

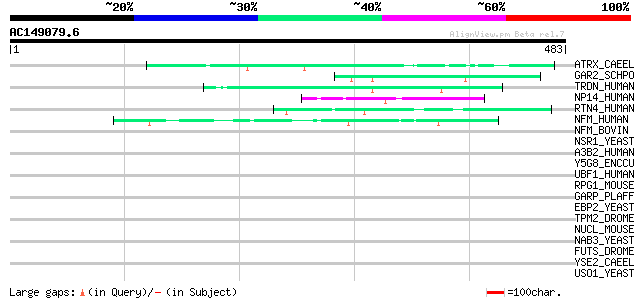
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC149079.6 - phase: 0
(483 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
ATRX_CAEEL (Q9U7E0) Transcriptional regulator ATRX homolog (X-li... 49 3e-05
GAR2_SCHPO (P41891) Protein gar2 48 6e-05
TRDN_HUMAN (Q13061) Triadin 46 2e-04
NP14_HUMAN (Q14978) Nucleolar phosphoprotein p130 (Nucleolar 130... 46 2e-04
RTN4_HUMAN (Q9NQC3) Reticulon 4 (Neurite outgrowth inhibitor) (N... 44 7e-04
NFM_HUMAN (P07197) Neurofilament triplet M protein (160 kDa neur... 44 7e-04
NFM_BOVIN (O77788) Neurofilament triplet M protein (160 kDa neur... 44 0.001
NSR1_YEAST (P27476) Nuclear localization sequence binding protei... 43 0.002
A3B2_HUMAN (Q13367) Adapter-related protein complex 3 beta 2 sub... 43 0.002
Y5G8_ENCCU (Q8STA9) Hypothetical protein ECU05_1680/ECU11_0050 42 0.003
UBF1_HUMAN (P17480) Nucleolar transcription factor 1 (Upstream b... 42 0.003
RPG1_MOUSE (Q9EPQ2) X-linked retinitis pigmentosa GTPase regulat... 42 0.003
GARP_PLAFF (P13816) Glutamic acid-rich protein precursor 42 0.003
EBP2_YEAST (P36049) rRNA processing protein EBP2 (EBNA1-binding ... 42 0.004
TPM2_DROME (P09491) Tropomyosin 2 (Tropomyosin I) 42 0.005
NUCL_MOUSE (P09405) Nucleolin (Protein C23) 42 0.005
NAB3_YEAST (P38996) Nuclear polyadenylated RNA-binding protein 3 42 0.005
FUTS_DROME (Q9W596) Microtubule-associated protein futsch 42 0.005
YSE2_CAEEL (Q09936) Hypothetical protein C53C9.2 in chromosome X 41 0.006
USO1_YEAST (P25386) Intracellular protein transport protein USO1 41 0.006
>ATRX_CAEEL (Q9U7E0) Transcriptional regulator ATRX homolog
(X-linked nuclear protein-1)
Length = 1359
Score = 48.9 bits (115), Expect = 3e-05
Identities = 75/362 (20%), Positives = 142/362 (38%), Gaps = 39/362 (10%)
Query: 120 EDEVDHQNQSESEDINKIQTWSNQNQHYRNKPMVIVAPQVQQLQNSVFEDEQSSFNENEK 179
+++++ Q E+E K + + KP P ++ +S ED+ +E E
Sbjct: 18 DEDLEMARQIENERKEKRAQKLKEKREREGKPPPKKRPAKKRKASSSEEDDD---DEEES 74
Query: 180 PLLLPVRSLKSRLSDDDCVVQSQSVD-----GLSKTTTKRFSSNSFNRVRNYAEFEGLVE 234
P +S K S+ + + D K K+ + R + +E E E
Sbjct: 75 PRKSSKKSRKRAKSESESDESDEEEDRKKSKSKKKVDQKKKEKSKKKRTTSSSEDEDSDE 134
Query: 235 DKLKQKEEENAVLPSPIPWRS--RSASARMMEPKQEAIEKALKASMVMEPKQEANENASK 292
++ ++ ++++ S S R ++ ++ EK++K + + +E SK
Sbjct: 135 EREQKSKKKSKKTKKQTSSESSEESEEERKVKKSKKNKEKSVKKRAETSEESDEDEKPSK 194
Query: 293 PSMVKTSSIKFTPSESVAKNSEDLIKKKSFYYKSYPPPPPPPPPTMFYKSTFLKSRYGEK 352
S + SES +++ +++ K K K K T EK
Sbjct: 195 KSKKGLKKKAKSESESESEDEKEVKKSKKKSKKVVKKESESEDEAPEKKKT-------EK 247
Query: 353 NGRNMSMGKSIEEGTNEKEDEDEEEKEYSTSSMQEQTDKERFIEKRVILEAEETDSDEDE 412
R S S E +EK DE+EEEKE SS + + K ++K L ++E E+
Sbjct: 248 --RKRSKTSSEESSESEKSDEEEEEKE---SSPKPKKKKPLAVKK---LSSDE--ESEES 297
Query: 413 DVGELKQSVKSVKTEEPCSSSLSGGIDTVSDEGPDVDKKADEFIAKFREQIRLQRIESIK 472
DV L Q K G + +SD + D+K++ + E++ ++ + +
Sbjct: 298 DVEVLPQKKK------------RGAVTLISDSEDEKDQKSESEASDVEEKVSKKKAKKQE 345
Query: 473 RS 474
S
Sbjct: 346 SS 347
Score = 40.8 bits (94), Expect = 0.008
Identities = 47/204 (23%), Positives = 81/204 (39%), Gaps = 36/204 (17%)
Query: 306 SESVAKNSEDLIKKKSFYYKSYPPPPPPPPPTMFYKSTFLKSRYGEKNGRNMSMGKSIEE 365
+E K ++ L +K+ + PPP P S+ + E+ S KS +
Sbjct: 29 NERKEKRAQKLKEKRE---REGKPPPKKRPAKKRKASSSEEDDDDEEESPRKSSKKSRKR 85
Query: 366 GTNEKE----DEDEEEKEYSTSSMQEQTDKERFIEKRVILEAEETDSDEDEDVGELKQSV 421
+E E DE+E+ K+ + +Q KE+ +KR +E+ DSDE+ E K
Sbjct: 86 AKSESESDESDEEEDRKKSKSKKKVDQKKKEKSKKKRTTSSSEDEDSDEER---EQKSKK 142
Query: 422 KSVKTEEPCSSSLS-----------------------GGIDTVSDEGPDVDKKADEFI-- 456
KS KT++ SS S SDE KK+ + +
Sbjct: 143 KSKKTKKQTSSESSEESEEERKVKKSKKNKEKSVKKRAETSEESDEDEKPSKKSKKGLKK 202
Query: 457 -AKFREQIRLQRIESIKRSTRVAR 479
AK + + + +K+S + ++
Sbjct: 203 KAKSESESESEDEKEVKKSKKKSK 226
Score = 40.8 bits (94), Expect = 0.008
Identities = 52/249 (20%), Positives = 92/249 (36%), Gaps = 24/249 (9%)
Query: 236 KLKQKEE-ENAVLPSPIPWRSRSASARMMEPKQEAIEKALKASMVMEPKQEANENASKPS 294
KLK+K E E P P + R AS+ + E E++ + S K+ +E+ S S
Sbjct: 38 KLKEKREREGKPPPKKRPAKKRKASSSEEDDDDE--EESPRKSSKKSRKRAKSESESDES 95
Query: 295 MVKTSSIKFTPSESVAKNSEDLIKKKSFYYKSYPPPPPPPPPTMFYKSTFLKSRYGEKNG 354
+ K + V + ++ KKK ++T
Sbjct: 96 DEEEDRKKSKSKKKVDQKKKEKSKKK--------------------RTTSSSEDEDSDEE 135
Query: 355 RNMSMGKSIEEGTNEKEDEDEEEKEYSTSSMQEQTDKERFIEKRVILEAEETDSDEDEDV 414
R K ++ + E EE E + + +KE+ ++KR +EE+D DE
Sbjct: 136 REQKSKKKSKKTKKQTSSESSEESEEERKVKKSKKNKEKSVKKRAET-SEESDEDEKPSK 194
Query: 415 GELKQSVKSVKTEEPCSSSLSGGIDTVSDEGPDVDKKADEFIAKFREQIRLQRIESIKRS 474
K K K+E S + + V KK E + E+ + ++ + K S
Sbjct: 195 KSKKGLKKKAKSESESESEDEKEVKKSKKKSKKVVKKESESEDEAPEKKKTEKRKRSKTS 254
Query: 475 TRVARNSSR 483
+ + S +
Sbjct: 255 SEESSESEK 263
Score = 38.1 bits (87), Expect = 0.051
Identities = 57/313 (18%), Positives = 123/313 (39%), Gaps = 47/313 (15%)
Query: 90 QEPDKDNNNNTKFDNAQSLVSRFLQVSSFFEDE-VDHQNQSESEDINKI---QTWSNQNQ 145
+E K + + K D + S+ + +S EDE D + + +S+ +K QT S ++
Sbjct: 98 EEDRKKSKSKKKVDQKKKEKSKKKRTTSSSEDEDSDEEREQKSKKKSKKTKKQTSSESSE 157
Query: 146 HYRNKPMVIVAPQVQQLQNSVFEDEQSSFNENEKPLLLPVRSLKSRLSDDDCVVQSQSVD 205
+ V + + ++ + +E+EKP + LK + + +S+S D
Sbjct: 158 ESEEERKVKKSKKNKEKSVKKRAETSEESDEDEKPSKKSKKGLKKKAKSES---ESESED 214
Query: 206 GLSKTTTKRFSSNSFNRVRNYAEFEGLVEDKLKQKEEENAVLPSPIPWRSRSASARMMEP 265
+ K+ S V+ +E E +K K ++ + RS+++S E
