

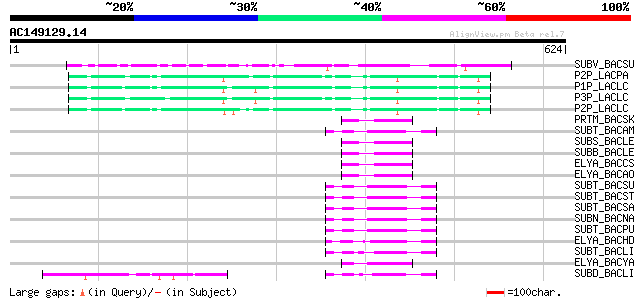
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC149129.14 - phase: 0 /pseudo
(624 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
SUBV_BACSU (P29141) Minor extracellular protease vpr precursor (... 105 5e-22
P2P_LACPA (Q02470) PII-type proteinase precursor (EC 3.4.21.96) ... 64 1e-09
P1P_LACLC (P16271) PI-type proteinase precursor (EC 3.4.21.-) (W... 61 1e-08
P3P_LACLC (P15292) PIII-type proteinase precursor (EC 3.4.21.96)... 60 2e-08
P2P_LACLC (P15293) PII-type proteinase precursor (EC 3.4.21.96) ... 59 3e-08
PRTM_BACSK (Q99405) M-protease (EC 3.4.21.-) 56 2e-07
SUBT_BACAM (P00782) Subtilisin BPN' precursor (EC 3.4.21.62) (Su... 55 4e-07
SUBS_BACLE (P29600) Subtilisin Savinase (EC 3.4.21.62) (Alkaline... 55 5e-07
SUBB_BACLE (P29599) Subtilisin BL (EC 3.4.21.62) (Alkaline prote... 55 5e-07
ELYA_BACCS (P41362) Alkaline protease precursor (EC 3.4.21.-) 55 5e-07
ELYA_BACAO (P27693) Alkaline protease precursor (EC 3.4.21.-) 55 5e-07
SUBT_BACSU (P04189) Subtilisin E precursor (EC 3.4.21.62) 54 1e-06
SUBT_BACST (P29142) Subtilisin J precursor (EC 3.4.21.62) 54 1e-06
SUBT_BACSA (P00783) Subtilisin amylosacchariticus precursor (EC ... 54 1e-06
SUBN_BACNA (P35835) Subtilisin NAT precursor (EC 3.4.21.62) 53 2e-06
SUBT_BACPU (P07518) Subtilisin (EC 3.4.21.62) (Alkaline mesenter... 52 5e-06
ELYA_BACHD (P41363) Thermostable alkaline protease precursor (EC... 51 8e-06
SUBT_BACLI (P00780) Subtilisin Carlsberg precursor (EC 3.4.21.62) 48 9e-05
ELYA_BACYA (P20724) Alkaline elastase YaB precursor (EC 3.4.21.-) 47 1e-04
SUBD_BACLI (P00781) Subtilisin DY (EC 3.4.21.62) 47 1e-04
>SUBV_BACSU (P29141) Minor extracellular protease vpr precursor (EC
3.4.21.-)
Length = 806
Score = 105 bits (261), Expect = 5e-22
Identities = 128/512 (25%), Positives = 217/512 (42%), Gaps = 97/512 (18%)
Query: 65 NGTIKGGSPKSRVVTYKVCWSQTIADGNSAVCYGADVLSAIDQAISDGVDIISVSVGGRS 124
NGTIKG +P + ++ Y+V + G S +V++ +++A+ DG D++++S+G +
Sbjct: 244 NGTIKGVAPDATLLAYRV-----LGPGGSGTT--ENVIAGVERAVQDGADVMNLSLG--N 294
Query: 125 SSNFEEIFTDEISIGAFQAFAKNILLVASAGNGGPTPGSVTNVAPWVFTVAASTIDRDFS 184
S N + T S A ++ ++ V S GN GP +V + P A S +
Sbjct: 295 SLNNPDWAT---STALDWAMSEGVVAVTSNGNSGPNGWTVGS--PGTSREAISV----GA 345
Query: 185 STITIGNKTVTGASLFVNLPPNQSFTLVDSIDAKFANVTNQDARFCKPGTLDPSKVSGKI 244
+ + + VT S + + D + A + N++ + G + GK
Sbjct: 346 TQLPLNEYAVTFGSY--SSAKVMGYNKEDDVKA----LNNKEVELVEAGIGEAKDFEGK- 398
Query: 245 VECVGEKITIKNTSEPVSGRLLGFATNSVSQGREALSAGAKGMILRNQPKFNGKTLLAES 304
+ G+ +K S + F V + A AGA GM++ N +G+ E+
Sbjct: 399 -DLTGKVAVVKRGS-------IAF----VDKADNAKKAGAIGMVVYNN--LSGEI---EA 441
Query: 305 NVLSTINYYDKHQLTRGHSIGISTTDTIKSVIKIRMSQPKTSYRRKPAPVM----ASFSS 360
NV +I +S D K V ++ + KT+++ + + A FSS
Sbjct: 442 NVPGM----------SVPTIKLSLEDGEKLVSALKAGETKTTFKLTVSKALGEQVADFSS 491
Query: 361 RGPNQVQPYILKPDVTAPGVNILAAYSLFASVSNLVT-DNRRGFPFNIQQGTSMSCPHVA 419
RGP + +++KPD++APGVNI VS + T D + + +QGTSM+ PH+A
Sbjct: 492 RGP-VMDTWMIKPDISAPGVNI---------VSTIPTHDPDHPYGYGSKQGTSMASPHIA 541
Query: 420 GTAGLIKTLHPNWSPAAIKSAIMTTA-TIRDNTNKLIRDAIDKTLANPFAYGSGHIQPNT 478
G +IK P WS IK+AIM TA T++D+ G + P+
Sbjct: 542 GAVAVIKQAKPKWSVEQIKAAIMNTAVTLKDS--------------------DGEVYPHN 581
Query: 479 AMDPGLVYDLSVVDYLNFLCAAGYSQRLISTLL------NPNMTFTCSGIHSINDLNYPS 532
A G ++ + + + YS T L N TFT SI
Sbjct: 582 AQGAGSARIMNAIKADSLVSPGSYS---YGTFLKENGNETKNETFTIENQSSIRKSYTLE 638
Query: 533 ITLPNLGLNAVNVTRIVTNVGPPSTYFAKVQL 564
+ G++ +R+V AKV++
Sbjct: 639 YSFNGSGISTSGTSRVVIPAHQTGKATAKVKV 670
>P2P_LACPA (Q02470) PII-type proteinase precursor (EC 3.4.21.96)
(Lactocepin) (Cell wall-associated serine proteinase)
(LP151)
Length = 1902
Score = 63.9 bits (154), Expect = 1e-09
