGKINNLTAISGIQSTTT--LGKYLGFPMLQGRPK--RSDFNFILEKMQ 652
KS A+ + N T I T KYLG + + + ++ + +++
Sbjct: 736 KSVAFIYT---NNNQAEKTVKDSIPFTVVPKKMKYLGVYLTKDVKDLYKENYETLRKEIA 792
Query: 653 TRLASWKNRLLNRTGRLTLATSVLSTIPTYYMQINWL----PQNICDSIDQTARNFLW 706
+ WKN + GR+ + +S +P N + P + +++ +F+W
Sbjct: 793 EDVNKWKNIPCSWLGRINIVK--MSILPKAIYNFNAIPIKAPLSYFKDLEKIILHFIW 848
>POLR_DROME (P16423) Retrovirus-related Pol polyprotein from type II
retrotransposable element R2DM [Contains: Protease (EC
3.4.23.-); Reverse transcriptase (EC 2.7.7.49);
Endonuclease]
Length = 1057
Score = 104 bits (260), Expect = 1e-21
Identities = 103/386 (26%), Positives = 168/386 (42%), Gaps = 35/386 (9%)
Query: 240 VIRRKWNKIHKLQLPNGISTSDSNILQEEALKYFKKFFCGSQIPYSRFFNEGRHPALD-- 297
V++R W+K + + ++ +D +++ + + +PY R P+
Sbjct: 279 VVQRNWDKHKGRCIKSLLNGTDESVMPSQEI----------MVPYWREVMTQPSPSSCSG 328
Query: 298 -----DTGKTSLTSPITKKEVFAALNSMKPYKAPGPDGFHCIFFKQYWHIVGDDVFHLVR 352
D + S IT++++ A+ S+ +PGPDG I K + + ++
Sbjct: 329 EVIQMDHSLERVWSAITEQDLRASRVSLS--SSPGPDG---ITPKSAREVPSGIMLRIMN 383
Query: 353 SAFLTGHFDPAISNTLIALIPKIDSPNTYKDFRPISLCNTLYKIITKVLVHRLRPFLNNL 412
G+ +I IPK + +DFRPIS+ + L + + +L RL +N
Sbjct: 384 LILWCGNLPHSIRLARTVFIPKTVTAKRPQDFRPISVPSVLVRQLNAILATRLNSSIN-- 441
Query: 413 IGPYQSSFLPGRGTADNSIILQEILHFMKRSKRKKGYVAFKLDLEKAFDNVNWDFLNSCL 472
P Q FLP G ADN+ I+ +L + R Y+A LD+ KAFD+++ + L
Sbjct: 442 WDPRQRGFLPTDGCADNATIVDLVLRHSHKHFRSC-YIA-NLDVSKAFDSLSHASIYDTL 499
Query: 473 LDFGFPDIIVKLIMHCVSSANYSLLWNGNKMPPFKPTHGLRQGDPLSPYLFILCMEKLSV 532
+G P V + + SL +G F P G++QGDPLSP LF L M++L
Sbjct: 500 RAYGAPKGFVDYVQNTYEGGGTSLNGDGWSSEEFVPARGVKQGDPLSPILFNLVMDRLLR 559
Query: 533 AIQDAVLQGSWEPIHIINDGPQISHLLFADDVLLFTKAKSSQLQFITNLFDRFSRASGLK 592
+ + G+ I N FADD++LF + + LQ + + F GLK
Sbjct: 560 TLPSEI--GAKVGNAITNAA------AFADDLVLFAETRMG-LQVLLDKTLDFLSIVGLK 610
Query: 593 INISKSRAYYSSGTPNGKINNLTAIS 618
+N K G P K L A S
Sbjct: 611 LNADKCFTVGIKGQPKQKCTVLEAQS 636
>PO22_POPJA (Q03274) Retrovirus-related Pol polyprotein from type I
retrotransposable element R2 [Contains: Reverse
transcriptase (EC 2.7.7.49); Endonuclease] (Fragment)
Length = 711
Score = 82.4 bits (202), Expect = 7e-15
Identities = 79/295 (26%), Positives = 132/295 (43%), Gaps = 22/295 (7%)
