

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148994.4 - phase: 0
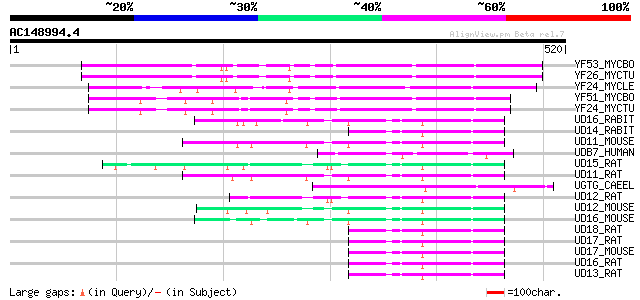
(520 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
YF53_MYCBO (P64870) Hypothetical protein Mb1553c 141 4e-33
YF26_MYCTU (P64869) Hypothetical protein Rv1526c/MT1577 141 4e-33
YF24_MYCLE (Q49929) Hypothetical glycosyl transferase ML2348 (EC... 131 4e-30
YF51_MYCBO (P64866) Hypothetical glycosyl transferase Mb1551 (EC... 128 3e-29
YF24_MYCTU (P64865) Hypothetical glycosyl transferase Rv1524/MT1... 128 3e-29
UD16_RABIT (Q28611) UDP-glucuronosyltransferase 1-6 precursor, m... 57 1e-07
UD14_RABIT (Q28612) UDP-glucuronosyltransferase 1-4 precursor, m... 52 3e-06
UD11_MOUSE (Q63886) UDP-glucuronosyltransferase 1-1 precursor, m... 52 3e-06
UDB7_HUMAN (P16662) UDP-glucuronosyltransferase 2B7 precursor, m... 52 4e-06
UD15_RAT (Q64638) UDP-glucuronosyltransferase 1-5 precursor, mic... 51 6e-06
UD11_RAT (Q64550) UDP-glucuronosyltransferase 1-1 precursor, mic... 50 1e-05
UGTG_CAEEL (Q22295) Putative UDP-glucuronosyltransferase UGT16 p... 50 2e-05
UD12_RAT (P20720) UDP-glucuronosyltransferase 1-2 precursor, mic... 50 2e-05
UD12_MOUSE (P70691) UDP-glucuronosyltransferase 1-2 precursor, m... 50 2e-05
UD16_MOUSE (Q64435) UDP-glucuronosyltransferase 1-6 precursor, m... 49 3e-05
UD18_RAT (Q64634) UDP-glucuronosyltransferase 1-8 precursor, mic... 48 7e-05
UD17_RAT (Q64633) UDP-glucuronosyltransferase 1-7 precursor, mic... 48 7e-05
UD17_MOUSE (Q62452) UDP-glucuronosyltransferase 1-7 precursor, m... 48 7e-05
UD16_RAT (P08430) UDP-glucuronosyltransferase 1-6 precursor, mic... 48 7e-05
UD13_RAT (Q64637) UDP-glucuronosyltransferase 1-3 precursor, mic... 48 7e-05
>YF53_MYCBO (P64870) Hypothetical protein Mb1553c
Length = 426
Score = 141 bits (356), Expect = 4e-33
Identities = 131/446 (29%), Positives = 188/446 (41%), Gaps = 37/446 (8%)
Query: 68 QIAMLIVGTRGDVQPFVAIGKRLQADGHRVRLATHKNFEDFVLSAGLEFYPLGGDPKVLA 127
+ + + GTRGDV+P A+G L+ GH V +A N +FV SAGL G D
Sbjct: 2 KFVLAVHGTRGDVEPCAAVGVELRRRGHAVHMAVPPNLIEFVESAGLTGVAYGPDSDEQI 61
Query: 128 EYMVKNKGFLPSGPSEIHLQRSQIRAIIHSLLPACNSRYPESNEPFKADAIIANPPAYG- 186
+ L + ++L R+ + + ++ AD ++ +G
Sbjct: 62 NTVAAFVRNLTRAQNPLNLARAVKELFVEGWAEMGTTLTTLADG---ADLVMTGQTYHGV 118
Query: 187 HTHVAEYLNVP---LHIF-------FTMPWTPTSDFPHPLSRVRQPIGYRL-SYQIVDAL 235
+VAEY ++P LH F +P PT P L R + +RL +Y DA
Sbjct: 119 AANVAEYYDIPAAALHHFPMQVNGQIAIPSIPT---PATLVRATMKVSWRLYAYVSKDA- 174
Query: 236 IWLGIRDLINEFRKKKLKLRAVTYL--RGSYTFPPDMPYGYIWSPHLVPKPKDWGPNIDI 293
R E AV L RG+ P Y ++ P L +W
Sbjct: 175 ----DRAQRRELGLPPAPAPAVRRLAERGA---PEIQAYDPVFFPGLAA---EWSDRRPF 224
Query: 294 VGFCFLDLASNYEPPKSLVDWLEEGENPIYVGFGSLPLQEPEKMTRIIVQALEQTGQRGI 353
VG ++L S EP + L W+ G PIY GFGS P+Q P + +I Q G+R +
Sbjct: 225 VGPLTMELHS--EPNEELESWIAAGTPPIYFGFGSTPVQTPVQTLAMISDVCAQLGERAL 282
Query: 354 INKGWGGLGNLAELNTSKSVYLLDNCPHDWLFPRCAAVVHHGGAGTTAAGLRAECPTTVV 413
I + + K V L++ + + P+C AVVHHGGAGTTAAGLRA PT ++
Sbjct: 283 IYSPAANSTRIRHADHVKRVGLVN---YSTILPKCRAVVHHGGAGTTAAGLRAGMPTLIL 339
Query: 414 PFFGDQPFWGERVHARGVGPAPIRVEEFTLERLVDAIRFMLNPEVKKRAVELANAMKNED 473
DQP W V VG A R T L+ +R +L PE RA E++ M
Sbjct: 340 WDVADQPIWAGAVQRLKVGSAK-RFTNITRGSLLKELRSILAPECAARAREISTRMTRPT 398
Query: 474 GVAGAVNAFYKHYPREKPDTEAEPRP 499
A + R+ P + P
