

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC149079.10 - phase: 0
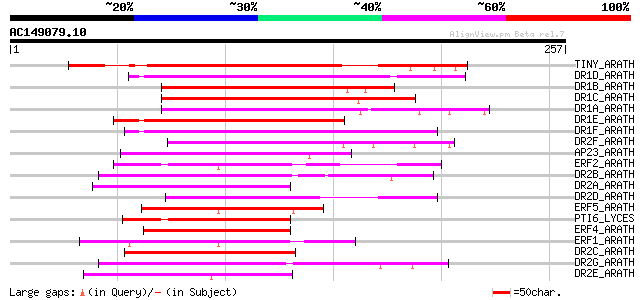
(257 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TINY_ARATH (Q39127) Transcriptional factor TINY 159 6e-39
DR1D_ARATH (Q9FJ93) Dehydration responsive element binding prote... 119 7e-27
DR1B_ARATH (P93835) Dehydration responsive element binding prote... 108 1e-23
DR1C_ARATH (Q9SYS6) Dehydration responsive element binding prote... 106 6e-23
DR1A_ARATH (Q9M0L0) Dehydration responsive element binding prote... 105 1e-22
DR1E_ARATH (Q9SGJ6) Dehydration responsive element binding prote... 102 7e-22
DR1F_ARATH (Q9LN86) Dehydration responsive element binding prote... 101 2e-21
DR2F_ARATH (Q9SVX5) Dehydration responsive element binding prote... 93 7e-19
AP23_ARATH (P42736) AP2 domain transcription factor RAP2.3 (Rela... 90 6e-18
ERF2_ARATH (O80338) Ethylene responsive element binding factor 2... 88 2e-17
DR2B_ARATH (O82133) Dehydration responsive element binding prote... 87 3e-17
DR2A_ARATH (O82132) Dehydration responsive element binding prote... 86 9e-17
DR2D_ARATH (Q9LQZ2) Putative dehydration responsive element bind... 86 1e-16
ERF5_ARATH (O80341) Ethylene responsive element binding factor 5... 79 1e-14
PTI6_LYCES (O04682) Pathogenesis-related genes transcriptional a... 78 2e-14
ERF4_ARATH (O80340) Ethylene responsive element binding factor 4... 78 2e-14
ERF1_ARATH (O80337) Ethylene responsive element binding factor 1... 77 4e-14
DR2C_ARATH (Q8LFR2) Dehydration responsive element binding prote... 76 7e-14
DR2G_ARATH (P61827) Putative dehydration responsive element bind... 76 9e-14
DR2E_ARATH (O80917) Dehydration responsive element binding prote... 74 3e-13
>TINY_ARATH (Q39127) Transcriptional factor TINY
Length = 218
Score = 159 bits (402), Expect = 6e-39
Identities = 101/192 (52%), Positives = 118/192 (60%), Gaps = 39/192 (20%)
Query: 28 STSSNSSSSLEEEEEEEEATNTKCKEKKKKKVGMVNKVNDRKNNPTYRGVRMRQWGKWVS 87
ST S +S++ +E EEE KKK V D +P YRGVR R WGKWVS
Sbjct: 6 STKSWEASAVRQENEEE-----------KKK-----PVKDSGKHPVYRGVRKRNWGKWVS 49
Query: 88 EIREPRKKSRIWLGTFPTPDMAARAHDVAALTIKGSSAYLNFPELAAVLPRAASASPKDI 147
EIREPRKKSRIWLGTFP+P+MAARAHDVAAL+IKG+SA LNFP+LA PR +S SP+DI
Sbjct: 50 EIREPRKKSRIWLGTFPSPEMAARAHDVAALSIKGASAILNFPDLAGSFPRPSSLSPRDI 109