Identities = 130/503 (25%), Positives = 197/503 (38%), Gaps = 68/503 (13%)
Query: 67 TIKGGSPKSRVVTYKVCWSQTIADGNSAVCYGADVLSAIDQAISDGVDIISVSVGGRSSS 126
++ G +P+++++ KV + SA A ++SAI+ + G D++++S+G S +
Sbjct: 301 SVVGVAPEAQLLAMKVFTNSD----TSATTGSATLVSAIEDSAKIGADVLNMSLGSDSGN 356
Query: 127 NFEEIFTDEISIGAFQ-AFAKNILLVASAGNGGPTPGSVTNVAPWVFTVAASTIDRDFSS 185
E + I A Q A V SAGN G T GS T V D +
Sbjct: 357 QTLE----DPEIAAVQNANESGTAAVISAGNSG-TSGSATQ---GVNKDYYGLQDNEMVG 408
Query: 186 TITIGNKTVTGASLFVNLPPNQSFTLVDSIDAKFANVTNQDARFCKPGTLDPSK------ 239
T T AS +Q+ T+ D D + T Q + G+ D K
Sbjct: 409 TPGTSRGATTVASAENTDVISQAVTITDGKDLQLGPETIQLSSNDFTGSFDQKKFYVVKD 468
Query: 240 VSGKIVECVGEKIT--IKNTSEPVSGRLLGFATNSVSQGREALSAGAKGMILRNQPKFNG 297
SG + + T K V L FA + + A +AGA G+I+ N
Sbjct: 469 ASGDLSKGAAADYTADAKGKIAIVKRGELNFA----DKQKYAQAAGAAGLIIVNND--GT 522
Query: 298 KTLLAESNVLSTINYYDKHQLTRGHSIGISTTDTIKSV-IKIRMS-QPKTSYRRKPAPVM 355
T L + +T + T + T S+ +KI ++ P Y M
Sbjct: 523 ATPLTSIRLTTTFPTFGLSSKTGQKLVDWVTAHPDDSLGVKIALTLLPNQKYTEDK---M 579
Query: 356 ASFSSRGPNQVQPYILKPDVTAPGVNILAAYSLFASVSNLVTDNRRGFPFNIQQGTSMSC 415
+ F+S GP V KPD+TAPG NI + T N G + GTSM+
Sbjct: 580 SDFTSYGP--VSNLSFKPDITAPGGNIWS------------TQNNNG--YTNMSGTSMAS 623
Query: 416 PHVAGTAGLIK-TLHPNWSP-----AAIKSAIMT--TATIRDNTNKLIRDA-IDKTLANP 466
P +AG+ L+K L+ +P +K +T T+ NT + I D + + +P
Sbjct: 624 PFIAGSQALLKQALNNKNNPFYADYKQLKGTALTDFLKTVEMNTAQPINDINYNNVIVSP 683
Query: 467 FAYGSGHIQPNTAMDPGLVYDLSVVDYLNFLCAAGYSQRLISTLLNPNMTFTCSGIHSI- 525
G+G + A+D L + S V N A ST +TFT H +
Sbjct: 684 RRQGAGLVDVKAAID-ALEKNPSTVVAENGYPAVELKD-FTSTDKTFKLTFTNRTTHELT 741
Query: 526 ------NDLN--YPSITLPNLGL 540
D N Y S T PN G+
Sbjct: 742 YQMDSNTDTNAVYTSATDPNSGV 764
>P1P_LACLC (P16271) PI-type proteinase precursor (EC 3.4.21.-)
(Wall-associated serine proteinase)
Length = 1902
Score = 60.8 bits (146), Expect = 1e-08
Identities = 119/501 (23%), Positives = 193/501 (37%), Gaps = 64/501 (12%)
Query: 67 TIKGGSPKSRVVTYKVCWSQTIADGNSAVCYGADVLSAIDQAISDGVDIISVSVGGRSSS 126
++ G +P+++++ KV + SA + ++SAI+ + G D++++S+G S +
Sbjct: 301 SVVGVAPEAQLLAMKVFTNSD----TSATTGSSTLVSAIEDSAKIGADVLNMSLGSDSGN 356
Query: 127 NFEEIFTDEISIGAFQAFAKNILLVASAGNGGPTPGSVTNVAPWVFTVAASTIDRDFSST 186
+ D A V SAGN G T GS T V D + T
Sbjct: 357 ---QTLEDPELAAVQNANESGTAAVISAGNSG-TSGSATE---GVNKDYYGLQDNEMVGT 409
Query: 187 ITIGNKTVTGASLFVNLPPNQSFTLVDSIDAKFANVTNQDARFCKPGTLDPSK------V 240
T AS Q+ T+ D + T Q + G+ D K
Sbjct: 410 PGTSRGATTVASAENTDVITQAVTITDGTGLQLGPGTIQLSSNDFTGSFDQKKFYVVKDA 469
Query: 241 SGKIVECVGEKITIKNTSEPVSGRLLGFATNSVS---QGREALSAGAKGMILRNQPKFNG 297
SG + K + + + G++ +S + + A +AGA G+I+ N +G
Sbjct: 470 SGNL-----SKGALADYTADAKGKIAIVKRGELSFDDKQKYAQAAGAAGLIIVNN---DG 521
Query: 298 KTLLAESNVLSTINYYDKHQLTRGHSIGISTTDTIKSVIKIRMSQPKTSYRRKPAPVMAS 357
S L+T G + T + ++++ ++ M+
Sbjct: 522 TATPVTSMALTTTFPTFGLSSVTGQKLVDWVTAHPDDSLGVKIALTLVPNQKYTEDKMSD 581
Query: 358 FSSRGPNQVQPYILKPDVTAPGVNILAAYSLFASVSNLVTDNRRGFPFNIQQGTSMSCPH 417
F+S GP V KPD+TAPG NI + T N G + GTSM+ P
Sbjct: 582 FTSYGP--VSNLSFKPDITAPGGNIWS------------TQNNNG--YTNMSGTSMASPF 625
Query: 418 VAGTAGLIK-TLHPNWSP-----AAIKSAIMT--TATIRDNTNKLIRDA-IDKTLANPFA 468
+AG+ L+K L+ +P +K +T T+ NT + I D + + +P
Sbjct: 626 IAGSQALLKQALNNKNNPFYAYYKQLKGTALTDFLKTVEMNTAQPINDINYNNVIVSPRR 685
Query: 469 YGSGHIQPNTAMDPGLVYDLSVVDYLNFLCAAGYSQRLISTLLNPNMTFTCSGIHSI--- 525
G+G + A+D L + S V N A ST +TFT S H +
Sbjct: 686 QGAGLVDVKAAID-ALEKNPSTVVAENGYPAVELKD-FTSTDKTFKLTFTNSTTHELTYQ 743
Query: 526 ----NDLN--YPSITLPNLGL 540
D N Y S T PN G+
Sbjct: 744 MDSNTDTNAVYTSATDPNSGV 764
>P3P_LACLC (P15292) PIII-type proteinase precursor (EC 3.4.21.96)
(Lactocepin) (Cell wall-associated serine proteinase)
Length = 1902
Score = 60.1 bits (144), Expect = 2e-08