Sbjct: 215 ---EKEVKKSKKKSKKVVKKESESEDEAPEKKKTEKRK----------RSKTSSEESSES 261
Query: 266 KQEAIEKALKASMVMEPKQEANENASKPSMVKTSSIKFTPSESVAKNSEDLIKKKSFYYK 325
++ + ++E E++ KP K ++K S+ ++ S+ +
Sbjct: 262 EKS------------DEEEEEKESSPKPKKKKPLAVKKLSSDEESEESDVEVL------- 302
Query: 326 SYPPPPPPPPPTMFYKSTFLKSRYGEKNGRNMSMGKSIEEGTNEKEDEDEEEKEYSTSSM 385
P T + EK+ ++ S +EE ++K+ + +E E + S
Sbjct: 303 --------PQKKKRGAVTLISDSEDEKDQKSESEASDVEEKVSKKKAKKQESSESGSDSS 354
Query: 386 QEQTDKERFIEKR 398
+ R +K+
Sbjct: 355 EGSITVNRKSKKK 367
>GAR2_SCHPO (P41891) Protein gar2
Length = 500
Score = 47.8 bits (112), Expect = 6e-05
Identities = 40/188 (21%), Positives = 75/188 (39%), Gaps = 8/188 (4%)
Query: 283 KQEANENASKPSMV----KTSSIKFTPSESVAKNSE--DLIKKKSFYYKSYPPPPPPPPP 336
K+ E ASK + K+ I ++ +AK S D+ KKS P P
Sbjct: 10 KKSVKETASKKGAIEKPSKSKKITKEAAKEIAKQSSKTDVSPKKSKKEAKRASSPEPSKK 69
Query: 337 TMFYKSTFLKSRYGEKNGRNMSMGKSIEEGTNEKEDEDEEEKEYSTSSMQEQTDKERFI- 395
++ + K + S E ++E E E + S+ S ++++E +
Sbjct: 70 SVKKQKKSKKKEESSSESESESSSSESESSSSESESSSSESESSSSESSSSESEEEVIVK 129
Query: 396 -EKRVILEAEETDSDEDEDVGELKQSVKSVKTEEPCSSSLSGGIDTVSDEGPDVDKKADE 454
E++ +E + S E E+ E ++ K SSS S ++ S+ ++ +E
Sbjct: 130 TEEKKESSSESSSSSESEEEEEAVVKIEEKKESSSDSSSESSSSESESESSSSESEEEEE 189
Query: 455 FIAKFREQ 462
+ K E+
Sbjct: 190 VVEKTEEK 197
Score = 40.4 bits (93), Expect = 0.010
Identities = 56/244 (22%), Positives = 99/244 (39%), Gaps = 48/244 (19%)
Query: 249 SPIPWRSRSASARMMEPKQEAIEKALKASMVMEPKQEA-NENASKPSMVKTSSIKFTPSE 307
SP + + A EP +++++K K+ E E+ +E++S S +S + + SE
Sbjct: 50 SPKKSKKEAKRASSPEPSKKSVKKQKKSKKKEESSSESESESSSSESESSSSESESSSSE 109
Query: 308 SVAKNSEDLIKKKSFYYKSYPPPPPPPPPTMFYKSTFLKSRYGEKNGRNMSMGKSIEEGT 367
S + +SE + + +K+ EK +S E +
Sbjct: 110 SESSSSESSSSESE-------------------EEVIVKTE--EKK-------ESSSESS 141
Query: 368 NEKEDEDEEE--------KEYSTSSMQEQTDKERFIEKRVILEAEETDSDEDEDVGELKQ 419
+ E E+EEE KE S+ S E + E E+ ++S+E+E+V E +
Sbjct: 142 SSSESEEEEEAVVKIEEKKESSSDSSSESSSSES------ESESSSSESEEEEEVVEKTE 195
Query: 420 SVKSVKTEEPCSSSLSGGIDTVSDEGPDVDKKADEFIAKFREQIRLQRIE--SIKRSTRV 477
K +E SSS S S E D D +D E + ++ E S +R ++
Sbjct: 196 EKKEGSSE---SSSDSESSSDSSSESGDSDSSSDSESESSSEDEKKRKAEPASEERPAKI 252
Query: 478 ARNS 481
+ S
Sbjct: 253 TKPS 256
Score = 30.8 bits (68), Expect = 8.1
Identities = 36/159 (22%), Positives = 65/159 (40%), Gaps = 2/159 (1%)
Query: 164 NSVFEDEQSSFNENEKPLLLPVRSLKSRLSDDDCVVQSQSVDGLSKTTTKRFSSN-SFNR 222
+S E E SS S +S S + + + KT K+ SS+ S +
Sbjct: 84 SSESESESSSSESESSSSESESSSSESESSSSESSSSESEEEVIVKTEEKKESSSESSSS 143
Query: 223 VRNYAEFEGLVEDKLKQKEEENAVLPSPIPWRSRSASARMMEPKQEAIEKALKASMVMEP 282
+ E E +V+ + K++ ++ S +S+ E ++E +EK +
Sbjct: 144 SESEEEEEAVVKIEEKKESSSDSSSESSSSESESESSSSESEEEEEVVEKTEEKKEGSSE 203
Query: 283 KQEANENASKPSMVKTSSIKFTPSESVAKNSEDLIKKKS 321
+E++S S S + SES +SED K+K+
Sbjct: 204 SSSDSESSSDSSSESGDSDSSSDSES-ESSSEDEKKRKA 241
>TRDN_HUMAN (Q13061) Triadin
Length = 728
Score = 45.8 bits (107), Expect = 2e-04
Identities = 55/273 (20%), Positives = 99/273 (36%), Gaps = 17/273 (6%)
Query: 169 DEQSSFNENEKPLLLPVRSLKSRLSDDDCVVQSQSVDGLSKTTTKRFSSNSFNRVRNYAE 228
DE + E ++P P+R K + D Q + + T + +VR +
Sbjct: 124 DEDTDKGEIDEP---PLR--KKEIHKDKTEKQEKPERKIQTKVTHKEKEKGKEKVREKEK 178
Query: 229 FEGLVEDKLKQKEEENAVLPSPIPWRSRSASARMMEPKQEAIEKALKASMVMEPKQEANE 288
E K K +++E + + ++ +A E K + K K V + + E
Sbjct: 179 PEKKATHKEKIEKKEKPETKTVAKEQKKAKTAEKSEEKTKKEVKGGKQEKVKQTAAKVKE 238
Query: 289 NASKPSMVKTSSIKFTPSESVAKNSE---------DLIKKKSFYYKSYPPPPPPPPPTMF 339
PS K K + S + + D+ P PPP P
Sbjct: 239 VQKTPSKPKEKEDKEKAAVSKHEQKDQYAFCRYMIDIFVHGDLKPGQSPAIPPPLPTEQA 298
Query: 340 YKSTFLKSRYGEKNGRNMSMGKSIEEGTNEKEDED---EEEKEYSTSSMQEQTDKERFIE 396
+ T EK G K + T +KE ED + EKE + +++ K +
Sbjct: 299 SRPTPASPALEEKEGEKKKAEKKVTSETKKKEKEDIKKKSEKETAIDVEKKEPGKASETK 358
Query: 397 KRVILEAEETDSDEDEDVGELKQSVKSVKTEEP 429
+ + A + + +DE + K++ K + E+P
Sbjct: 359 QGTVKIAAQAAAKKDEKKEDSKKTKKPAEVEQP 391
Score = 42.4 bits (98), Expect = 0.003
Identities = 53/245 (21%), Positives = 104/245 (41%), Gaps = 24/245 (9%)
Query: 234 EDKLKQKEEENAVLPSPIPWRSRSASARMMEPKQEAIEKALKASMVMEPKQEANENASKP 293
ED K+ E+E A+ + + E KQ ++ A +A+ + K+E ++ KP
Sbjct: 332 EDIKKKSEKETAIDVE------KKEPGKASETKQGTVKIAAQAAAKKDEKKEDSKKTKKP 385
Query: 294 SMVKTSSIKFTPSESVAKNSEDLIKKKSFYYKSYPPPPPPPPPTMFYKSTFLKSRYGEKN 353
+ V+ K E K+ E K + P + K+ K G +
Sbjct: 386 AEVEQPKGK--KQEKKEKHVEPAKSPKKEH-------SVPSDKQVKAKTERAKEEIGAVS 436
Query: 354 GRNMSMGKSIEEGTNEKEDEDEEEKEYSTSSMQEQTDKERFIEKRVILEAEETDSDEDED 413
+ GK E+ T E E +EK TSS+ + + + E++V +E + + +D
Sbjct: 437 SKKAVPGKKEEKTTKTVEQEIRKEKSGKTSSILKDKEPIKGKEEKVPASLKEKEPETKKD 496
Query: 414 VGELKQSVKSVKTEEPCSSSLSGGIDTVSDEGPDVDKKADEFIAKFREQIRLQRIESIKR 473
++ ++ K VK + P L G + + P + K+A I+ E++++ + + +K
Sbjct: 497 -EKMSKAGKEVKPKPP---QLQGKKEEKPE--PQIKKEAKPAIS---EKVQIHKQDIVKP 547
Query: 474 STRVA 478
V+
Sbjct: 548 EKTVS 552
>NP14_HUMAN (Q14978) Nucleolar phosphoprotein p130 (Nucleolar 130
kDa protein) (140 kDa nucleolar phosphoprotein)
(Nopp140) (Nucleolar and coiled-body phosphoprotein 1)
Length = 699
Score = 45.8 bits (107), Expect = 2e-04
Identities = 43/164 (26%), Positives = 72/164 (43%), Gaps = 14/164 (8%)
Query: 255 SRSASARMMEPKQEAIEKALKASMVMEPKQEANENASKPSMVKTSSIKFTPSESVAKNSE 314
S S+S+ + +E EKA PK++ A P T+ + + S + + E
Sbjct: 219 SSSSSSSSSDDSEE--EKAAATPKKTVPKKQVVAKA--PVKAATTPTRKSSSSEDSSSDE 274