Query: 148 QVAAAKAAATVYNHAITDQIELEAEAEPSQAVSSSSS---SSSSSSSSHNE--GSSLKGE 202
QVAA KA A E SQ+ SSSSS SSS SSSS SS G
Sbjct: 110 QVAALKA----------------AHMETSQSFSSSSSLTFSSSQSSSSLESLVSSSATGS 153
Query: 203 DDM--FLNLPDL 212
+++ + LP L
Sbjct: 154 EELGEIVELPSL 165
>DR1D_ARATH (Q9FJ93) Dehydration responsive element binding protein
1D (DREB1D protein) (C-repeat binding factor 4)
(C-repeat/dehydration responsive element binding factor
4) (CRT/DRE binding factor 4)
Length = 224
Score = 119 bits (298), Expect = 7e-27
Identities = 68/156 (43%), Positives = 88/156 (55%), Gaps = 5/156 (3%)
Query: 56 KKKVGMVNKVNDRKNNPTYRGVRMRQWGKWVSEIREPRKKSRIWLGTFPTPDMAARAHDV 115
KK+ G K +P YRGVR R GKWV E+REP KKSRIWLGTFPT +MAARAHDV
Sbjct: 38 KKRAG--RKKFRETRHPIYRGVRQRNSGKWVCEVREPNKKSRIWLGTFPTVEMAARAHDV 95
Query: 116 AALTIKGSSAYLNFPELAAVLPRAASASPKDIQVAAAKAAATVYNHAITDQIELEAEAEP 175
AAL ++G SA LNF + A L + PK+IQ AA++AA N T+ + AEAE
Sbjct: 96 AALALRGRSACLNFADSAWRLRIPETTCPKEIQKAASEAAMAFQNETTTEGSKTAAEAEE 155
Query: 176 SQAVSSSSSSSSSSSSSHNEGSSLKGEDDMFLNLPD 211
+ + + G +D+ L +P+
Sbjct: 156 A---AGEGVREGERRAEEQNGGVFYMDDEALLGMPN 188
>DR1B_ARATH (P93835) Dehydration responsive element binding protein
1B (DREB1B protein) (C-repeat binding factor 1)
(C-repeat/dehydration responsive element binding factor
1) (CRT/DRE binding factor 1)
Length = 213
Score = 108 bits (270), Expect = 1e-23
Identities = 62/118 (52%), Positives = 75/118 (63%), Gaps = 10/118 (8%)
Query: 71 NPTYRGVRMRQWGKWVSEIREPRKKSRIWLGTFPTPDMAARAHDVAALTIKGSSAYLNFP 130
+P YRGVR R GKWVSE+REP KK+RIWLGTF T +MAARAHDVAAL ++G SA LNF
Sbjct: 45 HPIYRGVRQRNSGKWVSEVREPNKKTRIWLGTFQTAEMAARAHDVAALALRGRSACLNFA 104
Query: 131 ELAAVLPRAASASPKDIQVAAAKAA---------ATVYNHAI-TDQIELEAEAEPSQA 178
+ A L S KDIQ AAA+AA T NH + ++ +EA P Q+
Sbjct: 105 DSAWRLRIPESTCAKDIQKAAAEAALAFQDETCDTTTTNHGLDMEETMVEAIYTPEQS 162
>DR1C_ARATH (Q9SYS6) Dehydration responsive element binding protein
1C (DREB1C protein) (C-repeat binding factor 2)
(C-repeat/dehydration responsive element binding factor
2) (CRT/DRE binding factor 2)
Length = 216
Score = 106 bits (264), Expect = 6e-23
Identities = 59/121 (48%), Positives = 75/121 (61%), Gaps = 3/121 (2%)
Query: 71 NPTYRGVRMRQWGKWVSEIREPRKKSRIWLGTFPTPDMAARAHDVAALTIKGSSAYLNFP 130