Sbjct: 399 AAVTAAADLLEATARQTPGSTPSSSP 424
>YF26_MYCTU (P64869) Hypothetical protein Rv1526c/MT1577
Length = 426
Score = 141 bits (356), Expect = 4e-33
Identities = 131/446 (29%), Positives = 188/446 (41%), Gaps = 37/446 (8%)
Query: 68 QIAMLIVGTRGDVQPFVAIGKRLQADGHRVRLATHKNFEDFVLSAGLEFYPLGGDPKVLA 127
+ + + GTRGDV+P A+G L+ GH V +A N +FV SAGL G D
Sbjct: 2 KFVLAVHGTRGDVEPCAAVGVELRRRGHAVHMAVPPNLIEFVESAGLTGVAYGPDSDEQI 61
Query: 128 EYMVKNKGFLPSGPSEIHLQRSQIRAIIHSLLPACNSRYPESNEPFKADAIIANPPAYG- 186
+ L + ++L R+ + + ++ AD ++ +G
Sbjct: 62 NTVAAFVRNLTRAQNPLNLARAVKELFVEGWAEMGTTLTTLADG---ADLVMTGQTYHGV 118
Query: 187 HTHVAEYLNVP---LHIF-------FTMPWTPTSDFPHPLSRVRQPIGYRL-SYQIVDAL 235
+VAEY ++P LH F +P PT P L R + +RL +Y DA
Sbjct: 119 AANVAEYYDIPAAALHHFPMQVNGQIAIPSIPT---PATLVRATMKVSWRLYAYVSKDA- 174
Query: 236 IWLGIRDLINEFRKKKLKLRAVTYL--RGSYTFPPDMPYGYIWSPHLVPKPKDWGPNIDI 293
R E AV L RG+ P Y ++ P L +W
Sbjct: 175 ----DRAQRRELGLPPAPAPAVRRLAERGA---PEIQAYDPVFFPGLAA---EWSDRRPF 224
Query: 294 VGFCFLDLASNYEPPKSLVDWLEEGENPIYVGFGSLPLQEPEKMTRIIVQALEQTGQRGI 353
VG ++L S EP + L W+ G PIY GFGS P+Q P + +I Q G+R +
Sbjct: 225 VGPLTMELHS--EPNEELESWIAAGTPPIYFGFGSTPVQTPVQTLAMISDVCAQLGERAL 282
Query: 354 INKGWGGLGNLAELNTSKSVYLLDNCPHDWLFPRCAAVVHHGGAGTTAAGLRAECPTTVV 413
I + + K V L++ + + P+C AVVHHGGAGTTAAGLRA PT ++
Sbjct: 283 IYSPAANSTRIRHADHVKRVGLVN---YSTILPKCRAVVHHGGAGTTAAGLRAGMPTLIL 339
Query: 414 PFFGDQPFWGERVHARGVGPAPIRVEEFTLERLVDAIRFMLNPEVKKRAVELANAMKNED 473
DQP W V VG A R T L+ +R +L PE RA E++ M
Sbjct: 340 WDVADQPIWAGAVQRLKVGSAK-RFTNITRGSLLKELRSILAPECAARAREISTRMTRPT 398
Query: 474 GVAGAVNAFYKHYPREKPDTEAEPRP 499
A + R+ P + P
Sbjct: 399 AAVTAAADLLEATARQTPGSTPSSSP 424
>YF24_MYCLE (Q49929) Hypothetical glycosyl transferase ML2348 (EC
2.-.-.-)
Length = 421
Score = 131 bits (330), Expect = 4e-30
Identities = 124/439 (28%), Positives = 189/439 (42%), Gaps = 51/439 (11%)
Query: 75 GTRGDVQPFVAIGKRLQADGHRVRLATHKNFEDFVLSAGLEFYPLGGDPKVLAEYMVKNK 134
G+RGDV+PF A+G LQ GH VR+ + FV SAGL G D + E++ +
Sbjct: 9 GSRGDVEPFAALGLELQRRGHEVRIGVPPDMLRFVESAGLAAVAYGPDTQ---EFLAR-- 63
Query: 135 GFLPSGPSEIHLQRSQIRAIIHSL--LPACNSRYPESNEPFK-----ADAIIANPPAYGH 187
+ + Q Q I+ + L + + K AD ++ G
Sbjct: 64 --------DTYSQWRQWWKILPPIKALQQLRQAWADMATDLKSLADGADLVMTGVVYQGV 115
Query: 188 T-HVAEYLNVPLHIFFTMPWTPTS----DFPHPLSRVRQPIGYRLSYQIVDALIWLGIRD 242
+VAEY +P + +P P PL+R +R WL +
Sbjct: 116 VANVAEYYGIPFGVLHFVPARVNGKIIPSLPSPLNRAILATVWRAH--------WL-LAK 166
Query: 243 LINEFRKKKLKLRAVTYL-------RGSYTFPPDMPYGYIWSPHLVPKPKDWGPNIDIVG 295
+ ++++L L T L RG+ Y + P L + ++G VG
Sbjct: 167 KPEDAQRRELGLPKATSLSTRRIVKRGALEI---QAYDELCFPGLAAEWAEYGDRRPFVG 223
Query: 296 FCFLDLASNYEPPKSLVDWLEEGENPIYVGFGSLPLQEPEKMTRIIVQALEQTGQRGIIN 355
L+L + + ++ W+ G PIY GFGS+P+ P +I A G+R +I+
Sbjct: 224 ALTLELPTAAD--NEVLSWIAAGTPPIYFGFGSMPIVSPTDTVAMIAAACADLGERALIS 281
Query: 356 KGWGGLGNLAELNTSKSVYLLDNCPHDWLFPRCAAVVHHGGAGTTAAGLRAECPTTVVPF 415
L + + + K ++ + H +FP C AVVHHGGAGTTAA LRA PT ++
Sbjct: 282 VKPKDLTQVPKFDHVK---IVTSVSHAAVFPACRAVVHHGGAGTTAASLRAGVPTLILWI 338
Query: 416 FGDQPFWGERVHARGVGPAPIRVEEFTLERLVDAIRFMLNPEVKKRAVELANAM-KNEDG 474
F +QP W ++ VG A R T L +R +L P+ RA E+AN M K ++
Sbjct: 339 FIEQPVWAAQIKRLKVG-AGRRFSATTQRSLAADLRTILAPQYATRAREVANRMSKPDES 397
Query: 475 VAGAVNAFYKHYPREKPDT 493
V A + R K T
Sbjct: 398 VNAAADLLEDKASRNKSRT 416