Query: 315 DLIKKKSFYYK----SYPPPPPPPPPTMFYKSTFLKSRYGEKNGRNMSMGKSIEEGTNEK 370
+ +KK K SY PPP PPP L ++ +K +S E+ ++E
Sbjct: 275 EEEQKKPMKNKPGPYSYAPPPSAPPP-----KKSLGTQPPKKAVEKQQPVESSEDSSDES 329
Query: 371 EDEDEEEKEYSTSS-MQEQTDKERFIEKRVILEAEETDSDEDED 413
+ EEEK+ T + + + T K +K ++ +DSD ED
Sbjct: 330 DSSSEEEKKPPTKAVVSKATTKPPPAKKAAESSSDSSDSDSSED 373
Score = 34.7 bits (78), Expect = 0.56
Identities = 50/236 (21%), Positives = 84/236 (35%), Gaps = 19/236 (8%)
Query: 254 RSRSASARMMEPKQEAIEKALKASMVMEPKQEANENASKPSMVKTSSIKFTPSESVAKNS 313
R +S+ E EK K +PK A S P+ + + S+S + +S
Sbjct: 419 RKADSSSSEEESSSSEEEKTKKMVATTKPKATAKAALSLPAKQAPQGSRDSSSDSDSSSS 478
Query: 314 EDLIKKKSFYYKSYPPPPPPPPPTMFYKSTFLKSRYGEKNGRNMSMGKSIEEGTNEKEDE 373
E+ +K S KS P S ++ G+ N S +++ E
Sbjct: 479 EEEEEKTS---KSAVKKKPQKVAGGAAPSKPASAKKGKAESSNSS--------SSDDSSE 527
Query: 374 DEEEKEYSTSSMQEQTDKERFIEKRVILEAEETDSDEDEDVGELKQ--------SVKSVK 425
+EEEK S + Q K + + E+E+ + K S+K K
Sbjct: 528 EEEEKLKGKGSPRPQAPKANGTSALTAQNGKAAKNSEEEEEEKKKAAVVVSKSGSLKKRK 587
Query: 426 TEEPCSSSLSGGIDTVSDEGPDVDKKADEFIAKFREQIRLQRIESIKRSTRVARNS 481
E + + + + P+ K + + R R E I+ +RVA NS
Sbjct: 588 QNEAAKEAETPQAKKIKLQTPNTFPKRKKGEKRASSPFRRVREEEIEVDSRVADNS 643
Score = 32.3 bits (72), Expect = 2.8
Identities = 32/138 (23%), Positives = 60/138 (43%), Gaps = 23/138 (16%)
Query: 274 LKASMVMEPKQEANENASKPSMVKTSSIKFTPSESVAKNSEDLIKKKSFYYKSYPPPPPP 333
LK++ V E K +AN +K + K SS + SE ++ E++
Sbjct: 58 LKSAKVPERKLQANGPVAKKAKKKASS---SDSEDSSEEEEEV----------------Q 98
Query: 334 PPPTMFYKSTFLKSRYGEKNGRNMSMGKSIEEGTNEKEDEDEEEKEYSTSSMQEQTDKER 393
PP K+ R G G+ + K+ E ++E+ +D++E++ +Q+ +
Sbjct: 99 GPPAK--KAAVPAKRVGLPPGK--AAAKASESSSSEESRDDDDEEDQKKQPVQKGVKPQA 154
Query: 394 FIEKRVILEAEETDSDED 411
K +A+ +DSD D
Sbjct: 155 KAAKAPPKKAKSSDSDSD 172
>RTN4_HUMAN (Q9NQC3) Reticulon 4 (Neurite outgrowth inhibitor) (Nogo
protein) (Foocen) (Neuroendocrine-specific protein)
(NSP) (Neuroendocrine specific protein C homolog)
(RTN-x) (Reticulon 5) (My043 protein)
Length = 1192
Score = 44.3 bits (103), Expect = 7e-04
Identities = 62/249 (24%), Positives = 101/249 (39%), Gaps = 24/249 (9%)
Query: 230 EGLVEDKLKQ--KEEENAVLPSPIPWRSRSASARMMEPKQEAIEKALKASMVMEPKQEAN 287
EGL D +++ + E N V + I + ++ + E QE++ A + E + EA
Sbjct: 548 EGLTPDLVQEACESELNEVTGTKIAYETKMDLVQTSEVMQESLYPAAQLCPSFE-ESEAT 606
Query: 288 ENASKPSMVKTSSIKFT-PSE--SVAKNSEDLIKKKSFYYKSYPPPPPPPPPTMFYKSTF 344
+ P +V + + PS SV + S ++ S Y+S P PPP S
Sbjct: 607 PSPVLPDIVMEAPLNSAVPSAGASVIQPSSSPLEASSVNYESIKHEPENPPPYEEAMSVS 666
Query: 345 LKSRYGEKNGRNMSMGKSIEEGTNEKEDEDEEEKEYSTSSMQEQTDKERFIEKRVILEAE 404
LK G K + I+E N E E Y + + + K L AE
Sbjct: 667 LKKVSGIK--------EEIKEPENINAALQETEAPYISIACD--------LIKETKLSAE 710
Query: 405 ET-DSDEDEDVGELKQSVKSVKTEEPCSSSLSGGIDTVSDEG-PDVDKKADEFIAKFREQ 462
D + ++ +++Q V SS S +D SD+ PDV +K DE + +E
Sbjct: 711 PAPDFSDYSEMAKVEQPVPDHSELVEDSSPDSEPVDLFSDDSIPDVPQKQDETVMLVKES 770
Query: 463 IRLQRIESI 471
+ ES+
Sbjct: 771 LTETSFESM 779
>NFM_HUMAN (P07197) Neurofilament triplet M protein (160 kDa
neurofilament protein) (Neurofilament medium
polypeptide) (NF-M) (Neurofilament 3)
Length = 915
Score = 44.3 bits (103), Expect = 7e-04
Identities = 76/348 (21%), Positives = 128/348 (35%), Gaps = 64/348 (18%)
Query: 91 EPDKDNNNNTKFDNAQSLVSRFLQVSSFFE-----DEVDHQNQSESEDINKIQTWSNQNQ 145
E K + ++ + A + ++ L S E +E + + ++E E++
Sbjct: 457 EETKVEDEKSEMEEALTAITEELAASMKEEKKEAAEEKEEEPEAEEEEVAA--------- 507
Query: 146 HYRNKPMVIVAPQVQQLQNSVFEDEQSSFNENEKPLLLPVRSLKSRLSDDDCVVQSQSVD 205
+ P+ AP+V++ + E+E E E D+ Q+ +
Sbjct: 508 --KKSPVKATAPEVKEEEGEKEEEEGQEEEEEE----------------DEGAKSDQAEE 549
Query: 206 GLSKTTTKRFSSNSFNRVRNYAEFEGLVEDKLKQKEEENAVLPSPIPWRSRSASARMMEP 265
G S+ K SS + E E E + + +EE V E
Sbjct: 550 GGSE---KEGSSEKEEGEQEEGETEAEAEGEEAEAKEEKKV-----------------EE 589
Query: 266 KQEAIEKALKASMVMEPKQEANENASKP---SMVKTSSIKFTPSESVAKNSEDLIKKKSF 322
K E E A K +V + K E E A P S V+ P V + + + K
Sbjct: 590 KSE--EVATKEELVADAKVEKPEKAKSPVPKSPVEEKGKSPVPKSPVEEKGKSPVPKSPV 647
Query: 323 YYKSYPPPPPPPPPTMFYKSTFLKSRYGEKNGRNMSMGKSIEEGTNEKE-----DEDEEE 377
K P P P KS KS EK ++ +EE ++ E ++EEE
Sbjct: 648 EEKGKSPVPKSPVEEKG-KSPVSKSPVEEK-AKSPVPKSPVEEAKSKAEVGKGEQKEEEE 705
Query: 378 KEYSTSSMQEQTDKERFIEKRVILEAEETDSDEDEDVGELKQSVKSVK 425
KE + +E+ +K+ K V + + ++E V E+ KSVK
Sbjct: 706 KEVKEAPKEEKVEKKEEKPKDVPEKKKAESPVKEEAVAEVVTITKSVK 753
Score = 31.6 bits (70), Expect = 4.8
Identities = 33/125 (26%), Positives = 54/125 (42%), Gaps = 15/125 (12%)
Query: 334 PPPTMFYKSTFLKSRYGEKNGRNMSMGKSIEEGTNEKEDEDEEEKEYSTSSMQEQTDKER 393
PP T+ K K + ++ + + IEE + EDE E +E T+ +E +
Sbjct: 427 PPITISSKIQKTKVEAPKLKVQHKFVEEIIEE--TKVEDEKSEMEEALTAITEELAASMK 484
Query: 394 FIEKRVILEAEETDSDEDEDVGELKQSVKS----VKTEEPCSSSLSGGIDTVSDEGPDVD 449
+K E EE E+E+V K VK+ VK EE + +EG + +
Sbjct: 485 EEKKEAAEEKEEEPEAEEEEVAAKKSPVKATAPEVKEEEG---------EKEEEEGQEEE 535
Query: 450 KKADE 454
++ DE
Sbjct: 536 EEEDE 540
Score = 30.8 bits (68), Expect = 8.1
Identities = 27/98 (27%), Positives = 42/98 (42%), Gaps = 4/98 (4%)
Query: 364 EEGTNEKEDEDEEEKEYSTSSMQEQTDKERFIEKRVILEAEETDSDEDEDVGEL---KQS 420
EEG E E+E +EE+E + +E EK E EE + +E E E +
Sbjct: 522 EEGEKE-EEEGQEEEEEEDEGAKSDQAEEGGSEKEGSSEKEEGEQEEGETEAEAEGEEAE 580
Query: 421 VKSVKTEEPCSSSLSGGIDTVSDEGPDVDKKADEFIAK 458
K K E S ++ + V+D + +KA + K
Sbjct: 581 AKEEKKVEEKSEEVATKEELVADAKVEKPEKAKSPVPK 618
Score = 30.8 bits (68), Expect = 8.1
