

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148995.10 - phase: 0
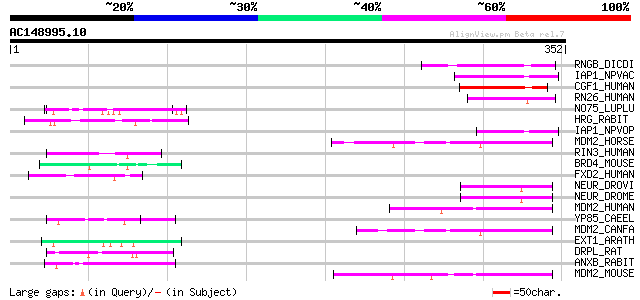
(352 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
RNGB_DICDI (Q7M3S9) Ring finger protein B (Protein rngB) 58 4e-08
IAP1_NPVAC (P41435) Apoptosis inhibitor 1 (IAP-1) 56 1e-07
CGF1_HUMAN (Q99675) Cell growth regulator with RING finger domai... 54 5e-07
RN26_HUMAN (Q9BY78) RING finger protein 26 53 1e-06
NO75_LUPLU (Q06841) Early nodulin 75 protein (N-75) (NGM-75) (Fr... 53 1e-06
HRG_RABIT (Q28640) Histidine-rich glycoprotein precursor (Histid... 52 2e-06
IAP1_NPVOP (O10296) Apoptosis inhibitor 1 (IAP-1) 51 4e-06
MDM2_HORSE (P56951) Ubiquitin-protein ligase E3 Mdm2 (EC 6.3.2.-... 51 5e-06
RIN3_HUMAN (Q8TB24) Ras and Rab interactor 3 (Ras interaction/in... 49 1e-05
BRD4_MOUSE (Q9ESU6) Bromodomain-containing protein 4 (Mitotic ch... 49 1e-05
FXD2_HUMAN (O60548) Forkhead box protein D2 (Forkhead-related pr... 49 2e-05
NEUR_DROVI (Q24746) Neuralized protein 48 3e-05
NEUR_DROME (P29503) Neuralized protein 48 3e-05
MDM2_HUMAN (Q00987) Ubiquitin-protein ligase E3 Mdm2 (EC 6.3.2.-... 48 3e-05
YP85_CAEEL (Q09442) Hypothetical RNA-binding protein C08B11.5 in... 48 4e-05
MDM2_CANFA (P56950) Ubiquitin-protein ligase E3 Mdm2 (EC 6.3.2.-... 48 4e-05
EXT1_ARATH (Q38913) Extensin 1 precursor (AtExt1) (AtExt4) 48 4e-05
DRPL_RAT (P54258) Atrophin-1 (Dentatorubral-pallidoluysian atrop... 48 4e-05
ANXB_RABIT (P33477) Annexin A11 (Annexin XI) (Calcyclin-associat... 48 4e-05
MDM2_MOUSE (P23804) Ubiquitin-protein ligase E3 Mdm2 (EC 6.3.2.-... 47 6e-05
>RNGB_DICDI (Q7M3S9) Ring finger protein B (Protein rngB)
Length = 943
Score = 57.8 bits (138), Expect = 4e-08
Identities = 30/85 (35%), Positives = 44/85 (51%), Gaps = 9/85 (10%)
Query: 262 VKQILSVNGMRYELQEIYGIGNSVESDVDDNEQGKECVICLSEPRDTIVHPCRHMCMCSG 321
+KQI S+ RY + + S+E + D + CVIC S P + ++ PCRH +CS
Sbjct: 866 LKQIGSIKDQRYLNRLV-----SLEKEKDQLKDQNSCVICASNPPNIVLLPCRHSSLCSD 920
Query: 322 CAKVLRFQTNRCPICRQPVERLLEI 346
C L +CPICR +E + I
Sbjct: 921 CCSKL----TKCPICRSHIENKISI 941
>IAP1_NPVAC (P41435) Apoptosis inhibitor 1 (IAP-1)
Length = 286
Score = 56.2 bits (134), Expect = 1e-07
Identities = 25/66 (37%), Positives = 37/66 (55%), Gaps = 3/66 (4%)
Query: 283 NSVESDVDDNEQGKECVICLSEPRDTIVHPCRHMCMCSGCAKVLRFQTNRCPICRQPVER 342
+++ + DD E+ EC +CL RD ++ PCRH C+C C L +CP CRQ V
Sbjct: 223 DNLNENADDIEEKYECKVCLERQRDAVLMPCRHFCVCVQCYFGL---DQKCPTCRQDVTD 279
Query: 343 LLEIKV 348
++I V
Sbjct: 280 FIKIFV 285
>CGF1_HUMAN (Q99675) Cell growth regulator with RING finger domain 1
(Cell growth regulatory gene 19 protein)
Length = 332
Score = 54.3 bits (129), Expect = 5e-07
Identities = 23/57 (40%), Positives = 38/57 (66%), Gaps = 5/57 (8%)
Query: 286 ESDVDDNEQG-KECVICLSEPRDTIVHPCRHMCMCSGCAKVLRFQTNRCPICRQPVE 341
ES+V+ +E+ K+CV+C + + ++ PCRH C+C GC K + +CP+CRQ V+
Sbjct: 261 ESEVEPSEENSKDCVVCQNGTVNWVLLPCRHTCLCDGCVKYFQ----QCPMCRQFVQ 313