Query: 307 PITKKEVFAALNSMKPYKAPGPDGFHCIFFKQYWHIVGDDVFHLVRSAFLTGHFDPAISN 366
PI ++E+ A+ KP APG DG + HL+R GH +
Sbjct: 5 PIAREEIQCAIKGWKP-SAPGSDGLTVQAITRTRLPRNFVQLHLLR-----GHVPTPWTA 58
Query: 367 TLIALIPKIDSPNTYKDFRPISLCNTLYKIITKVLVHRLRPFLNNLIGPYQSSFLPGRGT 426
LIPK ++RPI++ + L +++ ++L RL + + P Q + GT
Sbjct: 59 MRTTLIPKDGDLENPSNWRPITIASALQRLLHRILAKRLEAAVE--LHPAQKGYARIDGT 116
Query: 427 ADNSIILQEILHFMKRSKRKKGYVAFKLDLEKAFDNVNWDFLNSCLLDFGFPDIIVKLIM 486
NS++L + R +++K Y LD+ KAFD V+ + L G + I
Sbjct: 117 LVNSLLLDT--YISSRREQRKTYNVVSLDVRKAFDTVSHSSICRALQRLGIDEGTSNYIT 174
Query: 487 HCVSSANYSL-LWNGNKMPPFKPTHGLRQGDPLSPYLFILCMEKLSVAIQDAVLQGSWEP 545
+S + ++ + G++ G++QGDPLSP+LF +++L ++Q G
Sbjct: 175 GSLSDSTTTIRVGPGSQTRKICIRRGVKQGDPLSPFLFNAVLDELLCSLQSTPGIGG--- 231
Query: 546 IHIINDGPQISHLLFADDVLLFTKAKSSQLQFITNL--FDRFSRASGLKINISKS 598
I + +I L FADD+LL + + + T L F R G+ +N KS
Sbjct: 232 --TIGE-EKIPVLAFADDLLLL---EDNDVLLPTTLATVANFFRLRGMSLNAKKS 280
>PO21_NASVI (Q03278) Retrovirus-related Pol polyprotein from type I
retrotransposable element R2 [Contains: Reverse
transcriptase (EC 2.7.7.49); Endonuclease] (Fragment)
Length = 1025
Score = 75.9 bits (185), Expect = 6e-13
Identities = 83/318 (26%), Positives = 136/318 (42%), Gaps = 30/318 (9%)
Query: 325 APGPDGFHCIFFKQYWHIVGDDVFHLVRSAFLTGHFDPAISNTLIALIPKIDSPNTYKDF 384
A GPDG W+ + + + L G ++ LIPK F
Sbjct: 334 AAGPDGMTTTA----WNSIDECIKSLFNMIMYHGQCPRRYLDSRTVLIPKEPGTMDPACF 389
Query: 385 RPISLCNTLYKIITKVLVHRLRPFLNNLIGPYQSSFLPGRGTADNSIILQEILHFMKRSK 444
RP+S+ + + ++L +R+ + L+ Q +F+ G A+N+ +L ++ R K
Sbjct: 390 RPLSIASVALRHFHRILANRIGE--HGLLDTRQRAFIVADGVAENTSLLSAMIK-EARMK 446
Query: 445 RKKGYVAFKLDLEKAFDNVNWDFLNSCLLDFGFPDIIVKLIMHCVSSANYSLLWNGNKMP 504
K Y+A LD++KAFD+V + L P + IM ++ L K
Sbjct: 447 IKGLYIAI-LDVKKAFDSVEHRSILDALRRKKLPLEMRNYIMWVYRNSKTRLEVVKTKGR 505
Query: 505 PFKPTHGLRQGDPLSPYLFILCMEKLSVAIQDAVLQGSWEPIHIINDGPQISHLLFADDV 564
+P G+RQGDPLSP LF + DAVL+ E + +I L+FADD+
Sbjct: 506 WIRPARGVRQGDPLSPLLF--------NCVMDAVLRRLPENTGFLMGAEKIGALVFADDL 557
Query: 565 LLFTKAKSSQLQFITNLFDRFSRASGLKINISKSRAYYSSGTPNGKINNLTAIS------ 618
+L + + LQ + + + GL+ + + + + P+GK + +
Sbjct: 558 VLLAETREG-LQASLSRIEAGLQEQGLE--MMPRKCHTLALVPSGKEKKIKVETHKPFTV 614
Query: 619 GIQSTTTLG-----KYLG 631
G Q T LG KYLG