Identities = 28/113 (24%), Positives = 50/113 (43%), Gaps = 7/113 (6%)
Query: 358 SMGKSIEEGTNEKEDE--DEEEKEYSTSSMQEQTDKERFIEKRVILEAEETDSDEDEDVG 415
SM + +E EKE+E EEE+ + S + T E E+ E E + +E+ED G
Sbjct: 482 SMKEEKKEAAEEKEEEPEAEEEEVAAKKSPVKATAPEVKEEEGEKEEEEGQEEEEEEDEG 541
Query: 416 ELKQSVKSVKTEEPCSSSLSGGIDTVSD-----EGPDVDKKADEFIAKFREQI 463
+ +E+ SS G + EG + + K ++ + + E++
Sbjct: 542 AKSDQAEEGGSEKEGSSEKEEGEQEEGETEAEAEGEEAEAKEEKKVEEKSEEV 594
>NFM_BOVIN (O77788) Neurofilament triplet M protein (160 kDa
neurofilament protein) (Neurofilament medium
polypeptide) (NF-M) (Fragment)
Length = 810
Score = 43.5 bits (101), Expect = 0.001
Identities = 76/362 (20%), Positives = 141/362 (37%), Gaps = 27/362 (7%)
Query: 113 LQVSSFFEDEVDHQNQSESEDINKIQTWSNQNQHYRNKPMVIVAPQVQQLQNSVFEDEQS 172
L+V F +E+ + + E E + + + V V +V++ + E+++
Sbjct: 331 LKVQHKFVEEIIEETKVEDEKSEMEEALTAITEELA----VSVKEEVKEEEAEEKEEKEE 386
Query: 173 SFNENEKPLLLPVRSLKSRLSDDDCVVQSQSVDGLSKTTTKRFSSNSFNRVRNYAEFEGL 232
+ E PV++ L +++ + + +G + + ++ S +E EG
Sbjct: 387 AEEEVVAAKKSPVKATAPELKEEEG--EKEEEEGQEEEEEEEEAAKSDQAEEGGSEKEGS 444
Query: 233 VEDKLKQKEEENAVLPSPIPWRSRSASARMMEPK---QEAIEKALKASMVMEPKQEANEN 289
E + ++EEE + A+A E K ++A E A K + E K E E
Sbjct: 445 SEKEEGEQEEEGE---TEAEGEGEEAAAEAKEEKKMEEKAEEVAPKEELAAEAKVEKPEK 501
Query: 290 ASKPSMVKTSSIKFTPSESVAKNSEDLIKKKSFYYKSYPPPPPPPPPTMFYKSTFLKSRY 349
A P ++ T AK+ E KS KS P P + KS KS
Sbjct: 502 AKSPVAKSPTTKSPTAKSPEAKSPE----AKSPTAKS-PTAKSPVAKSPTAKSPEAKSPE 556
Query: 350 GEKNGRNMSMGKSIEEGTNEKEDEDEEEKEYSTSSMQEQTDKERFIEKRVILEAEETDSD 409
+ KS + + EE K + + ++ KE+ E++ EA+E+ +
Sbjct: 557 AKSPTAKSPTAKSPAAKSPAPKSPVEEVKPKAEAGAEKGEQKEKVEEEKK--EAKESPKE 614
Query: 410 ED--------EDVGELKQSVKSVKTEEPCSSSLSGGIDTVSDEGPDVDKKADEFIAKFRE 461
E +DV E K++ VK E P + VS E +++ + K +E
Sbjct: 615 EKAEKKEEKPKDVPEKKKAESPVKAESPVKEEVPAKPVKVSPEKEAKEEEKPQEKEKEKE 674
Query: 462 QI 463
++
Sbjct: 675 KV 676
Score = 42.0 bits (97), Expect = 0.004
Identities = 43/203 (21%), Positives = 82/203 (40%), Gaps = 23/203 (11%)
Query: 226 YAEFEGLVEDKLKQKEEENAVLPSPIPWRSRSASARMMEPKQEAIEKALKASMVMEPKQE 285
++ F G + L + + + S I + A ++ + + +E+ ++ + V + K E
Sbjct: 296 FSTFAGSITGPLYTHRQPSIAISSKI--QKTKVEAPKLKVQHKFVEEIIEETKVEDEKSE 353
Query: 286 ANENASKPSMVKTSSIKFTPSESVAKNSEDLIKKKSFYYKSYPPPPPPPPPTMFYKSTFL 345
E + + S+K E A+ E+ + + + P P L
Sbjct: 354 MEEALTAITEELAVSVKEEVKEEEAEEKEEKEEAEEEVVAAKKSPVKATAPE-------L 406
Query: 346 KSRYGEKNGRNMSMGKSIEEGTNEKEDEDEEEKEYSTSSMQEQTDKERFIEKRVILEAEE 405
K GEK EE ++E+E+EEE S + + ++KE EK E E+
Sbjct: 407 KEEEGEK-----------EEEEGQEEEEEEEEAAKSDQAEEGGSEKEGSSEKE---EGEQ 452
Query: 406 TDSDEDEDVGELKQSVKSVKTEE 428
+ E E GE +++ K E+
Sbjct: 453 EEEGETEAEGEGEEAAAEAKEEK 475
Score = 32.7 bits (73), Expect = 2.1
Identities = 58/345 (16%), Positives = 131/345 (37%), Gaps = 20/345 (5%)
Query: 91 EPDKDNNNNTKFDNAQSLVSRFLQVSSFFEDEVDHQNQSESEDINKIQTWSNQNQHYRNK 150
E K + ++ + A + ++ L VS ++EV + E E+ + + + +
Sbjct: 343 EETKVEDEKSEMEEALTAITEELAVS--VKEEVKEEEAEEKEEKEEAE---EEVVAAKKS 397
Query: 151 PMVIVAPQVQQLQNSVFEDEQSSFNENEKPLLLPVRSLKSRLSDDDCVVQSQSVDGLSKT 210
P+ AP++++ + E+E E E+ +S ++ + S+ +G +
Sbjct: 398 PVKATAPELKEEEGEKEEEEGQEEEEEEEEA---AKSDQAEEGGSEKEGSSEKEEGEQEE 454
Query: 211 TTKRFSSNSFNRVRNYAEFEGLVEDKLKQ---KEEENAVLPSPIPWRSRSASARMMEPKQ 267
+ + A+ E +E+K ++ KEE A P +++S A+ K
Sbjct: 455 EGETEAEGEGEEAAAEAKEEKKMEEKAEEVAPKEELAAEAKVEKPEKAKSPVAKSPTTKS 514
Query: 268 EAIEKALKASMVMEPKQEANENASKPSMVKTSSIKFTPSESVAKNSEDLIKKKSFYYKSY 327
+ S E K ++ + S V S +P + K + +
Sbjct: 515 PTAKSPEAKSP--EAKSPTAKSPTAKSPVAKSPTAKSPEAKSPEAKSPTAKSPTAKSPAA 572
Query: 328 PPPPPPPPPTMFYKSTFLKSRYGEKNGRNMSMGKSIEEGTNEKEDE-------DEEEKEY 380
P P P + GE+ + K +E E++ E D EK+
Sbjct: 573 KSPAPKSPVEEVKPKAEAGAEKGEQKEKVEEEKKEAKESPKEEKAEKKEEKPKDVPEKKK 632
Query: 381 STSSMQEQTDKERFIEKRVILEAEETDSDEDEDVGELKQSVKSVK 425
+ S ++ ++ + + + + + E ++ E+E E ++ + V+
Sbjct: 633 AESPVKAESPVKEEVPAKPVKVSPEKEAKEEEKPQEKEKEKEKVE 677
Score = 31.2 bits (69), Expect = 6.2
Identities = 40/192 (20%), Positives = 74/192 (37%), Gaps = 38/192 (19%)
Query: 234 EDKLKQKEEENAVLPSPIPWRSRSASARMMEP--KQEAIEKALKASMVMEPKQEANENAS 291
E+K ++KEE+ P +P + ++ S E K+E K +K S E K+E
Sbjct: 614 EEKAEKKEEK----PKDVPEKKKAESPVKAESPVKEEVPAKPVKVSPEKEAKEEEKPQEK 669
Query: 292 KPSMVKTSSI--------KFTPSESVAKNSEDLIKKKSFYYKSYPPPPPPPPPTMFYKST 343
+ K + K + E +A N E K++ + T
Sbjct: 670 EKEKEKVEEVGGKEEGGLKESRKEDIAINGEVEGKEEE-------------------QET 710
Query: 344 FLKSRYGEKNGRNMSMGKSIEEGTNEKEDEDEEEKEYSTSSMQEQTDK-----ERFIEKR 398
K GE+ ++ G + G +K + EEK T +++ T + ++I K
Sbjct: 711 KEKGSGGEEEKGVVTNGLDVSPGDEKKGGDKSEEKVVVTKMVEKITSEGGDGATKYITKS 770
Query: 399 VILEAEETDSDE 410
V + + + +E
Sbjct: 771 VTVTQKVEEHEE 782
>NSR1_YEAST (P27476) Nuclear localization sequence binding protein
(P67)
Length = 414
Score = 43.1 bits (100), Expect = 0.002
Identities = 44/188 (23%), Positives = 73/188 (38%), Gaps = 29/188 (15%)
Query: 258 ASARMMEPKQEAIEKALKASMVMEPKQEANENASKPSMVKTSSIKFTPSESVAKNSEDLI 317
AS + E K +A+ + S +E+ S+ +SS + SES + +S D
Sbjct: 16 ASKQAKEEKAKAVSSSSSESSSSSSSSSESESESESESESSSSSSSSDSESSSSSSSDSE 75
Query: 318 KKKSFYYKSYPPPPPPPPPTMFYKSTFLKSRYGEKNGRNMSMGKSIEEGTNEKED---ED 374
+ K + + + S S EE EKE+ E+
Sbjct: 76 SEAE-----------------------TKKEESKDSSSSSSDSSSDEEEEEEKEETKKEE 112
Query: 375 EEEKEYSTSSMQEQTDKERFIEKRVILEAEETDSDEDEDVGELKQSVKSVKTEEPCS--- 431
+E S SS +D E E+ + + D++E+ED + K+ +TEEP +