>RN26_HUMAN (Q9BY78) RING finger protein 26
Length = 433
Score = 53.1 bits (126), Expect = 1e-06
Identities = 19/59 (32%), Positives = 35/59 (59%), Gaps = 3/59 (5%)
Query: 291 DNEQGKECVICLSEPRDTIVHPCRHMCMCSGCAKVLR---FQTNRCPICRQPVERLLEI 346
+ E+ K+CVIC + + ++ PCRH+C+C C ++L CP+CR+ + + L +
Sbjct: 373 EQEERKKCVICQDQSKTVLLLPCRHLCLCQACTEILMRHPVYHRNCPLCRRGILQTLNV 431
>NO75_LUPLU (Q06841) Early nodulin 75 protein (N-75) (NGM-75)
(Fragment)
Length = 434
Score = 52.8 bits (125), Expect = 1e-06
Identities = 37/100 (37%), Positives = 45/100 (45%), Gaps = 16/100 (16%)
Query: 23 HPPP---PPPVTPQPEIAPHQFVYPGAAPYPNPPMHYPQYHYPGYYPPPIPMPHHHHPHP 79
+ PP PPPV P P+ P YP P PP+H P Y P PP P PH P
Sbjct: 193 YEPPYEKPPPVHPPPDEKP-PIEYP--PPLEKPPVHEPPYEKP---PPAQPPPHDKPPIE 246
Query: 80 HPHPHMDPAWVSRYYPCGPVVNQPA----PFV---EHQKA 112
+P PH P Y P V+ P+ PFV H+K+
Sbjct: 247 YPPPHEKPPVYEPPYERSPPVHPPSHEKPPFVYPPPHEKS 286
Score = 48.5 bits (114), Expect = 2e-05
Identities = 33/84 (39%), Positives = 37/84 (43%), Gaps = 11/84 (13%)
Query: 24 PPP--PPPVTPQPEIAPHQFVYPGAAPYPNPPMHYPQYHY--PGYYPPPIPMPHHHHPHP 79
PPP PPPV P P P YP P+ PP+ YP H P YPP PH P
Sbjct: 33 PPPAHPPPVHPPPHEKP-PIEYP--PPHTKPPIEYPPPHVKPPIQYPP----PHEKPPIE 85
Query: 80 HPHPHMDPAWVSRYYPCGPVVNQP 103
+P PH P Y P V+ P
Sbjct: 86 YPLPHEKPPVYEPPYEKPPPVHPP 109
Score = 44.3 bits (103), Expect = 5e-04
Identities = 33/103 (32%), Positives = 40/103 (38%), Gaps = 25/103 (24%)
Query: 23 HPPP--PPPVTPQPEIAPHQFVYPGAAPYPNPPMHYP--------QYHYPGY-----YPP 67
HPPP PP+ P PH+ PY PP+ +P +YH P YPP
Sbjct: 107 HPPPHEKPPIEYSP---PHEKPPVHEPPYEKPPLVHPPPHDKPPIEYHPPHEKPPIEYPP 163
Query: 68 P-------IPMPHHHHPHPHPHPHMDPAWVSRYYPCGPVVNQP 103
P P PH P +P PH P Y P V+ P
Sbjct: 164 PHEKPPIEYPPPHEKPPIEYPPPHEKPPVYEPPYEKPPPVHPP 206
Score = 41.2 bits (95), Expect = 0.004
Identities = 32/100 (32%), Positives = 38/100 (38%), Gaps = 19/100 (19%)
Query: 23 HPPP--------PPPVTPQPEIAPHQFVYPGAA---PYPNPPMHYPQ-------YHYPGY 64
HPPP PPP T P P V P P+ PP+ YP Y P
Sbjct: 42 HPPPHEKPPIEYPPPHTKPPIEYPPPHVKPPIQYPPPHEKPPIEYPLPHEKPPVYEPPYE 101
Query: 65 YPPPIPMPHHHHPH-PHPHPHMDPAWVSRYYPCGPVVNQP 103
PPP+ P H P + PH P Y P+V+ P
Sbjct: 102 KPPPVHPPPHEKPPIEYSPPHEKPPVHEPPYEKPPLVHPP 141
Score = 40.8 bits (94), Expect = 0.005
Identities = 33/123 (26%), Positives = 42/123 (33%), Gaps = 20/123 (16%)
Query: 23 HPPP---PPPVTPQPEIAPHQFVYPGAAPYP----NPPMHYPQYHYPGYYPPPIPMPHHH 75
+ PP PP P P + P YP A P PP+ +P + P Y PP P
Sbjct: 311 YAPPHEKPPVEYPPPHVKP-PVEYPSPAEKPPTHEKPPIEFPPHEKPPVYEPPYEKPPSV 369
Query: 76 HPHPHPHPHMDPAWVSRYYPCGPVVNQPAPFVEHQKAVTIRNDVNIKKETIVISPDEENP 135
P PH P + Y P H+K + KK SP E P
Sbjct: 370 CPPPHEKPPHHEKRSALYPP------------HHRKPPNHHKPLPYKKPPFYKSPPVEKP 417
Query: 136 GFF 138
+
Sbjct: 418 SSY 420
Score = 39.7 bits (91), Expect = 0.012
Identities = 36/125 (28%), Positives = 48/125 (37%), Gaps = 20/125 (16%)
Query: 23 HPPP---PPPVTPQPEIAPHQFVYPGAAPYPNPPMHYPQY------HYPGYYPPPI--PM 71
H PP PPP P P P YP P+ PP++ P Y H P + PP P