Sbjct: 615 GNQEITQLGHADQWKYLG 632
>PO21_SCICO (Q03279) Retrovirus-related Pol polyprotein from type I
retrotransposable element R2 [Contains: Reverse
transcriptase (EC 2.7.7.49); Endonuclease] (Fragment)
Length = 869
Score = 72.8 bits (177), Expect = 5e-12
Identities = 84/317 (26%), Positives = 140/317 (43%), Gaps = 23/317 (7%)
Query: 303 SLTSPITKKEVFAALNSMKPYKAPGPDGFHCIFFKQYWHIVGDDVFHLVRSAFLT-GHFD 361
+L P++ E+ +A S + K GPDG + W+ + D L+ + F+ G
Sbjct: 154 TLWDPVSLIEIKSARASNE--KGAGPDGVT----PRSWNALDDRYKRLLYNIFVFYGRVP 207
Query: 362 PAISNTLIALIPKIDSPNTYKDFRPISLCNTLYKIITKVLVHRLRPFLNNLIGPYQSSFL 421
I + PKI+ FRP+S+C+ + + K+L R Q+++L
Sbjct: 208 SPIKGSRTVFTPKIEGGPDPGVFRPLSICSVILREFNKILARRFVSCYT--YDERQTAYL 265
Query: 422 PGRGTADNSIILQEILHFMKRSKRKKGYVAFKLDLEKAFDNVNWDFLNSCLLDFGFPDII 481
P G N +L I+ KR RK+ ++A LDL KAF++V L + + G P +
Sbjct: 266 PIDGVCINVSMLTAIIAEAKR-LRKELHIAI-LDLVKAFNSVYHSALIDAITEAGCPPGV 323
Query: 482 VKLIMHCVSSANYSLLWNGNKMPPFKPTHGLRQGDPLSPYLFILCMEKLSVAIQDAVLQG 541
V I ++ + + G K G+ QGDPLS LF L EK A+ + +G
Sbjct: 324 VDYIADMYNNVITEMQFEG-KCELASILAGVYQGDPLSGPLFTLAYEKALRALNN---EG 379
Query: 542 SWEPIHIINDGPQISHLLFADDVLLFTKAKSSQLQFITNLFDRFSRASGLKINISKSRAY 601
++ + +++ ++DD LL LQ + F GL+IN KS+
Sbjct: 380 RFDIADV-----RVNASAYSDDGLLLAMTVIG-LQHNLDKFGETLAKIGLRINSRKSKTV 433
Query: 602 YSSGTPNGKINNLTAIS 618
S P+G+ + +S
Sbjct: 434 --SLVPSGREKKMKIVS 448
>MC50_ARATH (P92555) Hypothetical mitochondrial protein AtMg01250
(ORF102)
Length = 122
Score = 69.7 bits (169), Expect = 5e-11
Identities = 36/65 (55%), Positives = 42/65 (64%)
Query: 499 NGNKMPPFKPTHGLRQGDPLSPYLFILCMEKLSVAIQDAVLQGSWEPIHIINDGPQISHL 558
NG P+ GLRQGDPLSPYLFILC E LS + A QG I + N+ P+I+HL
Sbjct: 15 NGAPQGLVTPSRGLRQGDPLSPYLFILCTEVLSGLCRRAQEQGRLPGIRVSNNSPRINHL 74
Query: 559 LFADD 563
LFADD
Sbjct: 75 LFADD 79
>PO23_POPJA (Q05118) Retrovirus-related Pol polyprotein from type I
retrotransposable element R2 [Contains: Reverse
transcriptase (EC 2.7.7.49); Endonuclease] (Fragment)
Length = 606
Score = 66.6 bits (161), Expect = 4e-10
Identities = 55/187 (29%), Positives = 87/187 (46%), Gaps = 12/187 (6%)
Query: 425 GTADNSIILQEILHFMKRSKRK-KGYVAFKLDLEKAFDNVNWDFLNSCLLDFGFPDIIVK 483
GT N I+L+ H++K + K K Y LD+ KAFD V+ + + FG D +