Sbjct: 113 SKESSSSDSSSSSSSDSESEKEESNDKKRKSEDAEEEEDEESSNKKQKNEETEEPATIFV 172
Query: 432 SSLSGGID 439
LS ID
Sbjct: 173 GRLSWSID 180
Score = 37.0 bits (84), Expect = 0.11
Identities = 26/122 (21%), Positives = 49/122 (39%), Gaps = 8/122 (6%)
Query: 341 KSTFLKSRYGEKNGRNMSMGKSIEEGTNEKE--------DEDEEEKEYSTSSMQEQTDKE 392
K+ + S E + + S +S E +E E D + S S + +T KE
Sbjct: 24 KAKAVSSSSSESSSSSSSSSESESESESESESSSSSSSSDSESSSSSSSDSESEAETKKE 83
Query: 393 RFIEKRVILEAEETDSDEDEDVGELKQSVKSVKTEEPCSSSLSGGIDTVSDEGPDVDKKA 452
+ +D +E+E+ E K+ + SSS S ++ +E D +K+
Sbjct: 84 ESKDSSSSSSDSSSDEEEEEEKEETKKEESKESSSSDSSSSSSSDSESEKEESNDKKRKS 143
Query: 453 DE 454
++
Sbjct: 144 ED 145
>A3B2_HUMAN (Q13367) Adapter-related protein complex 3 beta 2
subunit (Beta3B-adaptin) (Adaptor protein complex AP-3
beta-2 subunit) (AP-3 complex beta-2 subunit) (Clathrin
assembly protein complex 3 beta-2 large chain)
(Neuron-specific vesicle coat prote
Length = 1082
Score = 43.1 bits (100), Expect = 0.002
Identities = 36/126 (28%), Positives = 52/126 (40%), Gaps = 6/126 (4%)
Query: 288 ENASKPSMVKTSSIKFTPSESVAKNSEDLIKKKSFYYKSYPPPPPPPPPTMFYKSTFLKS 347
E A PS+ ++T + K E K+K FY S P +S +S
Sbjct: 646 EEAPDPSVRNVEVPEWTKCSNREKRKE---KEKPFYSDSEGESGPTESADSDPES---ES 699
Query: 348 RYGEKNGRNMSMGKSIEEGTNEKEDEDEEEKEYSTSSMQEQTDKERFIEKRVILEAEETD 407
K+ G+S E NE +DEDEE+ S S E+ K + +K + E +
Sbjct: 700 ESDSKSSSESGSGESSSESDNEDQDEDEEKGRGSESEQSEEDGKRKTKKKVPERKGEASS 759
Query: 408 SDEDED 413
SDE D
Sbjct: 760 SDEGSD 765
Score = 35.4 bits (80), Expect = 0.33
Identities = 47/177 (26%), Positives = 66/177 (36%), Gaps = 21/177 (11%)
Query: 290 ASKPSMVKTSSIKFTPSESVAKNSEDLIKKKSFYYKSYPPPPPPPP---------PTMFY 340
A KP+ V SS K + S L+ K+ Y+ P P P P
Sbjct: 605 APKPAPVLESSFKDRDHFQLGSLSH-LLNAKATGYQELPDWPEEAPDPSVRNVEVPEWTK 663
Query: 341 KSTFLKSRYGEKNGRNMSMGKSIEEGTNEKEDEDEEEKEYSTSSMQEQTDKERFIEKRVI 400
S K + EK + S G+S G E D D E + S S ++
Sbjct: 664 CSNREKRKEKEKPFYSDSEGES---GPTESADSDPESESESDSKSSSESGSGESSS---- 716
Query: 401 LEAEETDSDEDEDVG---ELKQSVKSVKTEEPCSSSLSGGIDTVSDEGPDVDKKADE 454
E++ D DEDE+ G E +QS + K + G + SDEG D + E
Sbjct: 717 -ESDNEDQDEDEEKGRGSESEQSEEDGKRKTKKKVPERKGEASSSDEGSDSSSSSSE 772
>Y5G8_ENCCU (Q8STA9) Hypothetical protein ECU05_1680/ECU11_0050
Length = 612
Score = 42.4 bits (98), Expect = 0.003
Identities = 28/98 (28%), Positives = 44/98 (44%)
Query: 352 KNGRNMSMGKSIEEGTNEKEDEDEEEKEYSTSSMQEQTDKERFIEKRVILEAEETDSDED 411
K + M K EE E+E + EEEK +++ KE +K+ + E+ + ++
Sbjct: 302 KERQRREMEKKEEEKKKEEEKKKEEEKRKEEKKKKKEEKKEEKKKKKEEKKEEKKEEKKE 361
Query: 412 EDVGELKQSVKSVKTEEPCSSSLSGGIDTVSDEGPDVD 449
E E K+ K K EE SL G + E P V+
Sbjct: 362 EKKEEKKEEKKEEKKEEKSGKSLREGEASEEAEMPSVE 399
>UBF1_HUMAN (P17480) Nucleolar transcription factor 1 (Upstream
binding factor 1) (UBF-1) (Autoantigen NOR-90)
Length = 764
Score = 42.4 bits (98), Expect = 0.003
Identities = 55/227 (24%), Positives = 87/227 (38%), Gaps = 17/227 (7%)
Query: 234 EDKLKQKEEENAVLPSPIPWRSRSASARMMEPKQEAIEKALKASMVMEPKQEANENASKP 293
ED+ + + E + + P S EPK+ + K S + E N K
Sbjct: 536 EDQKRYERELSEMRAPPAATNSSKKMKFQGEPKKPPMNGYQKFSQELLSNGELNHLPLKE 595
Query: 294 SMVKTSSIKFTPSESVAKNSEDLIKKKSFYYKSYPP---PPPPPPPTMFYKSTFLKSRYG 350
MV+ S S+S ++ + L +++ YK + P YK R
Sbjct: 596 RMVEIGSRWQRISQSQKEHYKKLAEEQQKQYKVHLDLWVKSLSPQDRAAYKEYISNKRKS 655
Query: 351 --------EKNGRN--MSMGKSIEEGTNEKEDEDEEEKEYSTSSMQEQTDKERFIEKRVI 400
K+ R S +S E+ +++DEDE+E+E + D E
Sbjct: 656 MTKLRGPNPKSSRTTLQSKSESEEDDEEDEDDEDEDEEEEDDENGDSSEDGGDSSESSSE 715
Query: 401 LEAEETD----SDEDEDVGELKQSVKSVKTEEPCSSSLSGGIDTVSD 443
E+E+ D DEDED E + ++E SSS S G + SD
Sbjct: 716 DESEDGDENEEDDEDEDDDEDDDEDEDNESEGSSSSSSSSGDSSDSD 762
>RPG1_MOUSE (Q9EPQ2) X-linked retinitis pigmentosa GTPase
regulator-interacting protein 1 (RPGR-interacting
protein 1)
Length = 1331
Score = 42.4 bits (98), Expect = 0.003
Identities = 25/65 (38%), Positives = 38/65 (58%), Gaps = 2/65 (3%)
Query: 364 EEGTNEKEDEDEEEKEYSTSSMQEQTDKERFIEKRVILEAEETDSDEDEDVGELKQSVKS 423
EE EKE+E+EEE+E +E+ ++E EK E EE + +E+ED E K +++
Sbjct: 937 EEVKEEKEEEEEEEREEEEEKEEEKEEEEEEDEKEEEEEEEEEEEEEEED--ENKDVLEA 994
Query: 424 VKTEE 428
TEE
Sbjct: 995 SFTEE 999
Score = 38.1 bits (87), Expect = 0.051
Identities = 29/117 (24%), Positives = 50/117 (41%), Gaps = 7/117 (5%)
Query: 346 KSRYGEKNGRNMSMGKSIEEGTNEKEDEDEEEKEYSTSSMQEQTDKERFIEKRVILEAEE 405
KS Y G MS + E E+E +EE KE +E+ ++E +++ E EE
Sbjct: 890 KSHYLAPEGFQMSEAEKPEGEEKEEEGGEEEVKEEEVEEEEEEEEEEEEVKEEKEEEEEE 949
Query: 406 TDSDEDEDVGELKQSVKSVKTEEPCSSSLSGGIDTVSDEGPDVDKKADEFIAKFREQ 462
+E+E E ++ + + EE + +E + D+ D A F E+
Sbjct: 950 EREEEEEKEEEKEEEEEEDEKEEEEE-------EEEEEEEEEEDENKDVLEASFTEE 999
>GARP_PLAFF (P13816) Glutamic acid-rich protein precursor
Length = 678
Score = 42.4 bits (98), Expect = 0.003
Identities = 31/133 (23%), Positives = 58/133 (43%), Gaps = 2/133 (1%)
Query: 346 KSRYGEKNGRNMSMGKSIEEGTNEKEDEDEEEKEYSTSSMQEQTDKERFIEKRVILEAEE 405
K ++ +K K ++E + E ++++EE +E +E+ ++E E+ E EE
Sbjct: 541 KQKHVDKEEDKKEESKEVQEESKEVQEDEEEVEEDEEEEEEEEEEEEEEEEEEEEEEEEE 600
Query: 406 TDSDEDEDVGELKQSVKSVKTEEPCSSSLSGGIDTVSD--EGPDVDKKADEFIAKFREQI 463
+ +EDED + + + E D D E D D+ DE + E+
Sbjct: 601 EEEEEDEDEEDEDDAEEDEDDAEEDEDDAEEDDDEEDDDEEDDDEDEDEDEEDEEEEEEE 660
Query: 464 RLQRIESIKRSTR 476
+ + IKR+ R
Sbjct: 661 EEESEKKIKRNLR 673
>EBP2_YEAST (P36049) rRNA processing protein EBP2 (EBNA1-binding
protein homolog)