Identities = 120/504 (23%), Positives = 197/504 (38%), Gaps = 70/504 (13%)
Query: 67 TIKGGSPKSRVVTYKVCWSQTIADGNSAVCYGADVLSAIDQAISDGVDIISVSVGGRSSS 126
++ G +P+++++ KV + SA A V+SAI+ + G D++++S+G S +
Sbjct: 301 SVVGVAPEAQLLAMKVFSNSD----TSAKTGSATVVSAIEDSAKIGADVLNMSLGSNSGN 356
Query: 127 NFEEIFTDEISIGAFQ-AFAKNILLVASAGNGGPTPGSVTNVAPWVFTVAASTIDRDFSS 185
E + + A Q A V SAGN G + + V + + D +
Sbjct: 357 QTLE----DPELAAVQNANESGTAAVISAGNSGTSGSATEGVNKDYYGLQ----DNEMVG 408
Query: 186 TITIGNKTVTGASLFVNLPPNQSFTLVDSIDAKFANVTNQDARFCKPGTLDPSK------ 239
+ T AS Q+ T+ D + T Q + G+ D K
Sbjct: 409 SPGTSRGATTVASAENTDVITQAVTITDGTGLQLGPETIQLSSHDFTGSFDQKKFYIVKD 468
Query: 240 VSGKIVECVGEKITIKNTSEPVSGRLLGFATNSVS---QGREALSAGAKGMILRNQPKFN 296
SG + K + + + G++ S + + A +AGA G+I+ N
Sbjct: 469 ASGNL-----SKGALADYTADAKGKIAIVKRGEFSFDDKQKYAQAAGAAGLIIVNTD--G 521
Query: 297 GKTLLAESNVLSTINYYDKHQLTRGHSIGISTTDTIKSV-IKIRMSQ-PKTSYRRKPAPV 354
T + + +T + +T + T S+ +KI ++ P Y
Sbjct: 522 TATPMTSIALTTTFPTFGLSSVTGQKLVDWVTAHPDDSLGVKITLAMLPNQKYTEDK--- 578
Query: 355 MASFSSRGPNQVQPYILKPDVTAPGVNILAAYSLFASVSNLVTDNRRGFPFNIQQGTSMS 414
M+ F+S GP V KPD+TAPG NI + T N G + GTSM+
Sbjct: 579 MSDFTSYGP--VSNLSFKPDITAPGGNIWS------------TQNNNG--YTNMSGTSMA 622
Query: 415 CPHVAGTAGLIK-TLHPNWSP-----AAIKSAIMT--TATIRDNTNKLIRDA-IDKTLAN 465
P +AG+ L+K L+ +P +K +T T+ NT + I D + + +
Sbjct: 623 SPFIAGSQALLKQALNNKNNPFYAYYKQLKGTALTDFLKTVEMNTAQPINDINYNNVIVS 682
Query: 466 PFAYGSGHIQPNTAMDPGLVYDLSVVDYLNFLCAAGYSQRLISTLLNPNMTFTCSGIHSI 525
P G+G + A+D L + S V N A ST +TFT H +
Sbjct: 683 PRRQGAGLVDVKAAID-ALEKNPSTVVAENGYPAVELKD-FTSTDKTFKLTFTNRTTHEL 740
Query: 526 -------NDLN--YPSITLPNLGL 540
D N Y S T PN G+
Sbjct: 741 TYQMDSNTDTNAVYTSATDPNSGV 764
>P2P_LACLC (P15293) PII-type proteinase precursor (EC 3.4.21.96)
(Lactocepin) (Cell wall-associated serine proteinase)
(LP151)
Length = 1902
Score = 59.3 bits (142), Expect = 3e-08
Identities = 122/506 (24%), Positives = 191/506 (37%), Gaps = 74/506 (14%)
Query: 67 TIKGGSPKSRVVTYKVCWSQTIADGNSAVCYGADVLSAIDQAISDGVDIISVSVGGRSSS 126
++ G +P+++++ KV + SA A ++SAI+ + G D++++S+G S +
Sbjct: 301 SVVGVAPEAQLLAMKVFTNSD----TSATTGSATLVSAIEDSAKIGADVLNMSLGSDSGN 356
Query: 127 NFEEIFTDEISIGAFQAFAKNILLVASAGNGGPTPGSVTNVAPWVFTVAASTIDRDFSST 186
+ D A V SAGN G T GS T V D + T
Sbjct: 357 ---QTLEDPELAAVQNANESGTAAVISAGNSG-TSGSATE---GVNKDYYGLQDNEMVGT 409
Query: 187 ITIGNKTVTGASLFVNLPPNQSFTLVDSIDAKFANVTNQDARFCKPGTLDPSKV------ 240
T AS Q+ T+ D + T Q + G+ D K
Sbjct: 410 PGTSRGATTVASAENTDVITQAVTITDGTGLQLGPETIQLSSNDFTGSFDQKKFYVVKDA 469
Query: 241 -----SGKIVECVGE---KITIKNTSEPVSGRLLGFATNSVSQGREALSAGAKGMILRNQ 292
GK+ + + KI I E L FA + + A +AGA G+I+ N
Sbjct: 470 SGNLSKGKVADYTADAKGKIAIVKRGE------LTFA----DKQKYAQAAGAAGLIIVNN 519
Query: 293 PKFNGKTLLAESNVLSTINYYDKHQLTRGHSIGISTTDTIKSVIKIRMSQPKTSYRRKPA 352
+G S L+T G + + ++++ ++
Sbjct: 520 ---DGTATPVTSMALTTTFPTFGLSSVTGQKLVDWVAAHPDDSLGVKIALTLVPNQKYTE 576
Query: 353 PVMASFSSRGPNQVQPYILKPDVTAPGVNILAAYSLFASVSNLVTDNRRGFPFNIQQGTS 412
M+ F+S GP V KPD+TAPG NI + T N G + GTS
Sbjct: 577 DKMSDFTSYGP--VSNLSFKPDITAPGGNIWS------------TQNNNG--YTNMSGTS 620
Query: 413 MSCPHVAGTAGLIK-TLHPNWSP-----AAIKSAIMT--TATIRDNTNKLIRDA-IDKTL 463
M+ P +AG+ L+K L+ +P +K +T T+ NT + I D + +
Sbjct: 621 MASPFIAGSQALLKQALNNKNNPFYAYYKQLKGTALTDFLKTVEMNTAQPINDINYNNVI 680
Query: 464 ANPFAYGSGHIQPNTAMDPGLVYDLSVVDYLNFLCAAGYSQRLISTLLNPNMTFTCSGIH 523
+P G+G + A+D L + S V N A ST +TFT H
Sbjct: 681 VSPRRQGAGLVDVKAAID-ALEKNPSTVVAENGYPAVELKD-FTSTDKTFKLTFTNRTTH 738
Query: 524 SI-------NDLN--YPSITLPNLGL 540
+ D N Y S T PN G+
Sbjct: 739 ELTYQMDSNTDTNAVYTSATDPNSGV 764
>PRTM_BACSK (Q99405) M-protease (EC 3.4.21.-)
Length = 269
Score = 56.2 bits (134), Expect = 2e-07
Identities = 32/81 (39%), Positives = 45/81 (55%), Gaps = 18/81 (22%)