+P YRGVR R GKWV E+REP KK+RIWLGTF T +MAARAHDVAA+ ++G SA LNF
Sbjct: 48 HPIYRGVRQRNSGKWVCELREPNKKTRIWLGTFQTAEMAARAHDVAAIALRGRSACLNFA 107
Query: 131 ELAAVLPRAASASPKDIQVAAAKAAATVYN---HAITDQIELEAEAEPSQAVSSSSSSSS 187
+ A L S K+IQ AAA+AA + H TD L+ E +A+ + S
Sbjct: 108 DSAWRLRIPESTCAKEIQKAAAEAALNFQDEMCHMTTDAHGLDMEETLVEAIYTPEQSQD 167
Query: 188 S 188
+
Sbjct: 168 A 168
>DR1A_ARATH (Q9M0L0) Dehydration responsive element binding protein
1A (DREB1A protein) (C-repeat binding factor 3)
(C-repeat/dehydration responsive element binding factor
3) (CRT/DRE binding factor 3)
Length = 216
Score = 105 bits (261), Expect = 1e-22
Identities = 71/162 (43%), Positives = 91/162 (55%), Gaps = 11/162 (6%)
Query: 71 NPTYRGVRMRQWGKWVSEIREPRKKSRIWLGTFPTPDMAARAHDVAALTIKGSSAYLNFP 130
+P YRGVR R GKWV E+REP KK+RIWLGTF T +MAARAHDVAAL ++G SA LNF
Sbjct: 48 HPIYRGVRRRNSGKWVCEVREPNKKTRIWLGTFQTAEMAARAHDVAALALRGRSACLNFA 107
Query: 131 ELAAVLPRAASASPKDIQVAAAKAAATVYNH---AITDQIELEAEAEPSQAVSSSSSSSS 187
+ A L S KDIQ AAA+AA + A TD + E +A+ ++ S +
Sbjct: 108 DSAWRLRIPESTCAKDIQKAAAEAALAFQDEMCDATTDH-GFDMEETLVEAIYTAEQSEN 166
Query: 188 S---SSSSHNEGSSLKGE--DDMFLNLPDLTLDLRH--SGDD 222
+ + E SL + M L LP + + H GDD
Sbjct: 167 AFYMHDEAMFEMPSLLANMAEGMLLPLPSVQWNHNHEVDGDD 208
>DR1E_ARATH (Q9SGJ6) Dehydration responsive element binding protein
1E (DREB1E protein)
Length = 181
Score = 102 bits (255), Expect = 7e-22
Identities = 54/107 (50%), Positives = 68/107 (63%), Gaps = 2/107 (1%)
Query: 49 TKCKEKKKKKVGMVNKVNDRKNNPTYRGVRMRQWGKWVSEIREPRKKSRIWLGTFPTPDM 108
T + K KK+ G ++ +P YRGVR R KWV E+REP + R+WLGT+PT DM
Sbjct: 7 TVAEMKPKKRAG--RRIFKETRHPIYRGVRRRDGDKWVCEVREPIHQRRVWLGTYPTADM 64
Query: 109 AARAHDVAALTIKGSSAYLNFPELAAVLPRAASASPKDIQVAAAKAA 155
AARAHDVA L ++G SA LNF + A LP AS P I+ AA+AA
Sbjct: 65 AARAHDVAVLALRGRSACLNFSDSAWRLPVPASTDPDTIRRTAAEAA 111
>DR1F_ARATH (Q9LN86) Dehydration responsive element binding protein
1F (DREB1F protein)
Length = 209
Score = 101 bits (252), Expect = 2e-21
Identities = 61/145 (42%), Positives = 77/145 (53%), Gaps = 2/145 (1%)
Query: 54 KKKKKVGMVNKVNDRKNNPTYRGVRMRQWGKWVSEIREPRKKSRIWLGTFPTPDMAARAH 113
+ KK+ G +V +P YRG+R R KWV E+REP + RIWLGT+PT DMAARAH
Sbjct: 12 RPKKRAG--RRVFKETRHPVYRGIRRRNGDKWVCEVREPTHQRRIWLGTYPTADMAARAH 69