Length = 427
Score = 42.0 bits (97), Expect = 0.004
Identities = 44/183 (24%), Positives = 75/183 (40%), Gaps = 10/183 (5%)
Query: 267 QEAIEKALKASMVMEPKQEANENASKPSMVKTSSIKFTPSESVAKNSEDLIKKKSFYYKS 326
Q+ IEKA K ++ K+ +P++V S++K + + D+ K+ + +
Sbjct: 14 QKEIEKAEKLENDLKKKKSQELKKEEPTIVTASNLKKLEKK---EKKADVKKEVAADTEE 70
Query: 327 YPPPPPPPPPTMFYKSTFLKSRYGEKNGRNMSM-GKSIEEGTNEKEDEDEEEKEYSTSSM 385
Y K K + + M G E G + +E+E+EEE+E +
Sbjct: 71 YQSQALSKKEKRKLKKELKKMQEQDATEAQKHMSGDEDESGDDREEEEEEEEEEEGRLDL 130
Query: 386 QEQTDKERFIEKRVILEAEETDSDEDEDVGELKQSVKSVKTEEPCSSSLSGGIDTVSDEG 445
++ + E E DS+EDEDV ++S + + EE LS D D
Sbjct: 131 EKLAKSDSESEDD---SESENDSEEDEDVVAKEESEEKEEQEEEQDVPLS---DVEFDSD 184
Query: 446 PDV 448
DV
Sbjct: 185 ADV 187
>TPM2_DROME (P09491) Tropomyosin 2 (Tropomyosin I)
Length = 284
Score = 41.6 bits (96), Expect = 0.005
Identities = 55/242 (22%), Positives = 107/242 (43%), Gaps = 31/242 (12%)
Query: 89 NQEPDKDNNNNTKFDNAQSLVSRFLQVSSFF---EDEVDHQNQSESEDINKIQTWSNQNQ 145
NQ D ++ + + + L +F+QV +++++ N +E E+ K+ T +
Sbjct: 27 NQAKDANSRADKLNEEVRDLEKKFVQVEIDLVTAKEQLEKAN-TELEEKEKLLTATESEV 85
Query: 146 HYRNKPMVIVAPQVQQLQNSVFEDEQSSFNENEKPLLLPVRSLKSRLSDDDCVVQSQSVD 205
+N+ +VQQ++ + + E+ S +K L++ S D+ + ++
Sbjct: 86 ATQNR-------KVQQIEEDLEKSEERSTTAQQK-------LLEATQSADENNRMCKVLE 131
Query: 206 GLSKTTTKRFS--SNSFNRVRNYAEFEGLVEDKLKQK----EEENAVLPSPIPWRSRSAS 259
S+ +R +N R AE D++ +K E+E V R RS
Sbjct: 132 NRSQQDEERMDQLTNQLKEARMLAEDADTKSDEVSRKLAFVEDELEVAED----RVRSGE 187
Query: 260 ARMMEPKQE--AIEKALKASMVMEPKQEANENASKPSMVKTSSIKFTPSESVAKNSEDLI 317
+++ME ++E + +LK+ V E K K M KT SIK +E A+++E +
Sbjct: 188 SKIMELEEELKVVGNSLKSLEVSEEKANQRVEEFKREM-KTLSIKLKEAEQRAEHAEKQV 246
Query: 318 KK 319
K+
Sbjct: 247 KR 248
>NUCL_MOUSE (P09405) Nucleolin (Protein C23)
Length = 706
Score = 41.6 bits (96), Expect = 0.005
Identities = 46/201 (22%), Positives = 77/201 (37%), Gaps = 33/201 (16%)
Query: 223 VRNYAEFEGLVEDKLKQKEEENAVLPSPIPWRSRSASARMMEPKQEAIEKALKASMVMEP 282
V +E E + ED+ EE V+P + P ++ + K + V P
Sbjct: 23 VEEDSEDEEMSEDEDDSSGEEEVVIPQ------KKGKKATTTPAKKVVVSQTKKAAVPTP 76
Query: 283 KQEANENASKPSMVKTSSIKFTPSESVAKNSEDLIKKKSFYYKSYPPPPPPPPPTMFYKS 342
++A K ++ + TP++ + KK + K+ P P K
Sbjct: 77 AKKAAVTPGKKAVATPAKKNITPAKVIPTPG----KKGAAQAKALVPTPG--------KK 124
Query: 343 TFLKSRYGEKNGRNMSMGKSIEEGTNEKEDEDEEEKEYSTSSMQEQTDKERFIEKRV--- 399
G KNG+N S E+ E ED+ +E+++ +E+ + E I K V
Sbjct: 125 GAATPAKGAKNGKNAKKEDSDEDEDEEDEDDSDEDED-----DEEEDEFEPPIVKGVKPA 179
Query: 400 -------ILEAEETDSDEDED 413
E EE D DED++
Sbjct: 180 KAAPAAPASEDEEDDEDEDDE 200
Score = 32.0 bits (71), Expect = 3.7
Identities = 19/57 (33%), Positives = 32/57 (55%), Gaps = 7/57 (12%)
Query: 364 EEGTNEKEDEDEEEKEYSTSSMQE-QTDKERFIEKRVI------LEAEETDSDEDED 413
+E +++ED+DEEE++ S + E T K + +V+ + EE D +EDED
Sbjct: 194 DEDEDDEEDDDEEEEDDSEEEVMEITTAKGKKTPAKVVPMKAKSVAEEEDDEEEDED 250
>NAB3_YEAST (P38996) Nuclear polyadenylated RNA-binding protein 3
Length = 802
Score = 41.6 bits (96), Expect = 0.005
Identities = 35/138 (25%), Positives = 64/138 (46%), Gaps = 26/138 (18%)
Query: 346 KSRYGEKNGRNMSMGKSIEEGTNEKEDEDEEEKEYSTSSMQEQTDKERFIEKRVILEAEE 405
K GE+NG E N +E+E+EE ++ + + ++E E EE
Sbjct: 73 KEEKGEENG----------EVINTEEEEEEEHQQKGGNDDDDDDNEE---------EEEE 113
Query: 406 TDSDEDEDVGELKQSVKSVKTEEPCSSSLSGGIDTVSDEGPD----VDKKADEFIAKFRE 461
+ D+D+D + + + EE + + S G D+ +++G D DK D+ + RE
Sbjct: 114 EEDDDDDDDDDDDDEEEEEEEEEEGNDNSSVGSDSAAEDGEDEEDKKDKTKDKEVELRRE 173
Query: 462 QIRLQR---IESIKRSTR 476
+ ++ E+IK+ TR
Sbjct: 174 TLEKEQKDVDEAIKKITR 191
>FUTS_DROME (Q9W596) Microtubule-associated protein futsch
Length = 5412
Score = 41.6 bits (96), Expect = 0.005
Identities = 64/278 (23%), Positives = 114/278 (40%), Gaps = 24/278 (8%)
Query: 192 LSDDDCVVQSQSVDGLSKTTTKRFSSNSFNRVRNYAEFEGLVEDKLKQKEE---ENAVLP 248
LS + QS+ G K +S +R + AE + ++ K KEE E+
Sbjct: 1922 LSRPESTTQSKEA-GSIKDEKSPLASEEASRPASVAE--SVKDEAEKSKEESRRESVAEK 1978
Query: 249 SPIPWRSRSASARMMEPKQEAIEKALKAS---MVMEPKQEANENASKPSMVKTSSIKFTP 305
SP+P + S A + E ++ EK+ + S V E ++ AS+P+ V SIK
Sbjct: 1979 SPLPSKEASRPASVAESIKDEAEKSKEESRRESVAEKSPLPSKEASRPASV-AESIK--- 2034
Query: 306 SESVAKNSEDLIKKKSFYYKSYPPPPPPPPPTMFYKSTFLKSRYGEKNGRNMSMGKSIEE 365
A+ S++ +++S KS P P +S ++ ++ R S+ +
Sbjct: 2035 --DEAEKSKEESRRESVAEKSPLPSKEASRPASVAESIKDEAEKSKEESRRESVAEKSPL 2092
Query: 366 GTNE--------KEDEDEEEKEYSTSSMQEQTDKERFIEKRVILEAEETDSDEDEDVGEL 417
+ E + +DE EK S + +K K A +S +DE
Sbjct: 2093 PSKEASRPASVAESIKDEAEKSKEESRRESVAEKSPLPSKEASRPASVAESIKDEAEKSK 2152
Query: 418 KQSVK-SVKTEEPCSSSLSGGIDTVSDEGPDVDKKADE 454
++S + SV + P S + +V++ D +K+ E
Sbjct: 2153 EESRRESVAEKSPLPSKEASRPASVAESIKDEAEKSKE 2190
Score = 40.4 bits (93), Expect = 0.010
Identities = 58/261 (22%), Positives = 106/261 (40%), Gaps = 29/261 (11%)
Query: 230 EGLVEDKLKQKEE---ENAVLPSPIPWRSRSASARMMEPKQEAIEKALKASM---VMEPK 283
E + ++ K KEE E+ SP+P + S A + E ++ EK+ + S V E
Sbjct: 2105 ESIKDEAEKSKEESRRESVAEKSPLPSKEASRPASVAESIKDEAEKSKEESRRESVAEKS 2164
Query: 284 QEANENASKPSMVKTSSIKFTPSESVAKNSEDLIKKKSFYYKSYPPPPPPPPPTMFYKST 343
++ AS+P+ V SIK A+ S++ +++S KS P P +S
Sbjct: 2165 PLPSKEASRPASV-AESIK-----DEAEKSKEESRRESVAEKSPLPSKEASRPASVAESI 2218
Query: 344 FLKSRYGEKNGRNMSMGKSIEEGTNE--------KEDEDEEEKEYSTSSMQEQTDKERFI 395
++ ++ R S+ + + E + +DE EK S + +K
Sbjct: 2219 KDEAEKSKEETRRESVAEKSPLPSKEASRPASVAESIKDEAEKSKEESRRESAAEKSPLP 2278