>YF51_MYCBO (P64866) Hypothetical glycosyl transferase Mb1551 (EC
2.-.-.-)
Length = 414
Score = 128 bits (322), Expect = 3e-29
Identities = 117/408 (28%), Positives = 179/408 (43%), Gaps = 37/408 (9%)
Query: 75 GTRGDVQPFVAIGKRLQADGHRVRLATHKNFEDFVLSAGLEFYPLGG--DPKVLAEYMVK 132
GTRGD++P A+G LQ GH V LA N FV +AGL G + L E +
Sbjct: 9 GTRGDIEPCAAVGLELQRRGHDVCLAVPPNLIGFVETAGLSAVAYGSRDSQEQLDEQFLH 68
Query: 133 NKGFLPSGPSEIHLQRSQIRAIIHSLLPACN--SRYPESNEPFKADA-IIANPPAYGHT- 188
N L ++ I+ + ++ P + P A A ++ Y
Sbjct: 69 NAWKL----------QNPIKLLREAMAPVTEGWAELSAMLTPVAAGADLLLTGQIYQEVV 118
Query: 189 -HVAEYLNVPLHIFFTMPWTPTSDFPHPLSRVRQPIGYRLSYQIVDALIWLGIRDLINEF 247
+VAE+ +PL P + P +R+ P+ R + +D L W + + +
Sbjct: 119 ANVAEHHGIPLAALHFYPVRANGEIAFP-ARLPAPL-VRSTITAIDWLYWRMTKGVEDAQ 176
Query: 248 RKKKLKLRAVT------YLRGSYTFPPDMPYGYIWSPHLVPKPKDWGPNIDIVGFCFLDL 301
R++ +A T +RGS Y + P L +WG VG ++
Sbjct: 177 RRELGLPKASTPAPRRMAVRGSLEI---QAYDALCFPGLAA---EWGGRRPFVGALTMES 230
Query: 302 ASNYEPPKSLVDWLEEGENPIYVGFGSLPLQEPEKMTRIIVQALEQTGQRGIINKGWGGL 361
A++ + + W+ PIY GFGS+P+ +I A + G+R +I G
Sbjct: 231 ATDADD--EVASWIAADTPPIYFGFGSMPIGSLADRVAMISAACAELGERALICSGPSDA 288
Query: 362 GNLAELNTSKSVYLLDNCPHDWLFPRCAAVVHHGGAGTTAAGLRAECPTTVVPFFGDQPF 421
+ + + K V ++ H +FP C AVVHHGGAGTTAAGLRA PT ++ DQP
Sbjct: 289 TGIPQFDHVKVVRVVS---HAAVFPTCRAVVHHGGAGTTAAGLRAGIPTLILWVTSDQPI 345
Query: 422 WGERVHARGVGPAPIRVEEFTLERLVDAIRFMLNPEVKKRAVELANAM 469
W ++ VG R T E L+ +R +L P+ RA E+A+ M
Sbjct: 346 WAAQIKQLKVGRGR-RFSSATKESLIADLRTILAPDYVTRAREIASRM 392
>YF24_MYCTU (P64865) Hypothetical glycosyl transferase Rv1524/MT1575
(EC 2.-.-.-)
Length = 414
Score = 128 bits (322), Expect = 3e-29
Identities = 117/408 (28%), Positives = 179/408 (43%), Gaps = 37/408 (9%)
Query: 75 GTRGDVQPFVAIGKRLQADGHRVRLATHKNFEDFVLSAGLEFYPLGG--DPKVLAEYMVK 132
GTRGD++P A+G LQ GH V LA N FV +AGL G + L E +
Sbjct: 9 GTRGDIEPCAAVGLELQRRGHDVCLAVPPNLIGFVETAGLSAVAYGSRDSQEQLDEQFLH 68
Query: 133 NKGFLPSGPSEIHLQRSQIRAIIHSLLPACN--SRYPESNEPFKADA-IIANPPAYGHT- 188
N L ++ I+ + ++ P + P A A ++ Y
Sbjct: 69 NAWKL----------QNPIKLLREAMAPVTEGWAELSAMLTPVAAGADLLLTGQIYQEVV 118
Query: 189 -HVAEYLNVPLHIFFTMPWTPTSDFPHPLSRVRQPIGYRLSYQIVDALIWLGIRDLINEF 247
+VAE+ +PL P + P +R+ P+ R + +D L W + + +
Sbjct: 119 ANVAEHHGIPLAALHFYPVRANGEIAFP-ARLPAPL-VRSTITAIDWLYWRMTKGVEDAQ 176
Query: 248 RKKKLKLRAVT------YLRGSYTFPPDMPYGYIWSPHLVPKPKDWGPNIDIVGFCFLDL 301
R++ +A T +RGS Y + P L +WG VG ++
Sbjct: 177 RRELGLPKASTPAPRRMAVRGSLEI---QAYDALCFPGLAA---EWGGRRPFVGALTMES 230
Query: 302 ASNYEPPKSLVDWLEEGENPIYVGFGSLPLQEPEKMTRIIVQALEQTGQRGIINKGWGGL 361
A++ + + W+ PIY GFGS+P+ +I A + G+R +I G
Sbjct: 231 ATDADD--EVASWIAADTPPIYFGFGSMPIGSLADRVAMISAACAELGERALICSGPSDA 288
Query: 362 GNLAELNTSKSVYLLDNCPHDWLFPRCAAVVHHGGAGTTAAGLRAECPTTVVPFFGDQPF 421
+ + + K V ++ H +FP C AVVHHGGAGTTAAGLRA PT ++ DQP
Sbjct: 289 TGIPQFDHVKVVRVVS---HAAVFPTCRAVVHHGGAGTTAAGLRAGIPTLILWVTSDQPI 345
Query: 422 WGERVHARGVGPAPIRVEEFTLERLVDAIRFMLNPEVKKRAVELANAM 469
W ++ VG R T E L+ +R +L P+ RA E+A+ M
Sbjct: 346 WAAQIKQLKVGRGR-RFSSATKESLIADLRTILAPDYVTRAREIASRM 392
>UD16_RABIT (Q28611) UDP-glucuronosyltransferase 1-6 precursor,
microsomal (EC 2.4.1.17) (UDPGT) (UGT1*6) (UGT1-06)
(UGT1.6) (UGT1A6)
Length = 531
Score = 57.0 bits (136), Expect = 1e-07
Identities = 79/310 (25%), Positives = 131/310 (41%), Gaps = 33/310 (10%)