Query: 114 DVAALTIKGSSAYLNFPELAAVLPRAASASPKDIQVAAAKAAATVYNHAITDQIELEAEA 173
DVA L ++G SA LNF + A LP S P I+ AA+AA + I + A
Sbjct: 70 DVAVLALRGRSACLNFADSAWRLPVPESNDPDVIRRVAAEAAEMFRPVDLESGITVLPCA 129
Query: 174 EPSQAVSSSSSSSSSSSSSHNEGSS 198
+ S S S S S SS
Sbjct: 130 GDDVDLGFGSGSGSGSGSEERNSSS 154
>DR2F_ARATH (Q9SVX5) Dehydration responsive element binding protein
2F (DREB2F protein)
Length = 277
Score = 92.8 bits (229), Expect = 7e-19
Identities = 62/148 (41%), Positives = 85/148 (56%), Gaps = 15/148 (10%)
Query: 74 YRGVRMRQWGKWVSEIREPRKKSRIWLGTFPTPDMAARAHDVAALTIKGSSAYLNFPELA 133
YRGVR R WGKWV+EIREP+K++R+WLG+F T + AA A+D AAL + G AYLN P L
Sbjct: 28 YRGVRQRTWGKWVAEIREPKKRARLWLGSFATAEEAAMAYDEAALKLYGHDAYLNLPHLQ 87
Query: 134 AVLPRAASASPKDIQVAAAK------------AAATVYNHAITDQI-ELEAEAEPSQAVS 180
+ S S + V + K A H I ++ EL+ SQ+ S
Sbjct: 88 RNTRPSLSNSQRFKWVPSRKFISMFPSCGMLNVNAQPSVHIIQQRLEELKKTGLLSQSYS 147
Query: 181 SSSSSS-SSSSSSHNEGSSLKGE-DDMF 206
SSSSS+ S +++S + + KGE D+MF
Sbjct: 148 SSSSSTESKTNTSFLDEKTSKGETDNMF 175
>AP23_ARATH (P42736) AP2 domain transcription factor RAP2.3 (Related
to AP2 protein 3) (Cadmium-induced protein AS30)
Length = 248
Score = 89.7 bits (221), Expect = 6e-18
Identities = 49/110 (44%), Positives = 60/110 (54%), Gaps = 3/110 (2%)
Query: 52 KEKKKKKVGMVNKVNDRKNNPTYRGVRMRQWGKWVSEIREPRKKSRIWLGTFPTPDMAAR 111
KE+ KK RK YRG+R R WGKW +EIR+PRK R+WLGTF T + AA
Sbjct: 57 KEEAVKKEQATEPGKRRKRKNVYRGIRKRPWGKWAAEIRDPRKGVRVWLGTFNTAEEAAM 116
Query: 112 AHDVAALTIKGSSAYLNFPELAAVLP---RAASASPKDIQVAAAKAAATV 158
A+DVAA I+G A LNFP+L P +SP+ AK V
Sbjct: 117 AYDVAAKQIRGDKAKLNFPDLHHPPPPNYTPPPSSPRSTDQPPAKKVCVV 166
>ERF2_ARATH (O80338) Ethylene responsive element binding factor 2
(AtERF2)
Length = 243
Score = 87.8 bits (216), Expect = 2e-17
Identities = 58/153 (37%), Positives = 79/153 (50%), Gaps = 36/153 (23%)
Query: 49 TKCKEKKKKKVGMVNKVNDRKNNPTYRGVRMRQWGKWVSEIREPRKK-SRIWLGTFPTPD 107
T +EK KK + + K+ YRGVR R WGK+ +EIR+P K +R+WLGTF T +
Sbjct: 95 TAMEEKPKKAIPVTETAVKAKH---YRGVRQRPWGKFAAEIRDPAKNGARVWLGTFETAE 151
Query: 108 MAARAHDVAALTIKGSSAYLNFPELAAVLPRAASASPKDIQVAAAKAAATVYNHAITDQI 167
AA A+D+AA ++GS A LNFP R S P +++ + +
Sbjct: 152 DAALAYDIAAFRMRGSRALLNFPL------RVNSGEPDPVRITSKR-------------- 191
Query: 168 ELEAEAEPSQAVSSSSSSSSSSSSSHNEGSSLK 200