Sbjct: 225 HEPPYEKPPPAQPPPHDKP-PIEYP--PPHEKPPVYEPPYERSPPVHPPSHEKPPFVYPP 281
Query: 72 PHHHHP-HPHPHPHMDPAWVSRYYPCGPVVNQPAPFVEHQKAVTIRNDVNIKKETIVISP 130
PH P H P+ P + + P + P H+K ++K SP
Sbjct: 282 PHEKSPMHEPPYEKAPPVHLPPHEK--PPIEYAPP---HEKPPVEYPPPHVKPPVEYPSP 336
Query: 131 DEENP 135
E+ P
Sbjct: 337 AEKPP 341
Score = 30.4 bits (67), Expect = 7.0
Identities = 20/70 (28%), Positives = 26/70 (36%), Gaps = 12/70 (17%)
Query: 51 NPPMHYP-----------QYHYPGYYPPPIPMPHHHHPHP-HPHPHMDPAWVSRYYPCGP 98
+PP+H P + P + PP P HP P HP PH P P
Sbjct: 1 SPPVHPPPHEKPPIVHPPPHEKPPLFEPPFEKPPPAHPPPVHPPPHEKPPIEYPPPHTKP 60
Query: 99 VVNQPAPFVE 108
+ P P V+
Sbjct: 61 PIEYPPPHVK 70
>HRG_RABIT (Q28640) Histidine-rich glycoprotein precursor
(Histidine-proline rich glycoprotein) (HPRG) (Fragment)
Length = 526
Score = 52.0 bits (123), Expect = 2e-06
Identities = 37/112 (33%), Positives = 47/112 (41%), Gaps = 18/112 (16%)
Query: 10 HNRRRHATASRRTHP--PPP---PPVTPQPEIAPHQFVYPGAAPYPNPPMHYPQYHYPGY 64
H H HP PPP PP P P PH G P+ +PP P + +P +
Sbjct: 331 HGHHPHGPPPHGHHPHGPPPHGHPPHGPPPRHPPH-----GPPPHGHPPHGPPPHGHPPH 385
Query: 65 YPPPIPMPHHHHPH---PHPHPHMDPAWVSRYYPCGPVVNQPAPFVEHQKAV 113
PP PH H PH PH HP + + PC P ++ P HQ A+
Sbjct: 386 GPP----PHGHPPHGPPPHGHPPHGHGF-HDHGPCDPPSHKEGPQDLHQHAM 432
>IAP1_NPVOP (O10296) Apoptosis inhibitor 1 (IAP-1)
Length = 275
Score = 51.2 bits (121), Expect = 4e-06
Identities = 22/52 (42%), Positives = 29/52 (55%), Gaps = 3/52 (5%)
Query: 297 ECVICLSEPRDTIVHPCRHMCMCSGCAKVLRFQTNRCPICRQPVERLLEIKV 348
EC +CL RD ++ PCRH C+C C L +CP CRQ V ++I V
Sbjct: 226 ECKVCLERQRDAVLLPCRHFCVCMQCYFAL---DGKCPTCRQDVADFIKIFV 274
>MDM2_HORSE (P56951) Ubiquitin-protein ligase E3 Mdm2 (EC 6.3.2.-)
(p53-binding protein Mdm2) (Double minute 2 protein)
(Edm2)
Length = 491
Score = 50.8 bits (120), Expect = 5e-06
Identities = 42/158 (26%), Positives = 69/158 (43%), Gaps = 31/158 (19%)
Query: 205 SDLVKVGDVDVYPLAVKADASSDNHDGSNETETSSKPN------SQITQAVFEKEKGEFR 258
S+ K+GD +++ D D D T + S+ + +ITQA +E ++
Sbjct: 342 SEKAKLGD------SMQEDEGFDVPDCKKSTVSDSRESCVEENDDKITQASLSQESEDY- 394
Query: 259 VKVVKQILSVNGMRYELQEIYGIGNSVESDVDDNEQGKE----------CVICLSEPRD- 307
Q + N + Y QE + + D E+ E CVIC P++
Sbjct: 395 ----SQPSTSNSIIYSSQE--DVKEFEREETQDKEESMESSFPLNAIEPCVICQGRPKNG 448
Query: 308 TIVH-PCRHMCMCSGCAKVLRFQTNRCPICRQPVERLL 344
IVH H+ C CAK L+ + CP+CRQP++ ++
Sbjct: 449 CIVHGKTGHLMACFTCAKKLKKRNKPCPVCRQPIQMIV 486
>RIN3_HUMAN (Q8TB24) Ras and Rab interactor 3 (Ras
interaction/interference protein 3)
Length = 984
Score = 49.3 bits (116), Expect = 1e-05
Identities = 29/76 (38%), Positives = 33/76 (43%), Gaps = 12/76 (15%)
Query: 24 PPPPPPVTPQPEIAPHQF-VYPGAAPYPNPPMHYPQYHYPGYYPPPIPMPH--HHHPHPH 80
PPPPPPV P +P Q V P AP P P+ PP+P PH H P P
Sbjct: 280 PPPPPPVLPLQPCSPAQPPVLPALAPAPACPLPTS---------PPVPAPHVTPHAPGPP 330
Query: 81 PHPHMDPAWVSRYYPC 96
HP+ P PC
Sbjct: 331 DHPNQPPMMTCERLPC 346
>BRD4_MOUSE (Q9ESU6) Bromodomain-containing protein 4 (Mitotic