Query: 174 KADAIIANPPAYGHTHVAEYLNVP-LHIFFTMPWTPTSDF---PHPLS---RVRQPIGYR 226
K DA+ +P +AEYL +P +++F P + F P+P+S R +
Sbjct: 143 KFDALFTDPALPCGVILAEYLGLPSVYLFRGFPCSLEHGFGGSPNPVSYIPRCYTKFSDQ 202
Query: 227 LSY--QIVDALIWLGIRDLINEFRKKKLKLRAVTYLRGSYTFPPDMPYGYIWS---PHLV 281
+S+ ++V+ L+ L L K L AV L+ P +W +
Sbjct: 203 MSFPQRVVNFLVNLLEVPLFYCLYSKYEDL-AVELLKREVDLPTLFQKDPVWLLRYDFVF 261
Query: 282 PKPKDWGPNIDIVGFCFLDLASNYEPPKSLVDWLE-----EGENPIYV-GFGSLPLQEPE 335
P+ PN+ ++G N + P L E GE+ I V GS+ + PE
Sbjct: 262 EYPRPVMPNMVLIG------GINCKKPDVLSQEFEAYVNASGEHGIVVFSLGSMVSEIPE 315
Query: 336 KMTRIIVQALEQTGQRGIINKGWGGLGNLAELNTSKSVYLLDNCPHDWLF--PRCAAVVH 393
K I AL + Q + W G+ N +K+ YL+ P + L P+ A +
Sbjct: 316 KKAMEIADALGKIPQTVL----WRYTGSRPS-NLAKNTYLVKWLPQNVLLGHPKTRAFIT 370
Query: 394 HGGAGTTAAGLRAECPTTVVPFFGDQPFWGERVHARGVGPAPIRVEEFTLERLVDAIRFM 453
H G+ G+ P ++P FGDQ +R+ RG G + V E T + L +A++ +
Sbjct: 371 HSGSHGIYEGICNGVPMVMLPLFGDQMDNAKRIETRGAG-VTLNVLEMTSDDLANALKTV 429
Query: 454 LNPEVKKRAV 463
+N + K +
Sbjct: 430 INDKSYKENI 439
>UD14_RABIT (Q28612) UDP-glucuronosyltransferase 1-4 precursor,
microsomal (EC 2.4.1.17) (UDPGT) (UGT1*4) (UGT1-04)
(UGT1.4) (UGT1A4)
Length = 532
Score = 52.4 bits (124), Expect = 3e-06
Identities = 43/149 (28%), Positives = 70/149 (46%), Gaps = 9/149 (6%)
Query: 318 GENPIYV-GFGSLPLQEPEKMTRIIVQALEQTGQRGIINKGWGGLGNLAELNTSKSVYLL 376
GE+ I V GS+ + PEK I AL + Q + W G+ N +K+ YL+
Sbjct: 298 GEHGIVVFSLGSMVSEIPEKKAMEIADALGKIPQTVL----WRYTGSRPS-NLAKNTYLV 352
Query: 377 DNCPHDWLF--PRCAAVVHHGGAGTTAAGLRAECPTTVVPFFGDQPFWGERVHARGVGPA 434
P + L P+ A + H G+ G+ P ++P FGDQ +R+ RG G
Sbjct: 353 KWLPQNVLLGHPKTRAFITHSGSHGIYEGICNGVPMVMLPLFGDQMDNAKRIETRGAG-V 411
Query: 435 PIRVEEFTLERLVDAIRFMLNPEVKKRAV 463
+ V E T + L +A++ ++N + K +
Sbjct: 412 TLNVLEMTSDDLANALKTVINDKSYKENI 440
>UD11_MOUSE (Q63886) UDP-glucuronosyltransferase 1-1 precursor,
microsomal (EC 2.4.1.17) (UDPGT) (UGT1*1) (UGT1-01)
(UGT1.1) (UGT1A1) (UGTBR1)
Length = 535
Score = 52.4 bits (124), Expect = 3e-06
Identities = 79/320 (24%), Positives = 132/320 (40%), Gaps = 31/320 (9%)
Query: 163 NSRYPESNEPFKADAIIANPPAYGHTHVAEYLNVPLHIFFT-MPWTPTSDF---PHPLSR 218
N+ + S E DA++ +P + VA+YL VP F +P + S+ P PLS
Sbjct: 135 NAEFMASLEESHFDALLTDPFLPCGSIVAQYLTVPTVYFLNKLPCSLDSEATQCPVPLSY 194
Query: 219 VRQPIGY---RLSY-QIVDALIWLGIRDLINEFRKKKLKLRAVTYLRGSYTFPPDMPYGY 274
V + + + R+++ Q V ++ + + A L+ T +
Sbjct: 195 VPKSLSFNSDRMNFLQRVKNVLLAVSENFMCRVVYSPYGSLATEILQKEVTVQDLLSPAS 254
Query: 275 IW---SPHLVPKPKDWGPNIDIVGFCFLDLASNYEPPKSLVDWLE-----EGENPIYV-G 325
IW S + P+ PN+ +G N K L E GE+ I V
Sbjct: 255 IWLMRSDFVKDYPRPIMPNMVFIG------GINCLQKKPLSQEFEAYVNASGEHGIVVFS 308
Query: 326 FGSLPLQEPEKMTRIIVQALEQTGQRGIINKGWGGLGNLAELNTSKSVYLLDNCPHDWLF 385
GS+ + PEK I +AL + Q + W G N +K+ L+ P + L
Sbjct: 309 LGSMVSEIPEKKAMEIAEALGRIPQTVL----WRYTGTRPS-NLAKNTILVKWLPQNDLI 363
Query: 386 --PRCAAVVHHGGAGTTAAGLRAECPTTVVPFFGDQPFWGERVHARGVGPAPIRVEEFTL 443
P+ A + H G+ G+ P ++P FGDQ +R+ RG G + V E T
Sbjct: 364 GHPKTRAFITHSGSHGIYEGICNGVPMVMMPLFGDQMDNAKRMETRGAG-VTLNVLEMTA 422
Query: 444 ERLVDAIRFMLNPEVKKRAV 463
+ L +A++ ++N + K +
Sbjct: 423 DDLENALKTVINNKSYKENI 442
>UDB7_HUMAN (P16662) UDP-glucuronosyltransferase 2B7 precursor,
microsomal (EC 2.4.1.17) (UDPGT) (3,4-catechol estrogen
specific) (UDPGTH-2)
Length = 529
Score = 52.0 bits (123), Expect = 4e-06