SSSSSSSSSSS+S +E LK
Sbjct: 192 ------------SSSSSSSSSSSTSSSENGKLK 212
>DR2B_ARATH (O82133) Dehydration responsive element binding protein
2B (DREB2B protein)
Length = 330
Score = 87.4 bits (215), Expect = 3e-17
Identities = 57/156 (36%), Positives = 79/156 (50%), Gaps = 4/156 (2%)
Query: 42 EEEEATNTKCKEKKKKKVGMVNKVNDRKNNPTYRGVRMRQWGKWVSEIREPRKKSRIWLG 101
+E E K K KK M K ++ ++RGVR R WGKWV+EIREP+ +R+WLG
Sbjct: 46 KEGEKPKRKVPAKGSKKGCMKGKGGPDNSHCSFRGVRQRIWGKWVAEIREPKIGTRLWLG 105
Query: 102 TFPTPDMAARAHDVAALTIKGSSAYLNFPELAAVLPRAASASPKDIQVAAAKAAATVYNH 161
TFPT + AA A+D AA + GS A LNFP+ +V S S + +V + A V
Sbjct: 106 TFPTAEKAASAYDEAATAMYGSLARLNFPQ--SVGSEFTSTSSQS-EVCTVENKAVVCGD 162
Query: 162 AITDQIELEAEAEP-SQAVSSSSSSSSSSSSSHNEG 196
+ + E+ P SQ + S + S G
Sbjct: 163 VCVKHEDTDCESNPFSQILDVREESCGTRPDSCTVG 198
>DR2A_ARATH (O82132) Dehydration responsive element binding protein
2A (DREB2A protein)
Length = 335
Score = 85.9 bits (211), Expect = 9e-17
Identities = 45/92 (48%), Positives = 53/92 (56%)
Query: 39 EEEEEEEATNTKCKEKKKKKVGMVNKVNDRKNNPTYRGVRMRQWGKWVSEIREPRKKSRI 98
E EE K K KK M K + ++RGVR R WGKWV+EIREP + SR+
Sbjct: 44 ETVEEVSTKKRKVPAKGSKKGCMKGKGGPENSRCSFRGVRQRIWGKWVAEIREPNRGSRL 103
Query: 99 WLGTFPTPDMAARAHDVAALTIKGSSAYLNFP 130
WLGTFPT AA A+D AA + G A LNFP
Sbjct: 104 WLGTFPTAQEAASAYDEAAKAMYGPLARLNFP 135
>DR2D_ARATH (Q9LQZ2) Putative dehydration responsive element binding
protein 2D (DREB2D protein)
Length = 206
Score = 85.5 bits (210), Expect = 1e-16
Identities = 50/126 (39%), Positives = 62/126 (48%), Gaps = 26/126 (20%)
Query: 73 TYRGVRMRQWGKWVSEIREPRKKSRIWLGTFPTPDMAARAHDVAALTIKGSSAYLNFPEL 132
TY+GVR R WGKWV+EIREP + +R+WLGTF T AA A+D AA + G A+LN PE
Sbjct: 41 TYKGVRQRTWGKWVAEIREPNRGARLWLGTFDTSREAALAYDSAARKLYGPEAHLNLPES 100
Query: 133 AAVLPRAASASPKDIQVAAAKAAATVYNHAITDQIELEAEAEPSQAVSSSSSSSSSSSSS 192
P+ AS+ ++ PS SSS S S S
Sbjct: 101 LRSYPKTASSPA--------------------------SQTTPSSNTGGKSSSDSESPCS 134
Query: 193 HNEGSS 198
NE SS
Sbjct: 135 SNEMSS 140
>ERF5_ARATH (O80341) Ethylene responsive element binding factor 5
(AtERF5)
Length = 300
Score = 79.0 bits (193), Expect = 1e-14
Identities = 43/86 (50%), Positives = 53/86 (61%), Gaps = 2/86 (2%)
Query: 62 VNKVNDRKNNPTYRGVRMRQWGKWVSEIREPRKK-SRIWLGTFPTPDMAARAHDVAALTI 120