chromosome-associated protein) (MCAP)
Length = 1400
Score = 49.3 bits (116), Expect = 1e-05
Identities = 35/110 (31%), Positives = 42/110 (37%), Gaps = 28/110 (25%)
Query: 20 RRTHPPPPPPVTPQPEIAPHQFVYPGAAPY--------------PNPPMHYPQYHYPGYY 65
++ P PPPP PQP+ P Q P P P PP P + PG
Sbjct: 971 QQLQPQPPPPPPPQPQPPPQQQHQPPPRPVHLPSMPFSAHIQQPPPPPGQQPTHPPPGQQ 1030
Query: 66 PPPIPMPH------HHHPHPHPHPHMDPAWVSRYYPCGPVVNQPAPFVEH 109
PPP P P HHP P H DP Y G + P+P + H
Sbjct: 1031 PPP-PQPAKPQQVIQHHPSPR-HHKSDP------YSAGHLREAPSPLMIH 1072
Score = 38.5 bits (88), Expect = 0.026
Identities = 27/106 (25%), Positives = 39/106 (36%), Gaps = 18/106 (16%)
Query: 21 RTHPPPPPPVTPQPEIAPHQF-VYPGAAPYPNP-------------PMHYPQYHYPGYYP 66
++ PPPP P P P + Q P P P P P+H P + +
Sbjct: 953 QSQPPPPLPPPPHPSVQQQQLQPQPPPPPPPQPQPPPQQQHQPPPRPVHLPSMPFSAHIQ 1012
Query: 67 PPIPMPHHHHPHPHPHPHMDPAWVSRYYPCGPVVNQPAPFVEHQKA 112
P P P HP P P ++ P + + P+P H K+
Sbjct: 1013 QPPPPPGQQPTHPPPGQQPPPPQPAK--PQQVIQHHPSP--RHHKS 1054
Score = 35.0 bits (79), Expect = 0.29
Identities = 24/77 (31%), Positives = 30/77 (38%), Gaps = 16/77 (20%)
Query: 24 PPPPPPVTPQP-----EIAPHQFV----------YPGAAPYPNPPMHYPQYHYPG-YYPP 67
PPPPPP PQ + +P F+ PG+ P P H P PP
Sbjct: 780 PPPPPPSMPQQTAPAMKSSPPPFITAQVPVLEPQLPGSVFDPIGHFTQPILHLPQPELPP 839
Query: 68 PIPMPHHHHPHPHPHPH 84
+P P H PH + H
Sbjct: 840 HLPQPPEHSTPPHLNQH 856
Score = 35.0 bits (79), Expect = 0.29
Identities = 21/66 (31%), Positives = 27/66 (40%), Gaps = 3/66 (4%)
Query: 17 TASRRTHPPPPPPVT--PQPEIAPHQFVYPGAAPY-PNPPMHYPQYHYPGYYPPPIPMPH 73
T + + +PPPP T P P + P P P P+ P P PPP P+P
Sbjct: 208 TQTPQQNPPPPVQATTHPFPAVTPDLIAQPPVMTMVPPQPLQTPSPVPPQPPPPPAPVPQ 267
Query: 74 HHHPHP 79
HP
Sbjct: 268 PVQSHP 273
Score = 32.3 bits (72), Expect = 1.8
Identities = 34/132 (25%), Positives = 44/132 (32%), Gaps = 28/132 (21%)
Query: 7 TGAHNRRRHATASRRTHP---PPPPPVTPQPEIAPHQFVYPGAAPYPNPPMHYPQYHYPG 63
TG ++ H HP P P PV QP P Q P P PP Q P
Sbjct: 732 TGRDQKKHH----HHHHPQMQPAPAPVPQQPPPPPQQ-------PPPPPPPQ-QQQQQPP 779
Query: 64 YYPPPIPMPHHHHPHPHPHPHMDPAWVSRYYPCGPVVNQPAPFVEHQKAVTIRNDVNIKK 123
PPP MP P P P + P +E Q ++ + +
Sbjct: 780 PPPPPPSMPQQTAPAMKSSP-------------PPFITAQVPVLEPQLPGSVFDPIGHFT 826
Query: 124 ETIVISPDEENP 135
+ I+ P E P
Sbjct: 827 QPILHLPQPELP 838
>FXD2_HUMAN (O60548) Forkhead box protein D2 (Forkhead-related
protein FKHL17) (Forkhead-related transcription factor
9) (FREAC-9)
Length = 497
Score = 48.9 bits (115), Expect = 2e-05
Identities = 31/85 (36%), Positives = 36/85 (41%), Gaps = 20/85 (23%)
Query: 13 RRHATASRRTHPPPPPPVTPQPEIAPHQFVYPGAAPYPNPPMHYPQYHYPGYY------- 65
RR R+ PPP P P PE+ + GAA +P P + G Y
Sbjct: 218 RRRKRFKRQPLPPPHPHPHPHPEL----LLRGGAAAAGDPGAFLPGFAAYGAYGYGYGLA 273
Query: 66 ------PPPIPMPHHHHPHPHPHPH 84
PPP P PH PHPHPHPH
Sbjct: 274 LPAYGAPPPGPAPH---PHPHPHPH 295
Score = 31.2 bits (69), Expect = 4.1
Identities = 11/18 (61%), Positives = 13/18 (72%), Gaps = 3/18 (16%)
Query: 68 PIPMPHHHHPHPHPHPHM 85