Identities = 55/193 (28%), Positives = 89/193 (45%), Gaps = 21/193 (10%)
Query: 289 PNIDIVGFCFLDLASNYEPPKSLVDWLEE-GENPIYV-GFGSLPLQEPEKMTRIIVQALE 346
PN+D VG A PK + D+++ GEN + V GS+ E+ +I AL
Sbjct: 272 PNVDFVGGLHCKPAKPL--PKEMEDFVQSSGENGVVVFSLGSMVSNMTEERANVIASALA 329
Query: 347 QTGQRGIINKGWGGLGNLAE---LNTSKSVYLLDNCPHDWL-FPRCAAVVHHGGAGTTAA 402
Q Q+ + W GN + LNT ++ N D L P+ A + HGGA
Sbjct: 330 QIPQKVL----WRFDGNKPDTLGLNTRLYKWIPQN---DLLGHPKTRAFITHGGANGIYE 382
Query: 403 GLRAECPTTVVPFFGDQPFWGERVHARGVGPAPIRVEEFTLER--LVDAIRFMLN-PEVK 459
+ P +P F DQP + ARG A +RV+ T+ L++A++ ++N P K
Sbjct: 383 AIYHGIPMVGIPLFADQPDNIAHMKARG---AAVRVDFNTMSSTDLLNALKRVINDPSYK 439
Query: 460 KRAVELANAMKNE 472
+ ++L+ ++
Sbjct: 440 ENVMKLSRIQHDQ 452
>UD15_RAT (Q64638) UDP-glucuronosyltransferase 1-5 precursor,
microsomal (EC 2.4.1.17) (UDPGT) (UGT1*5) (UGT1-05)
(UGT1.5) (UGT1A5) (B5)
Length = 531
Score = 51.2 bits (121), Expect = 6e-06
Identities = 94/417 (22%), Positives = 165/417 (39%), Gaps = 65/417 (15%)
Query: 88 KRLQADGHRV-----RLATHKNFEDFVLSAGLEFYPLGGDPKVLAEYMVKNKG--FLPSG 140
+ L A GH+ + H EDF L+ YP+ + + M++N F
Sbjct: 46 RELHARGHQAVVLAPEVTVHIKEEDFFT---LQTYPVPYTRQGFRQQMMRNIKVVFETGN 102
Query: 141 PSEIHLQRSQIRAIIHS-LLPAC-----NSRYPESNEPFKADAIIANPPAYGHTHVAEYL 194
+ L+ S+I I + LL +C N + D ++ +P +A+YL
Sbjct: 103 YVKTFLETSEILKNISTVLLRSCMNLLHNGSLLQHLNSSSFDMVLTDPVIPCGQVLAKYL 162
Query: 195 NVPLHIFF-----------TMPWTPTSDFPHPLS----------RVRQPIGYRLSYQIVD 233
+P F T P+S P+ L+ RV+ + Y L+ + +
Sbjct: 163 GIPTVFFLRYIPCGIDSEATQCPKPSSYIPNLLTMLSDHMTFLQRVKNML-YPLALKYIC 221
Query: 234 ALIWLGIRDLINEFRKKKLKLRAVTYLRGSYTFPPDMPYGYIWSPHLVPKPKDWGPNIDI 293
+ L +E ++++ L V + F D + Y P+ PN+
Sbjct: 222 HFSFTRYESLASELLQREVSLVEVLSHASVWLFRGDFVFDY---------PRPVMPNMVF 272
Query: 294 VGF--CFLD--LASNYEPPKSLVDWLEEGENPIYV-GFGSLPLQEPEKMTRIIVQALEQT 348
+G C + L+ +E + GE+ I V GS+ + PEK I +AL +
Sbjct: 273 IGGINCVIKKPLSQEFEAYVNA-----SGEHGIVVFSLGSMVSEIPEKKAMEIAEALGRI 327
Query: 349 GQRGIINKGWGGLGNLAELNTSKSVYLLDNCPHDWLF--PRCAAVVHHGGAGTTAAGLRA 406
Q + W G N +K+ L+ P + L P+ A + H G+ G+
Sbjct: 328 PQTLL----WRYTGTRPS-NLAKNTILVKWLPQNDLLGHPKARAFITHSGSHGIYEGICN 382
Query: 407 ECPTTVVPFFGDQPFWGERVHARGVGPAPIRVEEFTLERLVDAIRFMLNPEVKKRAV 463
P ++P FGDQ +R+ RG G + V E T + L +A++ ++N + K +
Sbjct: 383 GVPMVMMPLFGDQMDNAKRMETRGAG-VTLNVLEMTADDLENALKTVINNKSYKENI 438
>UD11_RAT (Q64550) UDP-glucuronosyltransferase 1-1 precursor,
microsomal (EC 2.4.1.17) (UDPGT) (UGT1*1) (UGT1-01)
(UGT1.1) (UGT1A1) (B1)
Length = 535
Score = 50.4 bits (119), Expect = 1e-05
Identities = 77/320 (24%), Positives = 132/320 (41%), Gaps = 31/320 (9%)
Query: 163 NSRYPESNEPFKADAIIANPPAYGHTHVAEYLNVPLHIFFT-MPWT---PTSDFPHPLSR 218
N+ + S E DA++ +P + VA+YL++P F +P + + P PLS
Sbjct: 135 NAEFMASLEQSHFDALLTDPFLPCGSIVAQYLSLPAVYFLNALPCSLDLEATQCPAPLSY 194
Query: 219 VRQPIGY---RLSY-QIVDALIWLGIRDLINEFRKKKLKLRAVTYLRGSYTFPPDMPYGY 274
V + + R+++ Q V +I + + A L+ T +
Sbjct: 195 VPKSLSSNTDRMNFLQRVKNMIIALTENFLCRVVYSPYGSLATEILQKEVTVKDLLSPAS 254
Query: 275 IW---SPHLVPKPKDWGPNIDIVGFCFLDLASNYEPPKSLVDWLE-----EGENPIYV-G 325
IW + + P+ PN+ +G N K+L E GE+ I V
Sbjct: 255 IWLMRNDFVKDYPRPIMPNMVFIG------GINCLQKKALSQEFEAYVNASGEHGIVVFS 308
Query: 326 FGSLPLQEPEKMTRIIVQALEQTGQRGIINKGWGGLGNLAELNTSKSVYLLDNCPHDWLF 385
GS+ + PEK I +AL + Q + W G N +K+ L+ P + L