Sbjct: 1 GTLANIIMLE---HYIKLRRLKGKTYNVVSLDIRKAFDTVSHPAILRAMRAFGIDDGMQD 57
Query: 484 LIMHCVSSANYSLLWNGNKMPPFKPTHGLRQGDPLSPYLFILCMEKLSVAIQDAVLQGSW 543
IM ++ A +++ G +G++QGDPLSP LF + +++L + D S
Sbjct: 58 FIMSTITDAYTNIVVGGRTTNKIYIRNGVKQGDPLSPVLFNIVLDELVTRLNDEQPGASM 117
Query: 544 EPIHIINDGPQISHLLFADDVLLFTKAKSSQLQFITNLFDRFSRASGLKINISKSRAYYS 603
P +I+ L FADD+LL + F R G+ +N K A S
Sbjct: 118 TP------ACKIASLAFADDLLLLEDRDIDVPNSLATTCAYF-RTRGMTLNPEKC-ASIS 169
Query: 604 SGTPNGK 610
+ T +G+
Sbjct: 170 AATVSGR 176
>YMH5_CAEEL (P34472) Hypothetical protein F58A4.5 in chromosome III
Length = 1222
Score = 65.1 bits (157), Expect = 1e-09
Identities = 61/243 (25%), Positives = 107/243 (43%), Gaps = 25/243 (10%)
Query: 361 DPAISN----TLIALIPKIDSPNTYKDFRPISLCNTLYKIITKVLVHRLRPFLNNLIGPY 416
D AI N +I IPK +P++ ++RPISL + +I+ +++ R+R ++L+ P+
Sbjct: 658 DSAIPNRWKHAVIIPIPKKGNPSSPSNYRPISLTDPFARIMERIICSRIRSEYSHLLSPH 717
Query: 417 QSSFLPGRGTADNSIILQEILHFMKRSKRKKGYVAFKLDLEKAFDNVNWDFLNSCLLDFG 476
Q FL R + + + H + ++++ + F D KAFD V+ L L FG
Sbjct: 718 QHGFLNFRSCPSSLVRSISLYHSILKNEKSLDILFF--DFAKAFDKVSHPILLKKLALFG 775
Query: 477 FPDIIVKLIMHCVSSANYSLLWNGNKMPPFKP-THGLRQGDPLSPYLFILCMEKLSVAIQ 535
+ + +S+ N P + G+ QG P LFIL + L + ++
Sbjct: 776 LDKLTCSWFKEFLHLRTFSVKINKFVSSNAYPISSGVPQGSVSGPLLFILFINDLLIDLE 835
Query: 536 DAVLQGSWEPIHIINDGPQISHLLFADDVLLFTKAKSSQLQFITNLFDRFSRASGLKINI 595
P I FADD+ +F S LQ + ++S+ + L +
Sbjct: 836 -----------------PNIHVSCFADDIKIF-HHNPSTLQNSIDTIVKWSKKNKLPLAP 877
Query: 596 SKS 598
+KS
Sbjct: 878 AKS 880
>M310_ARATH (P93295) Hypothetical mitochondrial protein AtMg00310
(ORF154)
Length = 154
Score = 62.0 bits (149), Expect = 9e-09
Identities = 40/141 (28%), Positives = 62/141 (43%), Gaps = 3/141 (2%)
Query: 679 IPTYYMQINWLPQNICDSIDQTARNFLWKGSNNKG-IHLVNWKTITRPKSI-GGLGIRSA 736
+P Y M L + +C + F W NK I V W+ + + K GGLG R
Sbjct: 3 LPVYAMSCFRLSKLLCKKLTSAMTEFWWSSCENKRKISWVAWQKLCKSKEDDGGLGFRDL 62
Query: 737 RDANICLLGKLVWDMVQSTNKLWVNLLAKKYSSGTSLLEANVNSNSSPSWFSIIRAKDIL 796
N LL K + ++ + L LL +Y +S++E +V + S +W SII +++L
Sbjct: 63 GWFNQALLAKQSFRIIHQPHTLLSRLLRSRYFPHSSMMECSVGTRPSYAWRSIIHGRELL 122
Query: 797 KTGYSWRAGAGT-SSFWFSNW 816
G G G + W W
Sbjct: 123 SRGLLRTIGDGIHTKVWLDRW 143
>PO11_SCICO (Q03277) Retrovirus-related Pol polyprotein from type I