P+P PH PHPHPHP +
Sbjct: 227 PLPPPH---PHPHPHPEL 241
>NEUR_DROVI (Q24746) Neuralized protein
Length = 747
Score = 48.1 bits (113), Expect = 3e-05
Identities = 20/60 (33%), Positives = 31/60 (51%), Gaps = 2/60 (3%)
Query: 287 SDVDDNEQGKECVICLSEPRDTIVHPCRHMCMCSGCA--KVLRFQTNRCPICRQPVERLL 344
SD + EC IC P D++++ C HMCMC CA + +CP+CR + ++
Sbjct: 683 SDQQSTDSSAECTICYENPIDSVLYMCGHMCMCYDCAIEQWRGVGGGQCPLCRAVIRDVI 742
>NEUR_DROME (P29503) Neuralized protein
Length = 754
Score = 48.1 bits (113), Expect = 3e-05
Identities = 20/60 (33%), Positives = 31/60 (51%), Gaps = 2/60 (3%)
Query: 287 SDVDDNEQGKECVICLSEPRDTIVHPCRHMCMCSGCA--KVLRFQTNRCPICRQPVERLL 344
SD + EC IC P D++++ C HMCMC CA + +CP+CR + ++
Sbjct: 690 SDQQSTDSSAECTICYENPIDSVLYMCGHMCMCYDCAIEQWRGVGGGQCPLCRAVIRDVI 749
>MDM2_HUMAN (Q00987) Ubiquitin-protein ligase E3 Mdm2 (EC 6.3.2.-)
(p53-binding protein Mdm2) (Oncoprotein Mdm2) (Double
minute 2 protein) (Hdm2)
Length = 491
Score = 48.1 bits (113), Expect = 3e-05
Identities = 32/109 (29%), Positives = 53/109 (48%), Gaps = 7/109 (6%)
Query: 242 NSQITQAVFEKEKGEFRVKVVKQILSVNGMR----YELQEIYGIGNSVESDVDDNEQGKE 297
+ +ITQA +E ++ + + +E +E SVES + N +
Sbjct: 379 DDKITQASQSQESEDYSQPSTSSSIIYSSQEDVKEFEREETQDKEESVESSLPLNAI-EP 437
Query: 298 CVICLSEPRD-TIVH-PCRHMCMCSGCAKVLRFQTNRCPICRQPVERLL 344
CVIC P++ IVH H+ C CAK L+ + CP+CRQP++ ++
Sbjct: 438 CVICQGRPKNGCIVHGKTGHLMACFTCAKKLKKRNKPCPVCRQPIQMIV 486
>YP85_CAEEL (Q09442) Hypothetical RNA-binding protein C08B11.5 in
chromosome II
Length = 388
Score = 47.8 bits (112), Expect = 4e-05
Identities = 33/83 (39%), Positives = 35/83 (41%), Gaps = 10/83 (12%)
Query: 24 PPPPPPVTPQPEIAPHQFVYPGAAPYPNPPMHYPQYHYPGYYPPPIPMPHHHHPHPHPHP 83
PPPPP VTP P P PG P P PP G +PPP P P P P P
Sbjct: 264 PPPPPSVTPMPPPMPPT---PGMTPRPPPPPS------SGMWPPPPPPPPGRTPGPPGMP 314
Query: 84 HM-DPAWVSRYYPCGPVVNQPAP 105
M P SR+ P G P P
Sbjct: 315 GMPPPPPPSRFGPPGMGGMPPPP 337
Score = 43.5 bits (101), Expect = 8e-04
Identities = 29/72 (40%), Positives = 30/72 (41%), Gaps = 14/72 (19%)
Query: 24 PPPPPP-------VTPQPEIAPHQFVYPGAAPYPNPPMHYPQYHYPGYYPPPIPM----- 71
PPPPPP + P P YPG P P PP YP PG YPPP P
Sbjct: 317 PPPPPPSRFGPPGMGGMPPPPPPGMRYPGGMP-PPPPPRYPSAG-PGMYPPPPPSRPPAP 374
Query: 72 PHHHHPHPHPHP 83
P H P P P
Sbjct: 375 PSGHGMIPPPPP 386
Score = 42.7 bits (99), Expect = 0.001
Identities = 31/90 (34%), Positives = 34/90 (37%), Gaps = 6/90 (6%)
Query: 21 RTHPPP-----PPPVTPQPEIAPHQFVYPGAAPYPNPPMHYPQYHYPGYYPPPIPMPHHH 75
R PPP PPP P P P PG P P PP + G PPP P +
Sbjct: 286 RPPPPPSSGMWPPPPPPPPGRTPGPPGMPGMPP-PPPPSRFGPPGMGGMPPPPPPGMRYP 344
Query: 76 HPHPHPHPHMDPAWVSRYYPCGPVVNQPAP 105
P P P P+ YP P PAP
Sbjct: 345 GGMPPPPPPRYPSAGPGMYPPPPPSRPPAP 374
Score = 30.8 bits (68), Expect = 5.4
Identities = 20/48 (41%), Positives = 20/48 (41%), Gaps = 2/48 (4%)
Query: 36 IAPHQFVYPGAAPYPNPPMHYPQYHYPGYYPPPIPMPHHHHPHPHPHP 83
IA H PG P P M Q Y G YPP P P P P P P
Sbjct: 233 IAAHATGRPGYQP--PPLMGMAQSGYQGQYPPVPPPPPSVTPMPPPMP 278