Sbjct: 309 LGSMVSEIPEKKAMEIAEALGRIPQTVL----WRYTGTRPS-NLAKNTILVKWLPQNDLL 363
Query: 386 --PRCAAVVHHGGAGTTAAGLRAECPTTVVPFFGDQPFWGERVHARGVGPAPIRVEEFTL 443
P+ A + H G+ G+ P ++P FGDQ +R+ RG G + V E T
Sbjct: 364 GHPKARAFITHSGSHGIYEGICNGVPMVMMPLFGDQMDNAKRMETRGAG-VTLNVLEMTA 422
Query: 444 ERLVDAIRFMLNPEVKKRAV 463
+ L +A++ ++N + K +
Sbjct: 423 DDLENALKTVINNKSYKENI 442
>UGTG_CAEEL (Q22295) Putative UDP-glucuronosyltransferase UGT16
precursor (EC 2.4.1.17) (UDPGT)
Length = 525
Score = 49.7 bits (117), Expect = 2e-05
Identities = 57/238 (23%), Positives = 102/238 (41%), Gaps = 14/238 (5%)
Query: 284 PKDWGPNIDIVGFCFLDLASNYEPPKSLVDWLEEGEN-PIYVGFGSL-PLQE-PEKMTRI 340
P+ + VG N PK D+ ++G++ ++V FG++ P + PE++
Sbjct: 265 PRSYSSKFVYVGMLEAGKDENVTLPKKQDDYFKKGKSGSVFVSFGTVTPFRSLPERIQLS 324
Query: 341 IVQALEQTGQRGIINKGWGGLGNLAEL-NTSKSVYLLDNCPHDWLFPRC--AAVVHHGGA 397
I+ A+++ + K + A+ +T ++V L+D P + V HGG
Sbjct: 325 ILNAIQKLPDYHFVVKTTADDESSAQFFSTVQNVDLVDWVPQKAVLRHANLKLFVSHGGM 384
Query: 398 GTTAAGLRAECPTTVVPFFGDQPFWGERVHARGVGPAPIRVEEFTLERLVDAIRFMLNPE 457
+ + P ++P F DQ G V RG G +R E E DAI +L +
Sbjct: 385 NSVLETMYYGVPMVIMPVFTDQFRNGRNVERRGAGKMVLR-ETVVKETFFDAIHSVLEEK 443
Query: 458 VKKRAVE-LANAMKN-----EDGVAGAVNAFYKHYPREKPDTEAEPRPVPSVHKHLSI 509
+V+ +++ MKN E+ V ++ K+ E D E+ + H HL +
Sbjct: 444 SYSSSVKRISHLMKNKPFTSEERVTKWIDFVLKYETSEHFDLESNNLSIIE-HNHLDL 500
>UD12_RAT (P20720) UDP-glucuronosyltransferase 1-2 precursor,
microsomal (EC 2.4.1.17) (UDPGT) (UGT1*0) (UGT1-02)
(UGT1.2) (UGT1A2) (Bilirubin specific) (B2)
Length = 533
Score = 49.7 bits (117), Expect = 2e-05
Identities = 65/264 (24%), Positives = 111/264 (41%), Gaps = 28/264 (10%)
Query: 207 TPTSDFPHPLSRVRQPIGYRLSYQIVDALIWLGIRDLINEFRKKKLKLRAVTYLRGSYTF 266
T SD L RV+ + Y L+ + + L L +E ++++ L V + F
Sbjct: 198 TMLSDHMTFLQRVKNML-YPLTLKYICHLSITPYESLASELLQREMSLVEVLSHASVWLF 256
Query: 267 PPDMPYGYIWSPHLVPKPKDWGPNIDIVGF--CFLD--LASNYEPPKSLVDWLEEGENPI 322
D + Y P+ PN+ +G C + L+ +E + GE+ I
Sbjct: 257 RGDFVFDY---------PRPIMPNMVFIGGINCVIKKPLSQEFEAYVNA-----SGEHGI 302
Query: 323 YV-GFGSLPLQEPEKMTRIIVQALEQTGQRGIINKGWGGLGNLAELNTSKSVYLLDNCPH 381
V GS+ + PEK I +AL + Q + W G N +K+ L+ P
Sbjct: 303 VVFSLGSMVSEIPEKKAMEIAEALGRIPQTLL----WRYTGTRPS-NLAKNTILVKWLPQ 357
Query: 382 DWLF--PRCAAVVHHGGAGTTAAGLRAECPTTVVPFFGDQPFWGERVHARGVGPAPIRVE 439
+ L P+ A + H G+ G+ P ++P FGDQ +R+ RG G + V
Sbjct: 358 NDLLGHPKARAFITHSGSHGIYEGICNGVPMVMMPLFGDQMDNAKRMETRGAG-VTLNVL 416
Query: 440 EFTLERLVDAIRFMLNPEVKKRAV 463
E T + L +A++ ++N + K +
Sbjct: 417 EMTADDLENALKTVINNKSYKENI 440
>UD12_MOUSE (P70691) UDP-glucuronosyltransferase 1-2 precursor,
microsomal (EC 2.4.1.17) (UDPGT) (UGT1*0) (UGT1-02)
(UGT1.2) (UGT1A2) (Bilirubin specific)
Length = 533
Score = 49.7 bits (117), Expect = 2e-05
Identities = 75/316 (23%), Positives = 124/316 (38%), Gaps = 49/316 (15%)
Query: 176 DAIIANPPAYGHTHVAEYLNVPLHIFF-----------TMPWTPTSDFPHPLSRVR---- 220
D I+ +P +A+YL +P T P+S P+ L+R+
Sbjct: 146 DVILTDPIFPCGAVLAKYLQIPAVFILRSLSCGIEYEATQCPNPSSYIPNLLTRLSDHMD 205
Query: 221 --QPIGYRLSYQIVDALIWLGI---RDLINEFRKKKLKLRAVTYLRGSYTFPPDMPYGYI 275
Q + L Y ++ + L I L +E ++++ L V + F D Y
Sbjct: 206 FLQRVQNMLYYLVLKYICRLSITPYESLASELLQREVSLVEVLSHASVWLFRGDFVLDY- 264
Query: 276 WSPHLVPKPKDWGPNIDIVGFCFLDLASNYEPPKSLVDWLE-----EGENPIYV-GFGSL 329
P+ PN+ +G N K L E GE+ I V GS+