Query: 396 EKRVILEAEETDSDEDEDVGELKQSVKSVKTEEPCSSSLSGGIDTVSDEGPDVDKKADEF 455
K A +S +DE ++S + E + S+ G D+ P + E
Sbjct: 2279 SKEASRPASVAESVKDEADKSKEESRRESMAESGKAQSIKG------DQSPLKEVSRPES 2332
Query: 456 IAKFREQIRLQRIESIKRSTR 476
+A E ++ ++S + S R
Sbjct: 2333 VA---ESVKDDPVKSKEPSRR 2350
Score = 39.7 bits (91), Expect = 0.018
Identities = 54/240 (22%), Positives = 100/240 (41%), Gaps = 21/240 (8%)
Query: 230 EGLVEDKLKQKEE---ENAVLPSPIPWRSRSASARMMEPKQEAIEKALKAS---MVMEPK 283
E + ++ K KEE E+ SP+P + S A + E ++ EK+ + S V E
Sbjct: 2031 ESIKDEAEKSKEESRRESVAEKSPLPSKEASRPASVAESIKDEAEKSKEESRRESVAEKS 2090
Query: 284 QEANENASKPSMVKTSSIKFTPSESVAKNSEDLIKKKSFYYKSYPPPPPPPPPTMFYKST 343
++ AS+P+ V SIK A+ S++ +++S KS P P +S
Sbjct: 2091 PLPSKEASRPASV-AESIK-----DEAEKSKEESRRESVAEKSPLPSKEASRPASVAESI 2144
Query: 344 FLKSRYGEKNGRNMSMGKSIEEGTNE--------KEDEDEEEKEYSTSSMQEQTDKERFI 395
++ ++ R S+ + + E + +DE EK S + +K
Sbjct: 2145 KDEAEKSKEESRRESVAEKSPLPSKEASRPASVAESIKDEAEKSKEESRRESVAEKSPLP 2204
Query: 396 EKRVILEAEETDSDEDE-DVGELKQSVKSVKTEEPCSSSLSGGIDTVSDEGPDVDKKADE 454
K A +S +DE + + + +SV + P S + +V++ D +K+ E
Sbjct: 2205 SKEASRPASVAESIKDEAEKSKEETRRESVAEKSPLPSKEASRPASVAESIKDEAEKSKE 2264
Score = 39.3 bits (90), Expect = 0.023
Identities = 73/378 (19%), Positives = 142/378 (37%), Gaps = 32/378 (8%)
Query: 128 QSESEDINKIQTWSNQNQHYRNKPMVIV-APQVQQLQNSVFEDEQSSFNENEKPLLLPVR 186
+S +D K + S + P+ A + + SV ++ + S E+ + +
Sbjct: 3659 ESVKDDAEKSKEESRRESVAEKSPLASKEASRPASVAESVKDEAEKSKEESRRESVAEKS 3718
Query: 187 SLKSRLSDDDCVVQSQSVDGLSKT--------TTKRFSSNSFNRVRNYAEFEGLVEDKLK 238
L S+ + V D K+ ++ S S R + E + ++ K
Sbjct: 3719 PLPSKEASRPTSVAESVKDEAEKSKEESRRESVAEKSSLASKKASRPASVAESVKDEAEK 3778
Query: 239 QKEE---ENAVLPSPIPWRSRSASARMMEPKQEAIEKALKASM---VMEPKQEANENASK 292
KEE E+ SP+ + S A + E ++ EK+ + S V E ++ AS+
Sbjct: 3779 SKEESRRESVAEKSPLASKEASRPASVAESVKDEAEKSKEESRRESVAEKSPLPSKEASR 3838
Query: 293 PSMVKTSSIKFTPSESVAKNSEDLIKKKSFYYKSYPPPPPPPPPTMFYKSTFLKSRYGEK 352
P+ V S + A S++ +++S KS PT +S ++ ++
Sbjct: 3839 PTSVAESV------KDEADKSKEESRRESGAEKSPLASMEASRPTSVAESVKDETEKSKE 3892
Query: 353 NGRNMSM-------GKSIEEGTNEKED-EDEEEKEYSTSSMQEQTDKERFIEKRVILEAE 404
R S+ K T+ E +DE EK S + +K K A
Sbjct: 3893 ESRRESVTEKSPLPSKEASRPTSVAESVKDEAEKSKEESRRESVAEKSPLASKESSRPAS 3952
Query: 405 ETDSDEDEDVGELKQSVKSVKTEEPCSSSLSGGIDTVSDEGPDVDKKADEFIAKFREQIR 464
+S +DE G ++S + E + S+ G +++ + + D + +++
Sbjct: 3953 VAESIKDEAEGTKQESRRESMPESGKAESIKGDQSSLASK---ETSRPDSVVESVKDETE 4009
Query: 465 LQRIESIKRSTRVARNSS 482
+I +S +R S
Sbjct: 4010 KPEGSAIDKSQVASRPES 4027
Score = 36.6 bits (83), Expect = 0.15
Identities = 72/372 (19%), Positives = 149/372 (39%), Gaps = 61/372 (16%)
Query: 86 SRRNQEPDKDNNNNTKFDNAQSLVSRFLQVSSFFEDEVDHQNQSESEDINKIQTWSNQNQ 145
+ +++E + + +F VSR V+ +DE + +++ ES ++K
Sbjct: 3304 AEKSKEESRRESVAEQFPLVSKEVSRPASVAESVKDEAE-KSKEESPLMSK--------- 3353
Query: 146 HYRNKPMVIVAPQVQQLQNSVFEDEQSSFNENEKPLLLPVRSLKSRLSD------DDCVV 199
++P + + + S E + S E + PL S + +++ D
Sbjct: 3354 -EASRPASVAGSVKDEAEKSKEESRRESVAE-KSPLPSKEASRPASVAESVKDEADKSKE 3411
Query: 200 QSQSVDGLSKTTTKRFSSNSFNRVRNYAEFEGLVEDKLKQKEE---ENAVLPSPIPWRSR 256
+S+ G K+ +S +R + AE + ++ K KEE E+ SP+P +
Sbjct: 3412 ESRRESGAEKSP---LASKEASRPASVAE--SIKDEAEKSKEESRRESVAEKSPLPSKEA 3466
Query: 257 SASARMMEPKQEAIEKALKASM---VMEPKQEANENASKPSMVKTSSIKFTPSESVAKNS 313
S + E ++ EK+ + S V E A++ AS+P+ V S + A+ S
Sbjct: 3467 SRPTSVAESVKDEAEKSKEESRRDSVAEKSPLASKEASRPASVAESV------QDEAEKS 3520
Query: 314 EDLIKKKSFYYKSYPPPPPPPPPTMFYKSTFLKSRYGEKNGRNMSMGKSIEEGTNEKEDE 373
++ +++S KS ++ R S+ +SI++ + ++E
Sbjct: 3521 KEESRRESVAEKS--------------------PLASKEASRPASVAESIKDEAEKSKEE 3560
Query: 374 DEEEKEYSTSSM-QEQTDKERFIEKRVILEAEETDSDEDEDVGELKQSVKSVKTEEPCSS 432
E S + ++ + + + V EAE++ + D K + S + P S
Sbjct: 3561 SRRESVAEKSPLASKEASRPTSVAESVKDEAEKSKEESSRDSVAEKSPLASKEASRPASV 3620
Query: 433 SLSGGIDTVSDE 444
+ ++V DE
Sbjct: 3621 A-----ESVQDE 3627
Score = 33.1 bits (74), Expect = 1.6
Identities = 33/129 (25%), Positives = 59/129 (45%), Gaps = 14/129 (10%)
Query: 361 KSIEEGTNEKEDEDE---EEKEYSTSSMQEQTDK----ERFIEKRVILEAEETDSDEDED 413
KS ++ T+ D DE EEK ST+ +E++DK E+ +E V +E E S D+
Sbjct: 4271 KSDKDITDIIPDFDERQLEEKLKSTADTEEESDKSTRDEKSLEISVKVEIESEKSSPDQK 4330
Query: 414 VGELKQSVKSVKTEEPCSSSLSGGI------DTVSDEGPDVDKKADEFIAKFREQIRLQR 467
G + K K E+ + L GI ++V+ + V + + +++ L
Sbjct: 4331 SGPISIEEKD-KIEQSEKAQLRQGILTSSRPESVASQPESVPSPSQSAASHEHKEVELSE 4389
Query: 468 IESIKRSTR 476
++S+R
Sbjct: 4390 SHKAEKSSR 4398
Score = 30.8 bits (68), Expect = 8.1
Identities = 19/66 (28%), Positives = 31/66 (46%), Gaps = 2/66 (3%)
Query: 365 EGTNEKEDEDEEEKEYSTSSMQ--EQTDKERFIEKRVILEAEETDSDEDEDVGELKQSVK 422
EGT + E+E +EE+EY + EQ ++ +E+ + EE D E ++ K
Sbjct: 926 EGTGDGENEPDEEEEYLIIEKEEVEQYTEDSIVEQESSMTKEEEIQKHQRDSQESEKKRK 985
Query: 423 SVKTEE 428
EE
Sbjct: 986 KSAEEE 991
>YSE2_CAEEL (Q09936) Hypothetical protein C53C9.2 in chromosome X
Length = 397
Score = 41.2 bits (95), Expect = 0.006
Identities = 25/83 (30%), Positives = 45/83 (54%), Gaps = 5/83 (6%)
Query: 336 PTMFYKSTFLK---SRYGEKNGRNMSMGKSIEEGTNEKEDEDEEEK--EYSTSSMQEQTD 390
P YK F + SR E + +++ + +E E+E+E+EEEK E + +E+ +
Sbjct: 314 PFGHYKKKFEERESSRQSEIDSQSVKASEPVEPEPEEEEEEEEEEKIEEPAAKEEEEEEE 373
Query: 391 KERFIEKRVILEAEETDSDEDED 413
+E E+ + E EE + +E+ED
Sbjct: 374 EEEEEEEEELEEEEEEEEEEEED 396
>USO1_YEAST (P25386) Intracellular protein transport protein USO1