>MDM2_CANFA (P56950) Ubiquitin-protein ligase E3 Mdm2 (EC 6.3.2.-)
(p53-binding protein Mdm2) (Double minute 2 protein)
(Cdm2)
Length = 487
Score = 47.8 bits (112), Expect = 4e-05
Identities = 38/136 (27%), Positives = 62/136 (44%), Gaps = 24/136 (17%)
Query: 221 KADASSDNHDGSNETETSSKPNSQITQAVFEKEKGEFRVKVVKQILSVNGMRYELQEIYG 280
K A+SD+ + E + + +ITQA +E ++ Q + N + Y QE
Sbjct: 359 KKAAASDSRESCAE-----EIDDKITQASHSQESEDY-----SQPSTSNSIIYSSQE--D 406
Query: 281 IGNSVESDVDDNEQGKE----------CVICLSEPRD-TIVH-PCRHMCMCSGCAKVLRF 328
+ + D E+ E CVIC P++ IVH H+ C CAK L+
Sbjct: 407 VKEFEREETQDKEEIVESSFPLNAIEPCVICQGRPKNGCIVHGKTGHLMACFTCAKKLKK 466
Query: 329 QTNRCPICRQPVERLL 344
+ CP+CRQP++ ++
Sbjct: 467 RNKPCPVCRQPIQMIV 482
>EXT1_ARATH (Q38913) Extensin 1 precursor (AtExt1) (AtExt4)
Length = 373
Score = 47.8 bits (112), Expect = 4e-05
Identities = 37/113 (32%), Positives = 44/113 (38%), Gaps = 24/113 (21%)
Query: 21 RTHPPP-----PPPVTPQPEIAPHQFVYPGAAPYPNPPMHYPQ----YHYP----GYYPP 67
++ PPP PPPV P + P P PP+HY YH P Y PP
Sbjct: 208 KSPPPPVKHYSPPPVYKSPPPPVKYYSPPPVYKSPPPPVHYSPPPVVYHSPPPPVHYSPP 267
Query: 68 PI-----PMPHHHHP------HPHPHPHMDPAWVSRYYPCGPVVNQPAPFVEH 109
P+ P P H+ P P P H P V + P PV P P V H
Sbjct: 268 PVVYHSPPPPVHYSPPPVVYHSPPPPVHYSPPPVVYHSPPPPVHYSPPPVVYH 320
Score = 42.4 bits (98), Expect = 0.002
Identities = 30/98 (30%), Positives = 37/98 (37%), Gaps = 17/98 (17%)
Query: 23 HPPPPPPVTPQPEIAPHQFVYPGAAPYPNPPMHY-----------PQYHYPG----YYPP 67
H PPP V P H P P PP+HY P HY Y+ P
Sbjct: 263 HYSPPPVVYHSPPPPVHYSPPPVVYHSPPPPVHYSPPPVVYHSPPPPVHYSPPPVVYHSP 322
Query: 68 PIPMPHHHHPHPHPHPHMDPAWVSRYYPCGPVVNQPAP 105
P P H+ + P P H P V Y+ P V+ +P
Sbjct: 323 PPPKKHYEYKSPPPPVHYSPPTV--YHSPPPPVHHYSP 358
Score = 42.0 bits (97), Expect = 0.002
Identities = 30/94 (31%), Positives = 37/94 (38%), Gaps = 10/94 (10%)
Query: 21 RTHPPP-----PPPVTPQPEIAPHQFVYPGAAPYPNPPMHYPQYHYPGYYPPPIPMPHHH 75
++ PPP PPPV P P Y Y +PP Y P Y P P H+
Sbjct: 96 KSPPPPVKHYSPPPVYKSPP--PPVKHYSPPPVYKSPPPPVKHYSPPPVYKSPPPPVKHY 153
Query: 76 HPHPHPHPHMDPAWVSRYYPCGPVVNQPAPFVEH 109
P P + P +YY PV P P V+H
Sbjct: 154 SP---PPVYKSPPPPVKYYSPPPVYKSPPPPVKH 184
Score = 42.0 bits (97), Expect = 0.002
Identities = 33/101 (32%), Positives = 41/101 (39%), Gaps = 16/101 (15%)
Query: 21 RTHPPP-----PPPVTPQPEIAPHQFVYPGAAPYPNPPMHY----PQYHYPG---YYPPP 68
++ PPP PPPV P + P P PP+ Y P Y P Y PP
Sbjct: 40 KSPPPPVKHYSPPPVYKSPPPPVKHYSPPPVYKSPPPPVKYYSPPPVYKSPPPPVYKSPP 99
Query: 69 IPMPHHHHPHPHPHPHMDPAWVSRYYPCGPVVNQPAPFVEH 109
P+ H+ P + P P V Y P PV P P V+H
Sbjct: 100 PPVKHYSPPPVYKSP---PPPVKHYSP-PPVYKSPPPPVKH 136
Score = 41.6 bits (96), Expect = 0.003
Identities = 30/94 (31%), Positives = 37/94 (38%), Gaps = 10/94 (10%)
Query: 21 RTHPPP-----PPPVTPQPEIAPHQFVYPGAAPYPNPPMHYPQYHYPGYYPPPIPMPHHH 75
++ PPP PPPV P P Y Y +PP Y P Y P P H+
Sbjct: 128 KSPPPPVKHYSPPPVYKSPP--PPVKHYSPPPVYKSPPPPVKYYSPPPVYKSPPPPVKHY 185
Query: 76 HPHPHPHPHMDPAWVSRYYPCGPVVNQPAPFVEH 109