Sbjct: 265 --------PRPIMPNMVFIG------GINCVTKKPLSQEFEAYVNASGEHGIVVFSLGSM 310
Query: 330 PLQEPEKMTRIIVQALEQTGQRGIINKGWGGLGNLAELNTSKSVYLLDNCPHDWLF--PR 387
+ PEK I +AL + Q + W G N +K+ L+ P + L P+
Sbjct: 311 VSEIPEKKAMEIAEALGRIPQTVL----WRYTGTRPS-NLAKNTILVKWLPQNDLLGHPK 365
Query: 388 CAAVVHHGGAGTTAAGLRAECPTTVVPFFGDQPFWGERVHARGVGPAPIRVEEFTLERLV 447
A + H G+ G+ P ++P FGDQ +R+ RG G + V E T + L
Sbjct: 366 TRAFITHSGSHGIYEGICNGVPMVMMPLFGDQMDNAKRMETRGAG-VTLNVLEMTADDLE 424
Query: 448 DAIRFMLNPEVKKRAV 463
+A++ ++N + K +
Sbjct: 425 NALKTVINNKSYKENI 440
>UD16_MOUSE (Q64435) UDP-glucuronosyltransferase 1-6 precursor,
microsomal (EC 2.4.1.17) (UDPGT) (UGT1*6) (UGT1-06)
(UGT1.6) (UGT1A6) (UGP1A1) (Phenol
UDP-glucuronosyltransferase)
Length = 531
Score = 48.9 bits (115), Expect = 3e-05
Identities = 75/318 (23%), Positives = 127/318 (39%), Gaps = 50/318 (15%)
Query: 174 KADAIIANPPAYGHTHVAEYLNVP-LHIFFTMPWTPTSDFPHPLSRVRQPIGY------- 225
K DA+ +P +AEYLN+P +++F P + H L + P+ Y
Sbjct: 143 KFDALFTDPAMPCGVILAEYLNLPSVYLFRGFPCS----LEHMLGQSPSPVSYVPRFYTK 198
Query: 226 ---------RLSYQIVDALIWLGIRDLINEFRKKKLKLRAVTYLRGSYTFPPDMPYGYIW 276
RL+ IV+ L + + K ++ A L+ + P + +W
Sbjct: 199 FSDHMTFPQRLANFIVNIL-----ENYLYYCLYSKYEIIASDLLKRDVSLP-SLHQNSLW 252
Query: 277 S---PHLVPKPKDWGPNIDIVGFCFLDLASNYEPPKSLVDWLE-----EGENPIYV-GFG 327
+ P+ PN+ +G N + L E GE+ I V G
Sbjct: 253 LLRYDFVFEYPRPVMPNMIFLG------GINCKKKGKLTQEFEAYVNASGEHGIVVFSLG 306
Query: 328 SLPLQEPEKMTRIIVQALEQTGQRGIINKGWGGLGNLAELNTSKSVYLLDNCPHDWLF-- 385
S+ + PEK I +AL + Q + W G N +K+ L+ P + L
Sbjct: 307 SMVSEIPEKKAMEIAEALGRIPQTVL----WRYTGTRPS-NLAKNTILVKWLPQNDLLGH 361
Query: 386 PRCAAVVHHGGAGTTAAGLRAECPTTVVPFFGDQPFWGERVHARGVGPAPIRVEEFTLER 445
P+ A + H G+ G+ P ++P FGDQ +R+ RG G + V E T +
Sbjct: 362 PKTRAFITHSGSHGIYEGICNGVPMVMMPLFGDQMDNAKRMETRGAG-VTLNVLEMTADD 420
Query: 446 LVDAIRFMLNPEVKKRAV 463
L +A++ ++N + K +
Sbjct: 421 LENALKTVINNKSYKENI 438
>UD18_RAT (Q64634) UDP-glucuronosyltransferase 1-8 precursor,
microsomal (EC 2.4.1.17) (UDPGT) (UGT1*8) (UGT1-08)
(UGT1.8) (UGT1A8) (A3)
Length = 530
Score = 47.8 bits (112), Expect = 7e-05
Identities = 42/149 (28%), Positives = 69/149 (46%), Gaps = 9/149 (6%)
Query: 318 GENPIYV-GFGSLPLQEPEKMTRIIVQALEQTGQRGIINKGWGGLGNLAELNTSKSVYLL 376
GE+ I V GS+ + PEK I +AL + Q + W G N +K+ L+
Sbjct: 295 GEHGIVVFSLGSMVSEIPEKKAMEIAEALGRIPQTLL----WRYTGTRPS-NLAKNTILV 349
Query: 377 DNCPHDWLF--PRCAAVVHHGGAGTTAAGLRAECPTTVVPFFGDQPFWGERVHARGVGPA 434
P + L P+ A + H G+ G+ P ++P FGDQ +R+ RG G
Sbjct: 350 KWLPQNDLLGHPKARAFITHSGSHGIYEGICNGVPMVMMPLFGDQMDNAKRMETRGAG-V 408
Query: 435 PIRVEEFTLERLVDAIRFMLNPEVKKRAV 463
+ V E T + L +A++ ++N + K +
Sbjct: 409 TLNVLEMTADDLENALKTVINNKSYKENI 437
>UD17_RAT (Q64633) UDP-glucuronosyltransferase 1-7 precursor,
microsomal (EC 2.4.1.17) (UDPGT) (UGT1*7) (UGT1-07)
(UGT1.7) (UGT1A7) (A2)
Length = 531
Score = 47.8 bits (112), Expect = 7e-05
Identities = 42/149 (28%), Positives = 69/149 (46%), Gaps = 9/149 (6%)
Query: 318 GENPIYV-GFGSLPLQEPEKMTRIIVQALEQTGQRGIINKGWGGLGNLAELNTSKSVYLL 376
GE+ I V GS+ + PEK I +AL + Q + W G N +K+ L+
Sbjct: 296 GEHGIVVFSLGSMVSEIPEKKAMEIAEALGRIPQTLL----WRYTGTRPS-NLAKNTILV 350
Query: 377 DNCPHDWLF--PRCAAVVHHGGAGTTAAGLRAECPTTVVPFFGDQPFWGERVHARGVGPA 434
P + L P+ A + H G+ G+ P ++P FGDQ +R+ RG G
Sbjct: 351 KWLPQNDLLGHPKARAFITHSGSHGIYEGICNGVPMVMMPLFGDQMDNAKRMETRGAG-V 409