V K + YRGVR R WGK+ +EIR+P K+ SR+WLGTF T AARA+D AA +
Sbjct: 144 VTKPVSEEEKKHYRGVRQRPWGKFAAEIRDPNKRGSRVWLGTFDTAIEAARAYDEAAFRL 203
Query: 121 KGSSAYLNFP-ELAAVLPRAASASPK 145
+GS A LNFP E+ PRA K
Sbjct: 204 RGSKAILNFPLEVGKWKPRADEGEKK 229
>PTI6_LYCES (O04682) Pathogenesis-related genes transcriptional
activator PTI6
Length = 248
Score = 77.8 bits (190), Expect = 2e-14
Identities = 35/78 (44%), Positives = 51/78 (64%), Gaps = 3/78 (3%)
Query: 53 EKKKKKVGMVNKVNDRKNNPTYRGVRMRQWGKWVSEIREPRKKSRIWLGTFPTPDMAARA 112
++K++ V + V RK +RGVR R WG+W +EIR+P + R+WLGT+ TP+ AA
Sbjct: 80 DRKRRSVSPDSDVTRRKK---FRGVRQRPWGRWAAEIRDPTRGKRVWLGTYDTPEEAAVV 136
Query: 113 HDVAALTIKGSSAYLNFP 130
+D AA+ +KG A NFP
Sbjct: 137 YDKAAVKLKGPDAVTNFP 154
>ERF4_ARATH (O80340) Ethylene responsive element binding factor 4
(AtERF4)
Length = 222
Score = 77.8 bits (190), Expect = 2e-14
Identities = 34/68 (50%), Positives = 47/68 (69%)
Query: 63 NKVNDRKNNPTYRGVRMRQWGKWVSEIREPRKKSRIWLGTFPTPDMAARAHDVAALTIKG 122
N+ ++ YRGVR R WG++ +EIR+P KK+R+WLGTF T + AARA+D AA +G
Sbjct: 14 NQTHNNAKEIRYRGVRKRPWGRYAAEIRDPGKKTRVWLGTFDTAEEAARAYDTAARDFRG 73
Query: 123 SSAYLNFP 130
+ A NFP
Sbjct: 74 AKAKTNFP 81
>ERF1_ARATH (O80337) Ethylene responsive element binding factor 1
(AtERF1) (EREBP-2 protein)
Length = 268
Score = 77.0 bits (188), Expect = 4e-14
Identities = 48/138 (34%), Positives = 71/138 (50%), Gaps = 16/138 (11%)
Query: 33 SSSSLEEEEEEEEATNTKCKEK---------KKKKVGMVNKVNDRKNNPTYRGVRMRQWG 83
SSSS +E+ + + E KK+K V+ YRGVR R WG
Sbjct: 98 SSSSSDEDRSSFPSVKIETPESFAAVDSVPVKKEKTSPVSAAVTAAKGKHYRGVRQRPWG 157
Query: 84 KWVSEIREPRKK-SRIWLGTFPTPDMAARAHDVAALTIKGSSAYLNFPELAAVLPRAASA 142
K+ +EIR+P K +R+WLGTF T + AA A+D AA ++GS A LNFP R S
Sbjct: 158 KFAAEIRDPAKNGARVWLGTFETAEDAALAYDRAAFRMRGSRALLNFP------LRVNSG 211
Query: 143 SPKDIQVAAAKAAATVYN 160
P +++ + +++ + N
Sbjct: 212 EPDPVRIKSKRSSFSSSN 229
>DR2C_ARATH (Q8LFR2) Dehydration responsive element binding protein
2C (DREB2C protein)
Length = 341
Score = 76.3 bits (186), Expect = 7e-14
Identities = 40/79 (50%), Positives = 48/79 (60%)
Query: 54 KKKKKVGMVNKVNDRKNNPTYRGVRMRQWGKWVSEIREPRKKSRIWLGTFPTPDMAARAH 113
K +K M K YRGVR R+WGKWV+EIREP +R+WLGTF + AA A+
Sbjct: 52 KGSRKGCMKGKGGPENGICDYRGVRQRRWGKWVAEIREPDGGARLWLGTFSSSYEAALAY 111