Query: 374 DVTAPGVNILAAY--SLFASVSNLVTDNRRGFPFNIQQGTSMSCPHVAGTAGLIKTLHPN 431
D+ APGVN+ + Y S +AS++ GTSM+ PHVAG A L+K +P+
Sbjct: 191 DIVAPGVNVQSTYPGSTYASLN----------------GTSMATPHVAGVAALVKQKNPS 234
Query: 432 WSPAAIKSAIMTTATIRDNTN 452
WS I++ + TAT NTN
Sbjct: 235 WSNVQIRNHLKNTATGLGNTN 255
Score = 34.3 bits (77), Expect = 1.0
Identities = 36/152 (23%), Positives = 74/152 (48%), Gaps = 32/152 (21%)
Query: 47 HVTAGGNFVPG--ASIFGIGNGT--------------IKGGSPKSRVVTYKVCWSQTIAD 90
++ G +FVPG ++ G G+GT + G +P + + KV A
Sbjct: 42 NIRGGASFVPGEPSTQDGNGHGTHVAGTIAALNNSIGVLGVAPSAELYAVKVLG----AS 97
Query: 91 GNSAVCYGADVLSAIDQAISDGVDIISVSVGGRSSSNFEEIFTDEISIGAFQAFAKNILL 150
G+ +V + + ++ A ++G+ + ++S+G S S T E ++ + A ++ +L+
Sbjct: 98 GSGSV---SSIAQGLEWAGNNGMHVANLSLGSPSPS-----ATLEQAVNS--ATSRGVLV 147
Query: 151 VASAGNGGPTPGSVTNVAPWVFTVAASTIDRD 182
VA++GN G GS++ A + +A D++
Sbjct: 148 VAASGNSG--AGSISYPARYANAMAVGATDQN 177
>SUBT_BACAM (P00782) Subtilisin BPN' precursor (EC 3.4.21.62)
(Subtilisin Novo) (Subtilisin DFE) (Alkaline protease)
Length = 382
Score = 55.5 bits (132), Expect = 4e-07
Identities = 41/124 (33%), Positives = 52/124 (41%), Gaps = 37/124 (29%)
Query: 356 ASFSSRGPNQVQPYILKPDVTAPGVNILAAYSLFASVSNLVTDNRRGFPFNIQQGTSMSC 415
ASFSS GP DV APGV+I + G + GTSM+
Sbjct: 294 ASFSSVGPEL--------DVMAPGVSIQSTLP--------------GNKYGAYNGTSMAS 331
Query: 416 PHVAGTAGLIKTLHPNWSPAAIKSAIMTTATIRDNTNKLIRDAIDKTLANPFAYGSGHIQ 475
PHVAG A LI + HPNW+ ++S++ T T L + F YG G I
Sbjct: 332 PHVAGAAALILSKHPNWTNTQVRSSLENTTT---------------KLGDSFYYGKGLIN 376
Query: 476 PNTA 479
A
Sbjct: 377 VQAA 380
Score = 42.4 bits (98), Expect = 0.004
Identities = 41/148 (27%), Positives = 71/148 (47%), Gaps = 23/148 (15%)
Query: 68 IKGGSPKSRVVTYKVCWSQTIADGNSAVCYGADVLSAIDQAISDGVDIISVSVGGRSSSN 127
+ G +P + + KV ADG+ + +++ I+ AI++ +D+I++S+GG S S
Sbjct: 188 VLGVAPSASLYAVKVLG----ADGSGQYSW---IINGIEWAIANNMDVINMSLGGPSGSA 240
Query: 128 FEEIFTDEISIGAFQAFAKNILLVASAGNGGPTPGSVTNVA-----PWVFTVAASTIDRD 182
+ D +A A +++VA+AGN G T GS + V P V V A
Sbjct: 241 ALKAAVD-------KAVASGVVVVAAAGNEG-TSGSSSTVGYPGKYPSVIAVGAVDSSNQ 292
Query: 183 FSSTITIG---NKTVTGASLFVNLPPNQ 207
+S ++G + G S+ LP N+
Sbjct: 293 RASFSSVGPELDVMAPGVSIQSTLPGNK 320
>SUBS_BACLE (P29600) Subtilisin Savinase (EC 3.4.21.62) (Alkaline
protease)
Length = 269
Score = 55.1 bits (131), Expect = 5e-07
Identities = 31/81 (38%), Positives = 45/81 (55%), Gaps = 18/81 (22%)
Query: 374 DVTAPGVNILAAY--SLFASVSNLVTDNRRGFPFNIQQGTSMSCPHVAGTAGLIKTLHPN 431
D+ APGVN+ + Y S +AS++ GTSM+ PHVAG A L+K +P+
Sbjct: 191 DIVAPGVNVQSTYPGSTYASLN----------------GTSMATPHVAGAAALVKQKNPS 234
Query: 432 WSPAAIKSAIMTTATIRDNTN 452
WS I++ + TAT +TN
Sbjct: 235 WSNVQIRNHLKNTATSLGSTN 255
Score = 34.3 bits (77), Expect = 1.0
Identities = 36/152 (23%), Positives = 74/152 (48%), Gaps = 32/152 (21%)
Query: 47 HVTAGGNFVPG--ASIFGIGNGT--------------IKGGSPKSRVVTYKVCWSQTIAD 90
++ G +FVPG ++ G G+GT + G +P + + KV A
Sbjct: 42 NIRGGASFVPGEPSTQDGNGHGTHVAGTIAALNNSIGVLGVAPSAELYAVKVLG----AS 97
Query: 91 GNSAVCYGADVLSAIDQAISDGVDIISVSVGGRSSSNFEEIFTDEISIGAFQAFAKNILL 150
G+ +V + + ++ A ++G+ + ++S+G S S T E ++ + A ++ +L+
Sbjct: 98 GSGSV---SSIAQGLEWAGNNGMHVANLSLGSPSPS-----ATLEQAVNS--ATSRGVLV 147
Query: 151 VASAGNGGPTPGSVTNVAPWVFTVAASTIDRD 182
VA++GN G GS++ A + +A D++
Sbjct: 148 VAASGNSG--AGSISYPARYANAMAVGATDQN 177
>SUBB_BACLE (P29599) Subtilisin BL (EC 3.4.21.62) (Alkaline
protease)
Length = 269
Score = 55.1 bits (131), Expect = 5e-07
Identities = 31/81 (38%), Positives = 45/81 (55%), Gaps = 18/81 (22%)
Query: 374 DVTAPGVNILAAY--SLFASVSNLVTDNRRGFPFNIQQGTSMSCPHVAGTAGLIKTLHPN 431
D+ APGVN+ + Y S +AS++ GTSM+ PHVAG A L+K +P+
Sbjct: 191 DIVAPGVNVQSTYPGSTYASLN----------------GTSMATPHVAGAAALVKQKNPS 234