Length = 1790
Score = 41.2 bits (95), Expect = 0.006
Identities = 66/361 (18%), Positives = 144/361 (39%), Gaps = 32/361 (8%)
Query: 126 QNQSESEDINKIQTWSNQNQHYRNKPMVIVAPQVQQLQNSVFEDEQSSFNENEKPLLLPV 185
+ +SESE +++++ S++ + + + + ++Q ++N FE E+ NE +
Sbjct: 1326 KEKSESE-LSRLKKTSSEERKNAEEQLEKLKNEIQ-IKNQAFEKERKLLNEGSSTITQEY 1383
Query: 186 RSLKSRLSDDDCVVQSQS------VDGLSKTTTKRFSSN------SFNRVRNYAEFEGLV 233
+ L D+ +Q+++ +D K SN N +++ +
Sbjct: 1384 SEKINTLEDELIRLQNENELKAKEIDNTRSELEKVSLSNDELLEEKQNTIKSLQDEILSY 1443
Query: 234 EDKLKQKEEE--NAVLPSPIPWRSRSASARMMEPKQEAIEKALKASMVMEPKQEANENAS 291
+DK+ + +E+ + + S R + + +E+ LK K++A S
Sbjct: 1444 KDKITRNDEKLLSIERDNKRDLESLKEQLRAAQESKAKVEEGLKKLEEESSKEKAELEKS 1503
Query: 292 KPSMVKTSSIKFTPSESVAKNSEDLIKKKSFYYKSYPPPPPPPPPTMFYKSTFLKSRYGE 351
K M K S +E+ K+S + I+K + + ++ + L SR E
Sbjct: 1504 KEMMKKLES-TIESNETELKSSMETIRKSDEKLEQSKKSAEEDIKNLQHEKSDLISRINE 1562
Query: 352 KNGRNMSMGKSIEEGTNEKEDEDEEEKEYSTSSMQEQTDKERF---IEKRVILEAEETDS 408
K IEE ++ E + E T + +E+ E+ +L+++ D
Sbjct: 1563 SE-------KDIEELKSKLRIEAKSGSELETVKQELNNAQEKIRINAEENTVLKSKLEDI 1615
Query: 409 DEDEDVGELKQSVKSVKTEEPCSSSLSGGIDTVSDEGPDVDKKADEFIAKFREQIRLQRI 468
+ ELK +K+ + L+ + + E +KA + + R ++R ++
Sbjct: 1616 ER-----ELKDKQAEIKSNQEEKELLTSRLKELEQELDSTQQKAQKSEEERRAEVRKFQV 1670
Query: 469 E 469
E
Sbjct: 1671 E 1671
Score = 36.2 bits (82), Expect = 0.19
Identities = 101/506 (19%), Positives = 208/506 (40%), Gaps = 64/506 (12%)
Query: 9 ITKHQKPTLDNVKQNQQEPNKFHYNFAYKATIVLIFFVILPLLPSQAPEFISQNLLTRNW 68
+++ + +N K Q+E +K + N K T L + + +A I++NL
Sbjct: 872 LSREMQAVEENCKNLQKEKDKSNVNHQ-KETKSLKEDIAAKITEIKA---INENLEEMKI 927
Query: 69 ELLHLLFVGIAISYGLFSRRNQEPDKDNNNNTKFDNAQSLVSRFLQVSSFFEDEVDHQNQ 128
+ +L IS L +++ DN + +SL + + + + E + +
Sbjct: 928 QCNNLSKEKEHISKELVEYKSRFQSHDNLVAKLTEKLKSLANNYKDMQAENESLIKAVEE 987
Query: 129 SESEDI-------NKIQTWSNQNQHYRNKPMVIVAPQVQQLQNSVFEDEQ---------- 171
S++E NKI + S + ++++ + I ++QL+ ++ + EQ
Sbjct: 988 SKNESSIQLSNLQNKIDSMSQEKENFQIERGSI-EKNIEQLKKTISDLEQTKEEIISKSD 1046
Query: 172 SSFNENEKPLLLPVRSLKSRLSDDDCVVQSQSVDGLSKTTTKRFSSNS----FNRVRNYA 227
SS +E E + L L++ + +D ++V+ +S+ T R + + ++N
Sbjct: 1047 SSKDEYESQISLLKEKLETATTAND-----ENVNKISELTKTREELEAELAAYKNLKNEL 1101
Query: 228 EFEGLVEDK----LKQKEE---------ENAVLPSPIPWRSRSASARMMEPKQEAIEKAL 274
E + +K +K+ EE E + S A+ +E + E + L
Sbjct: 1102 ETKLETSEKALKEVKENEEHLKEEKIQLEKEATETKQQLNSLRANLESLEKEHEDLAAQL 1161
Query: 275 KA--SMVMEPKQEANENASKPSMVKTSSIKFTPSESVAKNSEDL---IKKKSFYYKSYPP 329
K + +++ NE S+ + TS+ + +ES+ K +++L +K +
Sbjct: 1162 KKYEEQIANKERQYNEEISQLNDEITSTQQ--ENESIKKKNDELEGEVKAMKSTSEEQSN 1219
Query: 330 PPPPPPPTMFYKSTFLKSRYGEKNGRNMSMGKSIEEGTNE-KEDEDE---EEKEYST--- 382
+ + LK + + KS+E T + KE +DE +EKE S
Sbjct: 1220 LKKSEIDALNLQIKELKKKNETNEASLLESIKSVESETVKIKELQDECNFKEKEVSELED 1279
Query: 383 --SSMQEQTDKERFIEKRVILEAEETDSDEDEDVGELKQSVKSVKTEEPCSSSLSGGIDT 440
+ +++ K ++K EE D+ E +L++ K +E S LS T
Sbjct: 1280 KLKASEDKNSKYLELQKESEKIKEELDAKTTELKIQLEKITNLSKAKEKSESELSRLKKT 1339
Query: 441 VSDEGPDVDKKADEFIAKFREQIRLQ 466
S+E K A+E + K + +I+++
Sbjct: 1340 SSEER----KNAEEQLEKLKNEIQIK 1361
Score = 35.0 bits (79), Expect = 0.43
Identities = 77/388 (19%), Positives = 146/388 (36%), Gaps = 59/388 (15%)
Query: 100 TKFDNAQSLVSRFLQ-VSSFFEDEVDHQNQSESEDINKIQTWSNQNQHYRNKPMVIVAPQ 158
++F + +LV++ + + S + D Q ++ES I ++ N++ + Q
Sbjct: 948 SRFQSHDNLVAKLTEKLKSLANNYKDMQAENESL-IKAVEESKNES-----------SIQ 995
Query: 159 VQQLQNSV--FEDEQSSFNENEKPLLLPVRSLKSRLSDDDCVVQSQSVDGLSKTTTKRFS 216
+ LQN + E+ +F + + LK +SD + T +
Sbjct: 996 LSNLQNKIDSMSQEKENFQIERGSIEKNIEQLKKTISD-------------LEQTKEEII 1042
Query: 217 SNSFNRVRNYAEFEGLVEDKLKQK---EEENAVLPSPIPWRSRSASARM---------ME 264
S S + Y L+++KL+ +EN S + A + +E
Sbjct: 1043 SKSDSSKDEYESQISLLKEKLETATTANDENVNKISELTKTREELEAELAAYKNLKNELE 1102
Query: 265 PKQEAIEKALKASMVMEPKQEANENASKPSMVKTSSIKFTPS-----ESVAKNSEDLIKK 319
K E EKALK V E ++ E + T + + S ES+ K EDL +
Sbjct: 1103 TKLETSEKALKE--VKENEEHLKEEKIQLEKEATETKQQLNSLRANLESLEKEHEDLAAQ 1160
Query: 320 KSFYYKSYPPPPPPPPPTMFYKSTFLKSRYGEKNGRNMSMGKSIEEGTNEKEDEDEEEKE 379
Y + + K + N S + E +K DE E E +
Sbjct: 1161 LKKYEEQ-----------IANKERQYNEEISQLNDEITSTQQE-NESIKKKNDELEGEVK 1208
Query: 380 YSTSSMQEQTDKERFIEKRVILEAEETDSDEDEDVGELKQSVKSVKTEEPCSSSLSGGID 439
S+ +EQ++ ++ + L+ +E + + L +S+KSV++E L +
Sbjct: 1209 AMKSTSEEQSNLKKSEIDALNLQIKELKKKNETNEASLLESIKSVESETVKIKELQDECN 1268
Query: 440 TVSDEGPDVDKKADEFIAKFREQIRLQR 467
E +++ K K + + LQ+
Sbjct: 1269 FKEKEVSELEDKLKASEDKNSKYLELQK 1296
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.309 0.126 0.342
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 56,211,684
Number of Sequences: 164201
Number of extensions: 2540907
Number of successful extensions: 25132
Number of sequences better than 10.0: 659
Number of HSP's better than 10.0 without gapping: 165
Number of HSP's successfully gapped in prelim test: 518
Number of HSP's that attempted gapping in prelim test: 19932
Number of HSP's gapped (non-prelim): 2729
length of query: 483
length of database: 59,974,054
effective HSP length: 114
effective length of query: 369
effective length of database: 41,255,140
effective search space: 15223146660
effective search space used: 15223146660
T: 11
A: 40
X1: 16 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.7 bits)
S2: 68 (30.8 bits)
Medicago: description of AC149079.6