Query: 114 DVAALTIKGSSAYLNFPEL 132
D AA I G SA LN PE+
Sbjct: 112 DEAAKAIYGQSARLNLPEI 130
>DR2G_ARATH (P61827) Putative dehydration responsive element binding
protein 2G (DREB2G protein)
Length = 307
Score = 75.9 bits (185), Expect = 9e-14
Identities = 55/167 (32%), Positives = 77/167 (45%), Gaps = 8/167 (4%)
Query: 42 EEEEATNTKCKEKKKKKVGMVNKVNDRKNNPTYRGVRMRQWGKWVSEIREPRKKSRIWLG 101
EEE+ K + +K M K T+RGVR R WGKWV+EIREP + +R+WLG
Sbjct: 2 EEEQPPAKKRNMGRSRKGCMKGKGGPENATCTFRGVRQRTWGKWVAEIREPNRGTRLWLG 61
Query: 102 TFPTPDMAARAHDVAALTIKGSSAYLNFPELAAVLPRAASASPKDIQVAAAKAAATVYNH 161
TF T AA A+D AA + G A LN L + +++ + + + YN
Sbjct: 62 TFNTSVEAAMAYDEAAKKLYGHEAKLN---LVHPQQQQQVVVNRNLSFSGHGSGSWAYNK 118
Query: 162 AITDQIELE---AEAEPSQAVSSSSSS--SSSSSSSHNEGSSLKGED 203
+ L+ +A S+ S SS SHN SS G +
Sbjct: 119 KLDMVHGLDLGLGQASCSRGSCSERSSFLQEDDDHSHNRCSSSSGSN 165
>DR2E_ARATH (O80917) Dehydration responsive element binding protein
2E (DREB2E protein)
Length = 244
Score = 73.9 bits (180), Expect = 3e-13
Identities = 45/105 (42%), Positives = 56/105 (52%), Gaps = 8/105 (7%)
Query: 35 SSLEEEEEEEEATNTKCKEKKKKKVGMVNKVNDRKNNPTYRGVRMRQWGKWVSEIREP-- 92
++L+ EEE A + + K KK M K +RGVR R WGKWV+EIREP
Sbjct: 31 ATLQRWEEEGLARARRVQAKGSKKGCMRGKGGPENPVCRFRGVRQRVWGKWVAEIREPVS 90
Query: 93 ------RKKSRIWLGTFPTPDMAARAHDVAALTIKGSSAYLNFPE 131
+ R+WLGTF T AA A+D AA + G A LNFPE
Sbjct: 91 HRGANSSRSKRLWLGTFATAAEAALAYDRAASVMYGPYARLNFPE 135
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.308 0.122 0.343
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 29,847,837
Number of Sequences: 164201
Number of extensions: 1220553
Number of successful extensions: 14923
Number of sequences better than 10.0: 276
Number of HSP's better than 10.0 without gapping: 179
Number of HSP's successfully gapped in prelim test: 103
Number of HSP's that attempted gapping in prelim test: 11324
Number of HSP's gapped (non-prelim): 1394
length of query: 257
length of database: 59,974,054
effective HSP length: 108
effective length of query: 149
effective length of database: 42,240,346
effective search space: 6293811554
effective search space used: 6293811554
T: 11
A: 40
X1: 16 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.7 bits)
S2: 64 (29.3 bits)
Medicago: description of AC149079.10