Query: 432 WSPAAIKSAIMTTATIRDNTN 452
WS I++ + TAT +TN
Sbjct: 235 WSNVQIRNHLKNTATSLGSTN 255
Score = 35.0 bits (79), Expect = 0.59
Identities = 36/152 (23%), Positives = 74/152 (48%), Gaps = 32/152 (21%)
Query: 47 HVTAGGNFVPG--ASIFGIGNGT--------------IKGGSPKSRVVTYKVCWSQTIAD 90
++ G +FVPG ++ G G+GT + G +P + + KV AD
Sbjct: 42 NIRGGASFVPGEPSTQDGNGHGTHVAGTIAALNNSIGVLGVAPSAELYAVKVLG----AD 97
Query: 91 GNSAVCYGADVLSAIDQAISDGVDIISVSVGGRSSSNFEEIFTDEISIGAFQAFAKNILL 150
G A+ + + ++ A ++G+ + ++S+G S S T E ++ + A ++ +L+
Sbjct: 98 GRGAI---SSIAQGLEWAGNNGMHVANLSLGSPSPS-----ATLEQAVNS--ATSRGVLV 147
Query: 151 VASAGNGGPTPGSVTNVAPWVFTVAASTIDRD 182
VA++GN G + S++ A + +A D++
Sbjct: 148 VAASGNSGAS--SISYPARYANAMAVGATDQN 177
>ELYA_BACCS (P41362) Alkaline protease precursor (EC 3.4.21.-)
Length = 380
Score = 55.1 bits (131), Expect = 5e-07
Identities = 31/81 (38%), Positives = 45/81 (55%), Gaps = 18/81 (22%)
Query: 374 DVTAPGVNILAAY--SLFASVSNLVTDNRRGFPFNIQQGTSMSCPHVAGTAGLIKTLHPN 431
D+ APGVN+ + Y S +AS++ GTSM+ PHVAG A L+K +P+
Sbjct: 302 DIVAPGVNVQSTYPGSTYASLN----------------GTSMATPHVAGAAALVKQKNPS 345
Query: 432 WSPAAIKSAIMTTATIRDNTN 452
WS I++ + TAT +TN
Sbjct: 346 WSNVQIRNHLKNTATSLGSTN 366
Score = 34.3 bits (77), Expect = 1.0
Identities = 36/152 (23%), Positives = 74/152 (48%), Gaps = 32/152 (21%)
Query: 47 HVTAGGNFVPG--ASIFGIGNGT--------------IKGGSPKSRVVTYKVCWSQTIAD 90
++ G +FVPG ++ G G+GT + G +P + + KV A
Sbjct: 153 NIRGGASFVPGEPSTQDGNGHGTHVAGTIAALNNSIGVLGVAPSAELYAVKVLG----AS 208
Query: 91 GNSAVCYGADVLSAIDQAISDGVDIISVSVGGRSSSNFEEIFTDEISIGAFQAFAKNILL 150
G+ +V + + ++ A ++G+ + ++S+G S S T E ++ + A ++ +L+
Sbjct: 209 GSGSV---SSIAQGLEWAGNNGMHVANLSLGSPSPS-----ATLEQAVNS--ATSRGVLV 258
Query: 151 VASAGNGGPTPGSVTNVAPWVFTVAASTIDRD 182
VA++GN G GS++ A + +A D++
Sbjct: 259 VAASGNSG--AGSISYPARYANAMAVGATDQN 288
>ELYA_BACAO (P27693) Alkaline protease precursor (EC 3.4.21.-)
Length = 380
Score = 55.1 bits (131), Expect = 5e-07
Identities = 31/81 (38%), Positives = 45/81 (55%), Gaps = 18/81 (22%)
Query: 374 DVTAPGVNILAAY--SLFASVSNLVTDNRRGFPFNIQQGTSMSCPHVAGTAGLIKTLHPN 431
D+ APGVN+ + Y S +AS++ GTSM+ PHVAG A L+K +P+
Sbjct: 302 DIVAPGVNVQSTYPGSTYASLN----------------GTSMATPHVAGAAALVKQKNPS 345
Query: 432 WSPAAIKSAIMTTATIRDNTN 452
WS I++ + TAT +TN
Sbjct: 346 WSNVQIRNHLKNTATSLGSTN 366
Score = 34.3 bits (77), Expect = 1.0
Identities = 36/152 (23%), Positives = 74/152 (48%), Gaps = 32/152 (21%)
Query: 47 HVTAGGNFVPG--ASIFGIGNGT--------------IKGGSPKSRVVTYKVCWSQTIAD 90
++ G +FVPG ++ G G+GT + G +P + + KV A
Sbjct: 153 NIRGGASFVPGEPSTQDGNGHGTHVAGTIAALNNSIGVLGVAPNAELYAVKVLG----AS 208
Query: 91 GNSAVCYGADVLSAIDQAISDGVDIISVSVGGRSSSNFEEIFTDEISIGAFQAFAKNILL 150
G+ +V + + ++ A ++G+ + ++S+G S S T E ++ + A ++ +L+
Sbjct: 209 GSGSV---SSIAQGLEWAGNNGMHVANLSLGSPSPS-----ATLEQAVNS--ATSRGVLV 258
Query: 151 VASAGNGGPTPGSVTNVAPWVFTVAASTIDRD 182
VA++GN G GS++ A + +A D++
Sbjct: 259 VAASGNSG--AGSISYPARYANAMAVGATDQN 288
>SUBT_BACSU (P04189) Subtilisin E precursor (EC 3.4.21.62)
Length = 381
Score = 53.9 bits (128), Expect = 1e-06
Identities = 41/124 (33%), Positives = 51/124 (41%), Gaps = 37/124 (29%)
Query: 356 ASFSSRGPNQVQPYILKPDVTAPGVNILAAYSLFASVSNLVTDNRRGFPFNIQQGTSMSC 415
ASFSS G DV APGV+I + G + GTSM+
Sbjct: 293 ASFSSAGSEL--------DVMAPGVSIQSTLP--------------GGTYGAYNGTSMAT 330
Query: 416 PHVAGTAGLIKTLHPNWSPAAIKSAIMTTATIRDNTNKLIRDAIDKTLANPFAYGSGHIQ 475
PHVAG A LI + HP W+ A ++ + +TAT L N F YG G I
Sbjct: 331 PHVAGAAALILSKHPTWTNAQVRDRLESTATY---------------LGNSFYYGKGLIN 375
Query: 476 PNTA 479
A
Sbjct: 376 VQAA 379
Score = 37.7 bits (86), Expect = 0.091
Identities = 47/193 (24%), Positives = 84/193 (43%), Gaps = 40/193 (20%)
Query: 37 NGVGEVQRKFHVTAGGNFVPGASI---FGIGNGT--------------IKGGSPKSRVVT 79