P P + P +YY PV P P V+H
Sbjct: 186 SP---PPVYKSPPPPVKYYSPPPVYKSPPPPVKH 216
Score = 40.0 bits (92), Expect = 0.009
Identities = 33/112 (29%), Positives = 42/112 (37%), Gaps = 13/112 (11%)
Query: 23 HPPPPPPVTPQPEIAPHQFVYPGAAPYPNPPMHYPQ----YHYPGYYPPPIPMPHHHHPH 78
H PPP V P H P P PP+HY YH P PPP+ H
Sbjct: 247 HYSPPPVVYHSPPPPVHYSPPPVVYHSPPPPVHYSPPPVVYHSP---PPPV------HYS 297
Query: 79 PHPHPHMDPAWVSRYYPCGPVVNQPAPFVEHQKAVTIRNDVNIKKETIVISP 130
P P + P Y P V + P P +H + + V+ T+ SP
Sbjct: 298 PPPVVYHSPPPPVHYSPPPVVYHSPPPPKKHYEYKSPPPPVHYSPPTVYHSP 349
Score = 38.9 bits (89), Expect = 0.020
Identities = 29/94 (30%), Positives = 36/94 (37%), Gaps = 10/94 (10%)
Query: 21 RTHPPP-----PPPVTPQPEIAPHQFVYPGAAPYPNPPMHYPQYHYPGYYPPPIPMPHHH 75
++ PPP PPPV P P Y Y +PP Y P Y P P H+
Sbjct: 160 KSPPPPVKYYSPPPVYKSPP--PPVKHYSPPPVYKSPPPPVKYYSPPPVYKSPPPPVKHY 217
Query: 76 HPHPHPHPHMDPAWVSRYYPCGPVVNQPAPFVEH 109
P P + P +YY PV P P V +
Sbjct: 218 SP---PPVYKSPPPPVKYYSPPPVYKSPPPPVHY 248
Score = 37.7 bits (86), Expect = 0.044
Identities = 27/83 (32%), Positives = 34/83 (40%), Gaps = 20/83 (24%)
Query: 23 HPPPPPPVTPQPEIAPHQFVYPGAAPYPNPPMHY-----PQYHY-------PGYYPPPI- 69
H PPPP P + H P Y PP+ Y P+ HY P +Y PP
Sbjct: 288 HSPPPPVHYSPPPVVYHS--PPPPVHYSPPPVVYHSPPPPKKHYEYKSPPPPVHYSPPTV 345
Query: 70 ----PMPHHHHPHPH-PHPHMDP 87
P P HH+ PH P+ + P
Sbjct: 346 YHSPPPPVHHYSPPHQPYLYKSP 368
Score = 35.4 bits (80), Expect = 0.22
Identities = 25/89 (28%), Positives = 31/89 (34%), Gaps = 13/89 (14%)
Query: 17 TASRRTHPPPPPPVTPQPEIAPHQFVYPGAAPYPNPPMHYPQYHYPGYYPPPIPMPHHHH 76
T + + PPPPV Y Y +PP Y P Y P P H+
Sbjct: 17 TTANYFYSSPPPPVKH----------YSPPPVYKSPPPPVKHYSPPPVYKSPPPPVKHYS 66
Query: 77 PHPHPHPHMDPAWVSRYYPCGPVVNQPAP 105
P P + P +YY PV P P
Sbjct: 67 P---PPVYKSPPPPVKYYSPPPVYKSPPP 92
Score = 31.2 bits (69), Expect = 4.1
Identities = 20/55 (36%), Positives = 24/55 (43%), Gaps = 4/55 (7%)
Query: 23 HPPPPPPVTPQPEIAPHQFVYPGAAPY--PNPPM-HYPQYHYPGYYPPPIPMPHH 74
H PPPP + + P Y Y P PP+ HY H P Y P P PH+
Sbjct: 320 HSPPPPKKHYEYKSPPPPVHYSPPTVYHSPPPPVHHYSPPHQPYLYKSP-PPPHY 373
>DRPL_RAT (P54258) Atrophin-1 (Dentatorubral-pallidoluysian atrophy
protein)
Length = 1183
Score = 47.8 bits (112), Expect = 4e-05
Identities = 36/110 (32%), Positives = 45/110 (40%), Gaps = 33/110 (30%)
Query: 24 PPPPPPVTPQPEIAPHQFVYPGAAP--------YPNPPMHYPQYHYPGYYPPPIPMPHHH 75
PPPPPP P + P+ +PG P +P P H+ +H P P P P H
Sbjct: 440 PPPPPP--PYGRLLPNNNTHPGPFPPTGGQSTAHPPAPAHH--HHQQQQQPQPQPQPQQH 495
Query: 76 H------------PHP-------HPHPH-MDPAWVS-RYYPCGPVVNQPA 104
H PHP H HP+ M P+ S R YP GP P+
Sbjct: 496 HHGNSGPPPPGAYPHPLESSNSHHAHPYNMSPSLGSLRPYPPGPAHLPPS 545
Score = 32.7 bits (73), Expect = 1.4
Identities = 28/87 (32%), Positives = 35/87 (40%), Gaps = 12/87 (13%)
Query: 19 SRRTHPPPPPPVT-PQPEIAPHQFVYPGAAPYPNPPMHYPQYHYPGYYPPPI------PM 71
+R HPPP P + P P+ P Q P + P+P + YH P PP P
Sbjct: 155 ARPYHPPPLFPPSPPPPDSIPRQ---PESGFEPHPSVPPTGYHAP--MEPPTSRLFQGPP 209