retrotransposable element R1 [Contains: Reverse
transcriptase (EC 2.7.7.49); Endonuclease] (Fragment)
Length = 1004
Score = 59.7 bits (143), Expect = 5e-08
Identities = 46/160 (28%), Positives = 75/160 (46%), Gaps = 13/160 (8%)
Query: 312 EVFAALNSMKPYKAPGPDGFHCIFFKQYWHIVGDDVFHLVRSAFLTGHFDP----AISNT 367
EV ++ K K+PGPDG + W + + +F L + L +F A
Sbjct: 409 EVSDSVRRCKVRKSPGPDGIVGEMVRAVWGAIPEYMFCLYKQCLLESYFPQKWKIASLVI 468
Query: 368 LIALIPKIDS-PNTYKDFRPISLCNTLYKIITKVLVHRLRPFLNNL-IGPYQSSFLPGRG 425
L+ L+ +I S P +Y RPI L + L K++ ++V RL L ++ + PYQ +F G+
Sbjct: 469 LLKLLDRIRSDPGSY---RPICLLDNLGKVLEGIMVKRLDQKLMDVEVSPYQFAFTYGKS 525
Query: 426 TADNSIILQEILHFMKRSKRKKGYVAFKLDLEKAFDNVNW 465
T D +Q + K + +D + AFDN+ W
Sbjct: 526 TEDAWRCVQRHV----ECSEMKYVIGLNIDFQGAFDNLGW 561
>RTJK_DROFU (P21329) RNA-directed DNA polymerase from mobile element
jockey (EC 2.7.7.49) (Reverse transcriptase)
Length = 916
Score = 54.3 bits (129), Expect = 2e-06
Identities = 135/649 (20%), Positives = 244/649 (36%), Gaps = 118/649 (18%)
Query: 90 VEVLCRLHSDHNPLLLRFGGLPLTRGPRPF---------RFEAAWIDHYDYGNVVKRSWS 140
V+ L L SDH PLL +P+ + PR RF+A H D ++ +
Sbjct: 204 VQTLHELSSDHTPLLADLHAMPINKPPRSCLLARGADIERFKAYLTQHIDLSVGIQGTDD 263
Query: 141 THTHNPTASLIKVMENSIIFNHDVFGNIFQRKSRVEWRLKGVQSYLERVDSYRHTLLEKE 200
++N+I D F +I +R + ++ S+ + V+S R L
Sbjct: 264 -------------IDNAI----DSFMDILKRAA-----IRSAPSHQQNVESSRQLQLPP- 300
Query: 201 LQDEYNHILFQEEMLWYQKSREQWVKLGDKNTAFFHAQTVIR--RKWNKIHKLQLPNGIS 258
+ + +K R ++ + GD H++ R + N+ + Q+ N +
Sbjct: 301 --------IVASLIRLKRKVRREYARTGDARIQQIHSRLANRLHKVLNRRKQSQIDNLLE 352
Query: 259 TSDSNILQEEALKYFKKFFCGSQIPYSRFFNEGRHPALDDTGKTSL-------------- 304
D++ +L K + P S N KT +
Sbjct: 353 NLDTDGSTNFSLWRITKRYKTQATPNSAIRNPAGGWCRTSREKTEVFANHLEQRFKALAF 412
Query: 305 ----------------------TSPITKKEVFAALNSMKPYKAPGPDGFHCIFFKQYWHI 342
P+T +EV ++ +KP KAPG D + + +
Sbjct: 413 APESHSLMVAESLQTPFQMALPADPVTLEEVKELVSKLKPKKAPGED----LLDNRTIRL 468
Query: 343 VGDD-VFHLV---RSAFLTGHFDPAISNTLIALIPKI-DSPNTYKDFRPISLCNTLYKII 397
+ D + +LV S G+F A I +I K P +RP SL +L K++
Sbjct: 469 LPDQALLYLVLIFNSILRVGYFPKARPTASIIMILKPGKQPLDVDSYRPTSLLPSLGKML 528
Query: 398 TKVLVHRL--RPFLNNLIGPYQSSFLPGRGTADNSIILQEILHFMKRSKRKKGYV-AFKL 454