+G+ +V G +FVP + G +GT + G SP + +
Sbjct: 139 SGIDSSHPDLNVRGGASFVPSETNPYQDGSSHGTHVAGTIAALNNSIGVLGVSPSASLYA 198
Query: 80 YKVCWSQTIADGNSAVCYGADVLSAIDQAISDGVDIISVSVGGRSSSNFEEIFTDEISIG 139
KV S G+ + +++ I+ AIS+ +D+I++S+GG + S + D
Sbjct: 199 VKVLDST----GSGQYSW---IINGIEWAISNNMDVINMSLGGPTGSTALKTVVD----- 246
Query: 140 AFQAFAKNILLVASAGNGGPTPGSVTNV---APWVFTVAASTID-----RDFSSTITIGN 191
+A + I++ A+AGN G + GS + V A + T+A ++ FSS + +
Sbjct: 247 --KAVSSGIVVAAAAGNEG-SSGSTSTVGYPAKYPSTIAVGAVNSSNQRASFSSAGSELD 303
Query: 192 KTVTGASLFVNLP 204
G S+ LP
Sbjct: 304 VMAPGVSIQSTLP 316
>SUBT_BACST (P29142) Subtilisin J precursor (EC 3.4.21.62)
Length = 381
Score = 53.9 bits (128), Expect = 1e-06
Identities = 41/124 (33%), Positives = 51/124 (41%), Gaps = 37/124 (29%)
Query: 356 ASFSSRGPNQVQPYILKPDVTAPGVNILAAYSLFASVSNLVTDNRRGFPFNIQQGTSMSC 415
ASFSS G DV APGV+I + G + GTSM+
Sbjct: 293 ASFSSAGSEL--------DVMAPGVSIQSTLP--------------GGTYGAYNGTSMAT 330
Query: 416 PHVAGTAGLIKTLHPNWSPAAIKSAIMTTATIRDNTNKLIRDAIDKTLANPFAYGSGHIQ 475
PHVAG A LI + HP W+ A ++ + +TAT L N F YG G I
Sbjct: 331 PHVAGAAALILSKHPTWTNAQVRDRLESTATY---------------LGNSFYYGKGLIN 375
Query: 476 PNTA 479
A
Sbjct: 376 VQAA 379
Score = 38.5 bits (88), Expect = 0.053
Identities = 47/192 (24%), Positives = 82/192 (42%), Gaps = 38/192 (19%)
Query: 37 NGVGEVQRKFHVTAGGNFVPGASI---FGIGNGT--------------IKGGSPKSRVVT 79
+G+ +V G +FVP + G +GT + G SP + +
Sbjct: 139 SGIDSSHPDLNVRGGASFVPSETNPYQDGSSHGTHVAGTIAALNNSIGVLGVSPSASLYA 198
Query: 80 YKVCWSQTIADGNSAVCYGADVLSAIDQAISDGVDIISVSVGGRSSSNFEEIFTDEISIG 139
KV S G+ + +++ I+ AIS+ +D+I++S+GG S S + D
Sbjct: 199 VKVLDST----GSGQYSW---IINGIEWAISNNMDVINMSLGGPSGSTALKTVVD----- 246
Query: 140 AFQAFAKNILLVASAGNGGPTPGSVT--NVAPWVFTVAASTID-----RDFSSTITIGNK 192
+A + I++ A+AGN G + S T A + T+A ++ FSS + +
Sbjct: 247 --KAVSSGIVVAAAAGNEGSSGSSSTVGYPAKYPSTIAVGAVNSSNQRASFSSAGSELDV 304
Query: 193 TVTGASLFVNLP 204
G S+ LP
Sbjct: 305 MAPGVSIQSTLP 316
>SUBT_BACSA (P00783) Subtilisin amylosacchariticus precursor (EC
3.4.21.62)
Length = 381
Score = 53.9 bits (128), Expect = 1e-06
Identities = 41/124 (33%), Positives = 51/124 (41%), Gaps = 37/124 (29%)
Query: 356 ASFSSRGPNQVQPYILKPDVTAPGVNILAAYSLFASVSNLVTDNRRGFPFNIQQGTSMSC 415
ASFSS G DV APGV+I + G + GTSM+
Sbjct: 293 ASFSSAGSEL--------DVMAPGVSIQSTLP--------------GGTYGAYNGTSMAT 330
Query: 416 PHVAGTAGLIKTLHPNWSPAAIKSAIMTTATIRDNTNKLIRDAIDKTLANPFAYGSGHIQ 475
PHVAG A LI + HP W+ A ++ + +TAT L N F YG G I
Sbjct: 331 PHVAGAAALILSKHPTWTNAQVRDRLESTATY---------------LGNSFYYGKGLIN 375
Query: 476 PNTA 479
A
Sbjct: 376 VQAA 379
Score = 38.5 bits (88), Expect = 0.053
Identities = 47/192 (24%), Positives = 82/192 (42%), Gaps = 38/192 (19%)
Query: 37 NGVGEVQRKFHVTAGGNFVPGASI---FGIGNGT--------------IKGGSPKSRVVT 79
+G+ +V G +FVP + G +GT + G SP + +
Sbjct: 139 SGIDSSHPDLNVRGGASFVPSETNPYQDGSSHGTHVAGTIAALNNSIGVLGVSPSASLYA 198
Query: 80 YKVCWSQTIADGNSAVCYGADVLSAIDQAISDGVDIISVSVGGRSSSNFEEIFTDEISIG 139
KV S G+ + +++ I+ AIS+ +D+I++S+GG S S + D
Sbjct: 199 VKVLDST----GSGQYSW---IINGIEWAISNNMDVINMSLGGPSGSTALKTVVD----- 246
Query: 140 AFQAFAKNILLVASAGNGGPTPGSVT--NVAPWVFTVAASTID-----RDFSSTITIGNK 192
+A + I++ A+AGN G + S T A + T+A ++ FSS + +
Sbjct: 247 --KAVSSGIVVAAAAGNEGSSGSSSTVGYPAKYPSTIAVGAVNSSNQRASFSSAGSELDV 304
Query: 193 TVTGASLFVNLP 204
G S+ LP
Sbjct: 305 MAPGVSIQSTLP 316
>SUBN_BACNA (P35835) Subtilisin NAT precursor (EC 3.4.21.62)
Length = 381
Score = 53.1 bits (126), Expect = 2e-06
Identities = 41/124 (33%), Positives = 51/124 (41%), Gaps = 37/124 (29%)
Query: 356 ASFSSRGPNQVQPYILKPDVTAPGVNILAAYSLFASVSNLVTDNRRGFPFNIQQGTSMSC 415
ASFSS G DV APGV+I + G + GTSM+
Sbjct: 293 ASFSSVGSEL--------DVMAPGVSIQSTLP--------------GGTYGAYNGTSMAT 330