Query: 72 PHHHHPHPHPHPHMDPAWVSRYYPCGP 98
P PHP +P V P GP
Sbjct: 210 PGAPPPHPQLYPGSAGGGVLSGPPMGP 236
Score = 32.3 bits (72), Expect = 1.8
Identities = 25/98 (25%), Positives = 33/98 (33%), Gaps = 28/98 (28%)
Query: 15 HATASRRTHPPPPPPVTPQPEIAPHQFVYPGAAPYPNPPMHYP----------------- 57
HA + PP P P +PH P ++ P PPM YP
Sbjct: 334 HAMGQGMSGLPPGPEKGPTLAPSPHPLP-PASSSAPGPPMRYPYSSCSSSSVAASSSSSA 392
Query: 58 ---QY-------HYPGYYPPPIPMPHHHHPHPHPHPHM 85
QY YP +PPP M + P + P +
Sbjct: 393 ATSQYPASQTLPSYPHSFPPPTSMSVSNQPPKYTQPSL 430
Score = 31.6 bits (70), Expect = 3.2
Identities = 17/63 (26%), Positives = 22/63 (33%)
Query: 24 PPPPPPVTPQPEIAPHQFVYPGAAPYPNPPMHYPQYHYPGYYPPPIPMPHHHHPHPHPHP 83
PPPP T P + Q A Y P P P P + +P H +
Sbjct: 702 PPPPAAPTTGPPLTATQIKQEPAEEYETPESPVPPARSPSPPPKVVDVPSHASQSARFNK 761
Query: 84 HMD 86
H+D
Sbjct: 762 HLD 764
>ANXB_RABIT (P33477) Annexin A11 (Annexin XI) (Calcyclin-associated
annexin 50) (CAP-50)
Length = 503
Score = 47.8 bits (112), Expect = 4e-05
Identities = 33/89 (37%), Positives = 38/89 (42%), Gaps = 9/89 (10%)
Query: 23 HPPPPP-----PVTPQPEIAPHQFVYPGAAPYPNPPMHYPQYH-YPGYYPPPIPMPHHHH 76
+PP PP P QP + P+ VYP P NPP P Y +PG P PMP H
Sbjct: 81 YPPVPPGGFGQPPPTQPSVPPYG-VYP--PPGGNPPSGVPSYPPFPGAPVPGQPMPPPGH 137
Query: 77 PHPHPHPHMDPAWVSRYYPCGPVVNQPAP 105
P P+P P P P QP P
Sbjct: 138 QPPGPYPGQLPVTYPGQSPVPPPGQQPMP 166
>MDM2_MOUSE (P23804) Ubiquitin-protein ligase E3 Mdm2 (EC 6.3.2.-)
(p53-binding protein Mdm2) (Oncoprotein Mdm2) (Double
minute 2 protein)
Length = 489
Score = 47.4 bits (111), Expect = 6e-05
Identities = 40/154 (25%), Positives = 66/154 (41%), Gaps = 19/154 (12%)
Query: 206 DLVKVGDVDVYPLAVKADASSDNHDGSNETETSSKP-------NSQITQAVFEKEKGEFR 258
D V++ + + +A+ D DG TE +K + Q +E ++
Sbjct: 335 DKVEISEKAKLENSAQAEEGLDVPDGKKLTENDAKEPCAEEDSEEKAEQTPLSQESDDYS 394
Query: 259 VKVVKQIL------SVNGMRYELQEIYGIGNSVESDVDDNEQGKECVICLSEPRD-TIVH 311
+ SV ++ E Q+ SVES N + CVIC P++ IVH
Sbjct: 395 QPSTSSSIVYSSQESVKELKEETQDK---DESVESSFSLNAI-EPCVICQGRPKNGCIVH 450
Query: 312 -PCRHMCMCSGCAKVLRFQTNRCPICRQPVERLL 344
H+ C CAK L+ + CP+CRQP++ ++
Sbjct: 451 GKTGHLMSCFTCAKKLKKRNKPCPVCRQPIQMIV 484
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.318 0.136 0.428
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 47,922,403
Number of Sequences: 164201
Number of extensions: 2436209
Number of successful extensions: 21979
Number of sequences better than 10.0: 780
Number of HSP's better than 10.0 without gapping: 290
Number of HSP's successfully gapped in prelim test: 516
Number of HSP's that attempted gapping in prelim test: 13385
Number of HSP's gapped (non-prelim): 3670
length of query: 352
length of database: 59,974,054
effective HSP length: 111
effective length of query: 241
effective length of database: 41,747,743
effective search space: 10061206063
effective search space used: 10061206063
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 66 (30.0 bits)
Medicago: description of AC148995.10