+++++R+ + I +Q F GT + L +++F + KK Y + L
Sbjct: 529 ERLILNRILTSEEVTRAIPKFQFGFRLQHGTPEQ---LHRVVNFALEALEKKEYAGSCFL 585
Query: 455 DLEKAFDNVNWDFLNSCLLDFGFPDIIVKLIMHCVSSANYSLLWNGNKMPPFKPTHGLRQ 514
D+++AFD V L P + +LI +S+ +G + G+ Q
Sbjct: 586 DIQQAFDRVWHPGLLYKAKSLLSPQLF-QLIKSFWEGRKFSVTADGCRSSVKFIEAGVPQ 644
Query: 515 GDPLSPYLFILCMEKLSVAIQDAVLQGSWEPIHIINDGPQISHLLFADDVLLFTKA---- 570
G L P L+ S+ D Q + + + +G ++ +ADD+ + TK+
Sbjct: 645 GSVLGPTLY-------SIFTADMPNQNA---VTGLAEG-EVLIATYADDIAVLTKSTCIV 693
Query: 571 -KSSQLQFITNLFDRFSRASGLKINISKSRAYYSSGTPNGKINN---LTAISGIQSTTTL 626
+ LQ + F ++ +K N+S + ++ T I + +T + S T
Sbjct: 694 EATDALQEYLDAFQEWA----VKWNVSINAGKCANVTFTNAIRDCPGVTINGSLLSHTHE 749
Query: 627 GKYLGFPMLQGRPKRSDFNFILEKMQTRLASWKNRLLNRTGRLTLATSV 675
KYLG + + R + +TR+ N LL +L+L V
Sbjct: 750 YKYLGVILDRSLTFRRHITSLQHSFRTRITK-MNWLLAARNKLSLDNKV 797
>AI2M_YEAST (P03876) Putative COX1/OXI3 intron 2 protein
Length = 789
Score = 47.4 bits (111), Expect = 2e-04
Identities = 44/171 (25%), Positives = 74/171 (42%), Gaps = 25/171 (14%)
Query: 372 IPKIDSPNTYKDFRPISLCNTLYKIITKVLVHRLRPFLNNLIGPYQSSFLPGRGTADNSI 431
+ +++ P T FRP+S+ N KI+ + + L NN Y F P N
Sbjct: 278 VRRVEIPKTSGGFRPLSVGNPREKIVQESMRMMLEIIYNNSFSYYSHGFRP------NLS 331
Query: 432 ILQEILHFMKRSKRKKGYVAFKLDLEKAFD----NVNWDFLNSCLLDFGFPDIIVKLIM- 486
L I+ + ++ K+DL K FD N+ + LN + D GF D++ KL+
Sbjct: 332 CLTAIIQCKNYMQYCNWFI--KVDLNKCFDTIPHNMLINVLNERIKDKGFMDLLYKLLRA 389
Query: 487 -HCVSSANYSLLWNGNKMPPFKPTHGLRQGDPLSPYLFILCMEKLSVAIQD 536
+ + NY T G+ QG +SP L + ++KL +++
Sbjct: 390 GYVDKNNNY-----------HNTTLGIPQGSVVSPILCNIFLDKLDKYLEN 429
>Y2R2_DROME (P16425) Hypothetical 115 kDa protein in type I
retrotransposable element R1DM (ORF 2)
Length = 1021
Score = 43.5 bits (101), Expect = 0.003
Identities = 58/266 (21%), Positives = 101/266 (37%), Gaps = 33/266 (12%)
Query: 312 EVFAALNSMKPYKAPGPDGFHCIFFKQYWHIVGDDVFHLVRSAFLTGHFD-----PAISN 366
EV + +K ++PG DG + K W + + + L G+F P + +
Sbjct: 438 EVDTCVARLKSRRSPGLDGINGTICKAVWRAIPEHLASLFSRCIRLGYFPAEWKCPRVVS 497
Query: 367 TLIALIPKIDSPNTYKDFRPISLCNTLYKIITKVLVHRLRPFLNNLIGPYQSSFLPGRGT 426
L P++Y R I L K++ ++V+R+R L +Q F GR
Sbjct: 498 LLKGPDKDKCEPSSY---RGICLLPVFGKVLEAIMVNRVREVLPEGC-RWQFGFRQGRCV 553
Query: 427 ADNSIILQEILHFMKRSKRKKGYVAFKLDLEKAFDNVNWDFLNSCLLDFGFPDIIVKLIM 486