Query: 416 PHVAGTAGLIKTLHPNWSPAAIKSAIMTTATIRDNTNKLIRDAIDKTLANPFAYGSGHIQ 475
PHVAG A LI + HP W+ A ++ + +TAT L N F YG G I
Sbjct: 331 PHVAGAAALILSKHPTWTNAQVRDRLESTATY---------------LGNSFYYGKGLIN 375
Query: 476 PNTA 479
A
Sbjct: 376 VQAA 379
Score = 37.4 bits (85), Expect = 0.12
Identities = 31/112 (27%), Positives = 57/112 (50%), Gaps = 16/112 (14%)
Query: 101 VLSAIDQAISDGVDIISVSVGGRSSSNFEEIFTDEISIGAFQAFAKNILLVASAGNGGPT 160
+++ I+ AIS+ +D+I++S+GG + S + D +A + I++ A+AGN G +
Sbjct: 213 IINGIEWAISNNMDVINMSLGGPTGSTALKTVVD-------KAVSSGIVVAAAAGNEG-S 264
Query: 161 PGSVTNV---APWVFTVAASTID-----RDFSSTITIGNKTVTGASLFVNLP 204
GS + V A + T+A ++ FSS + + G S+ LP
Sbjct: 265 SGSTSTVGYPAKYPSTIAVGAVNSSNQRASFSSVGSELDVMAPGVSIQSTLP 316
>SUBT_BACPU (P07518) Subtilisin (EC 3.4.21.62) (Alkaline
mesentericopeptidase)
Length = 275
Score = 52.0 bits (123), Expect = 5e-06
Identities = 40/124 (32%), Positives = 51/124 (40%), Gaps = 37/124 (29%)
Query: 356 ASFSSRGPNQVQPYILKPDVTAPGVNILAAYSLFASVSNLVTDNRRGFPFNIQQGTSMSC 415
ASFSS G DV APGV+I + G + GTSM+
Sbjct: 187 ASFSSAGSEL--------DVMAPGVSIQSTLP--------------GGTYGAYNGTSMAT 224
Query: 416 PHVAGTAGLIKTLHPNWSPAAIKSAIMTTATIRDNTNKLIRDAIDKTLANPFAYGSGHIQ 475
PHVAG A LI + HP W+ A ++ + +TAT L + F YG G I
Sbjct: 225 PHVAGAAALILSKHPTWTNAQVRDRLESTATY---------------LGSSFYYGKGLIN 269
Query: 476 PNTA 479
A
Sbjct: 270 VQAA 273
Score = 37.7 bits (86), Expect = 0.091
Identities = 44/187 (23%), Positives = 82/187 (43%), Gaps = 28/187 (14%)
Query: 37 NGVGEVQRKFHVTAGGNFVPGASI---FGIGNGTIKGGSPKSR--------VVTYKVCWS 85
+G+ +V G +FVP + G +GT G+ + V ++
Sbjct: 33 SGIDSSHPDLNVRGGASFVPSETNPYQDGSSHGTHVAGTIAALNNSIGVLGVAPSSALYA 92
Query: 86 QTIADGNSAVCYGADVLSAIDQAISDGVDIISVSVGGRSSSNFEEIFTDEISIGAFQAFA 145
+ D + Y +++ I+ AIS+ +D+I++S+GG + S + D +A +
Sbjct: 93 VKVLDSTGSGQYSW-IINGIEWAISNNMDVINMSLGGPTGSTALKTVVD-------KAVS 144
Query: 146 KNILLVASAGNGGPTPGSVTNV---APWVFTVAASTID-----RDFSSTITIGNKTVTGA 197
I++ A+AGN G + GS + V A + T+A ++ FSS + + G
Sbjct: 145 SGIVVAAAAGNEG-SSGSTSTVGYPAKYPSTIAVGAVNSANQRASFSSAGSELDVMAPGV 203
Query: 198 SLFVNLP 204
S+ LP
Sbjct: 204 SIQSTLP 210
>ELYA_BACHD (P41363) Thermostable alkaline protease precursor (EC
3.4.21.-)
Length = 361
Score = 51.2 bits (121), Expect = 8e-06
Identities = 39/124 (31%), Positives = 58/124 (46%), Gaps = 37/124 (29%)
Query: 356 ASFSSRGPNQVQPYILKPDVTAPGVNILAAYSLFASVSNLVTDNRRGFPFNIQQGTSMSC 415
ASFS+ GP + +++APGVN+ + Y T NR + GTSM+
Sbjct: 273 ASFSTYGP--------EIEISAPGVNVNSTY----------TGNR----YVSLSGTSMAT 310
Query: 416 PHVAGTAGLIKTLHPNWSPAAIKSAIMTTATIRDNTNKLIRDAIDKTLANPFAYGSGHIQ 475
PHVAG A L+K+ +P+++ I+ I TAT L +P YG+G +
Sbjct: 311 PHVAGVAALVKSRYPSYTNNQIRQRINQTATY---------------LGSPSLYGNGLVH 355
Query: 476 PNTA 479
A
Sbjct: 356 AGRA 359
>SUBT_BACLI (P00780) Subtilisin Carlsberg precursor (EC 3.4.21.62)
Length = 379
Score = 47.8 bits (112), Expect = 9e-05
Identities = 40/124 (32%), Positives = 52/124 (41%), Gaps = 37/124 (29%)
Query: 356 ASFSSRGPNQVQPYILKPDVTAPGVNILAAYSLFASVSNLVTDNRRGFPFNIQQGTSMSC 415
ASFSS G +V APG + + Y S T N GTSM+
Sbjct: 291 ASFSSVGAEL--------EVMAPGAGVYSTYP----TSTYATLN----------GTSMAS 328
Query: 416 PHVAGTAGLIKTLHPNWSPAAIKSAIMTTATIRDNTNKLIRDAIDKTLANPFAYGSGHIQ 475
PHVAG A LI + HPN S + +++ + +TAT L + F YG G I
Sbjct: 329 PHVAGAAALILSKHPNLSASQVRNRLSSTATY---------------LGSSFYYGKGLIN 373
Query: 476 PNTA 479
A
Sbjct: 374 VEAA 377
Score = 43.9 bits (102), Expect = 0.001
Identities = 46/191 (24%), Positives = 85/191 (44%), Gaps = 25/191 (13%)
Query: 38 GVGEVQRKFHVTAGGNFVPGASIF--GIGNGTIKGGSPKSRVVTYKVC--------WSQT 87
G+ +V G +FV G + G G+GT G+ + T V ++
Sbjct: 139 GIQASHPDLNVVGGASFVAGEAYNTDGNGHGTHVAGTVAALDNTTGVLGVAPSVSLYAVK 198
Query: 88 IADGNSAVCYGADVLSAIDQAISDGVDIISVSVGGRSSSNFEEIFTDEISIGAFQAFAKN 147