Query: 435 PIRVEEFTLERLVDAIRFMLNPEVKKRAV 463
+ V E T + L +A++ ++N + K +
Sbjct: 410 TLNVLEMTADDLENALKTVINNKSYKENI 438
>UD17_MOUSE (Q62452) UDP-glucuronosyltransferase 1-7 precursor,
microsomal (EC 2.4.1.17) (UDPGT) (UGT1*7) (UGT1-07)
(UGT1.7) (UGT1A7) (UGTP4) (Fragment)
Length = 520
Score = 47.8 bits (112), Expect = 7e-05
Identities = 42/149 (28%), Positives = 69/149 (46%), Gaps = 9/149 (6%)
Query: 318 GENPIYV-GFGSLPLQEPEKMTRIIVQALEQTGQRGIINKGWGGLGNLAELNTSKSVYLL 376
GE+ I V GS+ + PEK I +AL + Q + W G N +K+ L+
Sbjct: 285 GEHGIVVFSLGSMVSEIPEKKAMEIAEALGRIPQTVL----WRYTGTRPS-NLAKNTILV 339
Query: 377 DNCPHDWLF--PRCAAVVHHGGAGTTAAGLRAECPTTVVPFFGDQPFWGERVHARGVGPA 434
P + L P+ A + H G+ G+ P ++P FGDQ +R+ RG G
Sbjct: 340 KWLPQNDLIGHPKTRAFITHSGSHGIYEGICNGVPMVMMPLFGDQMDNAKRMETRGAG-V 398
Query: 435 PIRVEEFTLERLVDAIRFMLNPEVKKRAV 463
+ V E T + L +A++ ++N + K +
Sbjct: 399 TLNVLEMTADDLENALKTVINNKSYKENI 427
>UD16_RAT (P08430) UDP-glucuronosyltransferase 1-6 precursor,
microsomal (EC 2.4.1.17) (UDPGT) (UGT1*6) (UGT1-06)
(UGT1.6) (UGT1A6) (A1) (P-nitrophenol specific)
Length = 529
Score = 47.8 bits (112), Expect = 7e-05
Identities = 42/149 (28%), Positives = 69/149 (46%), Gaps = 9/149 (6%)
Query: 318 GENPIYV-GFGSLPLQEPEKMTRIIVQALEQTGQRGIINKGWGGLGNLAELNTSKSVYLL 376
GE+ I V GS+ + PEK I +AL + Q + W G N +K+ L+
Sbjct: 294 GEHGIVVFSLGSMVSEIPEKKAMEIAEALGRIPQTLL----WRYTGTRPS-NLAKNTILV 348
Query: 377 DNCPHDWLF--PRCAAVVHHGGAGTTAAGLRAECPTTVVPFFGDQPFWGERVHARGVGPA 434
P + L P+ A + H G+ G+ P ++P FGDQ +R+ RG G
Sbjct: 349 KWLPQNDLLGHPKARAFITHSGSHGIYEGICNGVPMVMMPLFGDQMDNAKRMETRGAG-V 407
Query: 435 PIRVEEFTLERLVDAIRFMLNPEVKKRAV 463
+ V E T + L +A++ ++N + K +
Sbjct: 408 TLNVLEMTADDLENALKTVINNKSYKENI 436
>UD13_RAT (Q64637) UDP-glucuronosyltransferase 1-3 precursor,
microsomal (EC 2.4.1.17) (UDPGT) (UGT1*3) (UGT1-03)
(UGT1.3) (UGT1A3) (B3)
Length = 531
Score = 47.8 bits (112), Expect = 7e-05
Identities = 42/149 (28%), Positives = 69/149 (46%), Gaps = 9/149 (6%)
Query: 318 GENPIYV-GFGSLPLQEPEKMTRIIVQALEQTGQRGIINKGWGGLGNLAELNTSKSVYLL 376
GE+ I V GS+ + PEK I +AL + Q + W G N +K+ L+
Sbjct: 296 GEHGIVVFSLGSMVSEIPEKKAMEIAEALGRIPQTLL----WRYTGTRPS-NLAKNTILV 350
Query: 377 DNCPHDWLF--PRCAAVVHHGGAGTTAAGLRAECPTTVVPFFGDQPFWGERVHARGVGPA 434
P + L P+ A + H G+ G+ P ++P FGDQ +R+ RG G
Sbjct: 351 KWLPQNDLLGHPKARAFITHSGSHGIYEGICNGVPMVMMPLFGDQMDNAKRMETRGAG-V 409
Query: 435 PIRVEEFTLERLVDAIRFMLNPEVKKRAV 463
+ V E T + L +A++ ++N + K +
Sbjct: 410 TLNVLEMTADDLENALKTVINNKSYKENI 438
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.321 0.141 0.443
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 69,530,521
Number of Sequences: 164201
Number of extensions: 3244426
Number of successful extensions: 7113
Number of sequences better than 10.0: 84
Number of HSP's better than 10.0 without gapping: 26
Number of HSP's successfully gapped in prelim test: 58
Number of HSP's that attempted gapping in prelim test: 7068
Number of HSP's gapped (non-prelim): 96
length of query: 520
length of database: 59,974,054
effective HSP length: 115
effective length of query: 405
effective length of database: 41,090,939
effective search space: 16641830295
effective search space used: 16641830295
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 68 (30.8 bits)
Medicago: description of AC148994.4