D + + + S + F +D + AFDNV W S L D G ++ +
Sbjct: 554 ED---AWRHVKSSVGASAAQYVLGTF-VDFKGAFDNVEWSAALSRLADLGCREM---GLW 606
Query: 487 HCVSSANYSLLWNGNKMPPFKPTHGLRQGDPLSPYLFILCMEKLSVAIQDAVLQGSWEPI 546
S +++ + + T G QG P+++ + M+ L +Q
Sbjct: 607 QSFFSGRRAVIRSSSGTVEVPVTRGCPQGSISGPFIWDILMDVLLQRLQ----------- 655
Query: 547 HIINDGPQISHLLFADDVLLFTKAKS 572
P +ADD+LL + S
Sbjct: 656 ------PYCQLSAYADDLLLLVEGNS 675
>AI1M_YEAST (P03875) COX1/OXI3 intron 1 protein
Length = 834
Score = 43.5 bits (101), Expect = 0.003
Identities = 73/343 (21%), Positives = 132/343 (38%), Gaps = 59/343 (17%)
Query: 375 IDSPNTYKDFRPISLCNTLYKIITKVLVHRLRPFLNNLIGPYQSSFLPGRGTADNSIILQ 434
++ P RP+S+ N KI+ +V+ L + + + F R +
Sbjct: 313 VNIPKPKGGMRPLSVGNPRDKIVQEVMRMILDTIFDKKMSTHSHGF---RKNMSCQTAIW 369
Query: 435 EILHFMKRSKRKKGYVAFKLDLEKAFDNVNWDF----LNSCLLDFGFPDIIVKLIMHCVS 490
E+ + S ++DL+K FD ++ D L + D GF D++ KL+
Sbjct: 370 EVRNMFGGSNW-----FIEVDLKKCFDTISHDLIIKELKRYISDKGFIDLVYKLLR---- 420
Query: 491 SANYSLLWNGNKMPPFKPTHGLRQGDPLSPYLFILCM----------------------- 527
A Y K KP GL QG +SP L + M
Sbjct: 421 -AGYI----DEKGTYHKPMLGLPQGSLISPILCNIVMTLVDNWLEDYINLYNKGKVKKQH 475
Query: 528 ---EKLSVAIQDAVL---------QGSWEPIHIINDG--PQISHLLFADDVLLFTKAKSS 573
+KLS I A + + + P I ND ++ ++ +ADD+L+ +
Sbjct: 476 PTYKKLSRMIAKAKMFSTRLKLHKERAKGPTFIYNDPNFKRMKYVRYADDILIGVLGSKN 535
Query: 574 QLQFITNLFDRFSRASGLKINISKSRAYYSSGTPNGKINNLTAISGIQSTTTLGKYLGFP 633
+ I + F + GL +N K+ ++ TP + +I+ ++ T+ K +
Sbjct: 536 DCKMIKRDLNNFLNSLGLTMNEEKTLITCATETPARFLGYNISITPLKRMPTVTKTIRGK 595
Query: 634 MLQGR-PKRSDFNFILEKMQTRLASWKNRLLNRTGRLTLATSV 675
++ R R N + + +LA+ N+ GR+ + T V
Sbjct: 596 TIRSRNTTRPIINAPIRDIINKLATNGYCKHNKNGRMGVPTRV 638
>CENE_HUMAN (Q02224) Centromeric protein E (CENP-E protein)
Length = 2663
Score = 41.2 bits (95), Expect = 0.017
Identities = 32/129 (24%), Positives = 60/129 (45%), Gaps = 8/129 (6%)
Query: 140 STHTHNPTASLIKVMENSIIFNHDVFGNIFQRKSRVEWRLKGVQSYLERVDSYRHTLLEK 199
+T TH + +L++ ++ S+ DVF N S +EW E ++S ++
Sbjct: 447 TTKTHKLSINLLREIDESVCSESDVFSNTLDTLSEIEWNPATKLLNQENIESELNS---- 502
Query: 200 ELQDEYNHILFQEEMLWYQKSREQWVKLGDKNTAFFHAQTVIRRKWNKIHKLQLPNGIST 259
L+ +Y++++ E L +K E +KL +KN + RK K ++QL + IS
Sbjct: 503 -LRADYDNLVLDYEQLRTEK-EEMELKLKEKND--LDEFEALERKTKKDQEMQLIHEISN 558