+ + + + Y ++S I+ A ++G+D+I++S+GG S S + D A+A+
Sbjct: 199 VLNSSGSGTYSG-IVSGIEWATTNGMDVINMSLGGPSGSTAMKQAVD-------NAYARG 250
Query: 148 ILLVASAGNGGPTPGSVTNVAPWVF--TVAASTIDRD-----FSSTITIGNKTVTGASLF 200
+++VA+AGN G + + T P + +A +D + FSS GA ++
Sbjct: 251 VVVVAAAGNSGSSGNTNTIGYPAKYDSVIAVGAVDSNSNRASFSSVGAELEVMAPGAGVY 310
Query: 201 VNLPPNQSFTL 211
P + TL
Sbjct: 311 STYPTSTYATL 321
>ELYA_BACYA (P20724) Alkaline elastase YaB precursor (EC 3.4.21.-)
Length = 378
Score = 47.4 bits (111), Expect = 1e-04
Identities = 27/80 (33%), Positives = 39/80 (48%), Gaps = 14/80 (17%)
Query: 374 DVTAPGVNILAAYSLFASVSNLVTDNRRGFPFNIQQGTSMSCPHVAGTAGLIKTLHPNWS 433
D+ APGV + + G + GTSM+ PHVAG A L+K +P+WS
Sbjct: 300 DIVAPGVGVQSTVP--------------GNGYASFNGTSMATPHVAGVAALVKQKNPSWS 345
Query: 434 PAAIKSAIMTTATIRDNTNK 453
I++ + TAT NT +
Sbjct: 346 NVQIRNHLKNTATNLGNTTQ 365
Score = 31.6 bits (70), Expect = 6.5
Identities = 35/151 (23%), Positives = 63/151 (41%), Gaps = 32/151 (21%)
Query: 48 VTAGGNFVPGASIFGIGNG----------------TIKGGSPKSRVVTYKVCWSQTIADG 91
+ G +FVPG GNG + G +P + KV A G
Sbjct: 152 IRGGASFVPGEPNISDGNGHGTQVAGTIAALNNSIGVLGVAPNVDLYGVKVLG----ASG 207
Query: 92 NSAVCYGADVLSAIDQAISDGVDIISVSVGGRSSSNFEEIFTDEISIGAFQAFAKNILLV 151
+ ++ + + + A ++G+ I ++S+G + S E + QA A +L+V
Sbjct: 208 SGSI---SGIAQGLQWAANNGMHIANMSLGSSAGSATMEQAVN-------QATASGVLVV 257
Query: 152 ASAGNGGPTPGSVTNVAPWVFTVAASTIDRD 182
A++GN G G+V A + +A D++
Sbjct: 258 AASGNSG--AGNVGFPARYANAMAVGATDQN 286
>SUBD_BACLI (P00781) Subtilisin DY (EC 3.4.21.62)
Length = 274
Score = 47.0 bits (110), Expect = 1e-04
Identities = 57/226 (25%), Positives = 99/226 (43%), Gaps = 29/226 (12%)
Query: 38 GVGEVQRKFHVTAGGNFVPGASIF--GIGNGTIKGGSPKSRVVTYKVC--------WSQT 87
G+ V G +FV G S G G+GT G+ + T V ++
Sbjct: 34 GIAASHTDLKVVGGASFVSGESYNTDGNGHGTHVAGTVAALDNTTGVLGVAPNVSLYAIK 93
Query: 88 IADGNSAVCYGADVLSAIDQAISDGVDIISVSVGGRSSSNFEEIFTDEISIGAFQAFAKN 147
+ + + + Y A ++S I+ A +G+D+I++S+GG S S + D +A+A
Sbjct: 94 VLNSSGSGTYSA-IVSGIEWATQNGLDVINMSLGGPSGSTALKQAVD-------KAYASG 145
Query: 148 ILLVASAGNGGPTPGSVTNV---APWVFTVAASTIDRD-----FSSTITIGNKTVTGASL 199
I++VA+AGN G + GS + A + +A +D + FSS G S+
Sbjct: 146 IVVVAAAGNSG-SSGSQNTIGYPAKYDSVIAVGAVDSNKNRASFSSVGAELEVMAPGVSV 204
Query: 200 FVNLPPNQSFTLVDSIDAKFANVTNQDAR-FCKPGTLDPSKVSGKI 244
+ P N ++T ++ +V A K TL S+V ++
Sbjct: 205 YSTYPSN-TYTSLNGTSMASPHVAGAAALILSKYPTLSASQVRNRL 249
Score = 45.8 bits (107), Expect = 3e-04
Identities = 39/124 (31%), Positives = 53/124 (42%), Gaps = 37/124 (29%)
Query: 356 ASFSSRGPNQVQPYILKPDVTAPGVNILAAYSLFASVSNLVTDNRRGFPFNIQQGTSMSC 415
ASFSS G +V APGV++ + Y SN T GTSM+
Sbjct: 186 ASFSSVGAEL--------EVMAPGVSVYSTYP-----SNTYTS---------LNGTSMAS 223
Query: 416 PHVAGTAGLIKTLHPNWSPAAIKSAIMTTATIRDNTNKLIRDAIDKTLANPFAYGSGHIQ 475
PHVAG A LI + +P S + +++ + +TAT L + F YG G I
Sbjct: 224 PHVAGAAALILSKYPTLSASQVRNRLSSTAT---------------NLGDSFYYGKGLIN 268
Query: 476 PNTA 479
A
Sbjct: 269 VEAA 272
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.321 0.136 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 71,776,766
Number of Sequences: 164201
Number of extensions: 3025715
Number of successful extensions: 7015
Number of sequences better than 10.0: 72
Number of HSP's better than 10.0 without gapping: 28
Number of HSP's successfully gapped in prelim test: 44
Number of HSP's that attempted gapping in prelim test: 6913
Number of HSP's gapped (non-prelim): 114
length of query: 624
length of database: 59,974,054
effective HSP length: 116
effective length of query: 508
effective length of database: 40,926,738
effective search space: 20790782904
effective search space used: 20790782904
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 69 (31.2 bits)
Medicago: description of AC149129.14