Query: 260 SDSNILQEE 268
+ + E
Sbjct: 559 LKNLVKHRE 567
>YO84_CAEEL (P34620) Hypothetical protein ZK1236.4 in chromosome III
Length = 364
Score = 34.7 bits (78), Expect = 1.6
Identities = 29/128 (22%), Positives = 50/128 (38%), Gaps = 17/128 (13%)
Query: 454 LDLEKAFDNVNWDFLNSCLLDFGFPDIIVKLIMHCVSSANYSLLWNGNKMPPFKPTHGLR 513
LD KAFD V+ D L L +++ + +++ ++ + P K G+
Sbjct: 23 LDFSKAFDKVSHDILLDKLTSIKINKHLIRWLDVFLTNRSFKVKVGNTLSEPKKTVCGVP 82
Query: 514 QGDPLSPYLFILCMEKLSVAIQDAVLQGSWEPIHIINDGPQISHLLFADDVLLFTKAKSS 573
QG +SP LF + + ++S + V FADD+ L+ +
Sbjct: 83 QGSVISPVLFGIFVNEISANLPVGVYCKQ-----------------FADDIKLYAAIPKN 125
Query: 574 QLQFITNL 581
Q Q L
Sbjct: 126 QSQISLQL 133
>EPA4_CHICK (Q07496) Ephrin type-A receptor 4 precursor (EC
2.7.1.112) (Tyrosine-protein kinase receptor CEK8)
Length = 986
Score = 34.3 bits (77), Expect = 2.1
Identities = 30/123 (24%), Positives = 60/123 (48%), Gaps = 15/123 (12%)
Query: 690 PQNICDSIDQTARNFLWKGSNNK-GIHLVNWKTITRPKSIG--------GLGIRSARDAN 740
PQN+ ++++T+ N W NK G +++ + + G G G+ + N
Sbjct: 332 PQNLISNVNETSVNLEWSAPQNKGGRDDISYNVVCKRCGAGEPSHCRSCGSGVHFSPQQN 391
Query: 741 ICLLGKL-VWDMVQSTN---KLW-VNLLAKKYSSGTSLLEANVNSN-SSPSWFSIIRAKD 794
K+ + D++ TN ++W VN ++K S + V +N ++PS ++I+AK+
Sbjct: 392 GLKTTKVSITDLLAHTNYTFEVWAVNGVSKHNPSQDQAVSVTVTTNQAAPSPIALIQAKE 451
Query: 795 ILK 797
I +
Sbjct: 452 ITR 454
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.331 0.143 0.457
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 140,609,696
Number of Sequences: 164201
Number of extensions: 5960541
Number of successful extensions: 14435
Number of sequences better than 10.0: 25
Number of HSP's better than 10.0 without gapping: 11
Number of HSP's successfully gapped in prelim test: 14
Number of HSP's that attempted gapping in prelim test: 14388
Number of HSP's gapped (non-prelim): 34
length of query: 1214
length of database: 59,974,054
effective HSP length: 122
effective length of query: 1092
effective length of database: 39,941,532
effective search space: 43616152944
effective search space used: 43616152944
T: 11
A: 40
X1: 15 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.9 bits)
S2: 72 (32.3 bits)
Medicago: description of AC149130.9


