

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148994.1 + phase: 0
(347 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
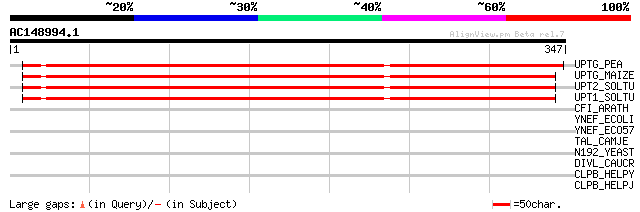
UPTG_PEA (O04300) Alpha-1,4-glucan-protein synthase [UDP-forming... 368 e-101
UPTG_MAIZE (P80607) Alpha-1,4-glucan-protein synthase [UDP-formi... 364 e-100
UPT2_SOLTU (Q8RU27) Alpha-1,4-glucan-protein synthase [UDP-formi... 358 1e-98
UPT1_SOLTU (Q9SC19) Alpha-1,4-glucan-protein synthase [UDP-formi... 353 3e-97
CFI_ARATH (P41088) Chalcone--flavonone isomerase (EC 5.5.1.6) (C... 30 6.9
YNEF_ECOLI (P76147) Hypothetical protein yneF 30 9.0
YNEF_ECO57 (Q8XAZ4) Hypothetical protein yneF 30 9.0
TAL_CAMJE (Q9PIL5) Transaldolase (EC 2.2.1.2) 30 9.0
N192_YEAST (P47054) Nucleoporin NUP192 (Nuclear pore protein NUP... 30 9.0
DIVL_CAUCR (Q9RQQ9) Sensor protein divL (EC 2.7.3.-) 30 9.0
CLPB_HELPY (P71404) Chaperone clpB 30 9.0
CLPB_HELPJ (Q9ZMH1) Chaperone clpB 30 9.0
>UPTG_PEA (O04300) Alpha-1,4-glucan-protein synthase [UDP-forming]
(EC 2.4.1.112) (UDP-glucose:protein transglucosylase)
(UPTG) (Reversibly glycosylated polypeptide)
Length = 364
Score = 368 bits (945), Expect = e-101
Identities = 168/339 (49%), Positives = 243/339 (71%), Gaps = 6/339 (1%)
Query: 9 NEVDIVIGALHSNLTPFMNEWKSIFSRFHLIIVKDPALKEELQIPEGFSADVYTNSEIER 68
+E+DIVI + + F+ W+ F ++HLIIV+D + +++PEGF ++Y ++I R
Sbjct: 13 DELDIVIPTIRN--LDFLEMWRPFFEQYHLIIVQDGDPSKVIKVPEGFDYELYNRNDINR 70
Query: 69 VVGSSTS-IRFSGYACRYFGFLVSKKKYVVCIDDDCVPAKDDAGNVVDAVAQHIVNLKTP 127
++G S I F ACR FG++VSKKKY+ IDDDC AKD G+ ++A+ QHI NL +P
Sbjct: 71 ILGPKASCISFKDSACRCFGYMVSKKKYIYTIDDDCFVAKDPTGHEINALEQHIKNLLSP 130
Query: 128 ATPFFFNTLYDPFRKGADFVRGYPFSLRSGVDCALSCGLWLNLADLDAPTQALKPTQRNS 187
+TPFFFNTLYDP+R+G DFVRGYPFSLR GV A+S GLWLN+ D DAPTQ +KP +RN+
Sbjct: 131 STPFFFNTLYDPYREGTDFVRGYPFSLREGVPTAVSHGLWLNIPDYDAPTQLVKPHERNT 190
Query: 188 RYVDAVLTVPTRAMLPVSGINIAFNRELVGPALVPALVLAGEGKLRWETVEDIWCGLCVK 247
R+VDAVLT+P ++ P+ G+N+AFNREL+GPA+ L+ G+ R+ +D+W G C+K
Sbjct: 191 RFVDAVLTIPKGSLFPMCGMNLAFNRELIGPAMYFGLMGDGQPIGRY---DDMWAGWCIK 247
Query: 248 IVCDHLSLGVKSGLPYVWRNERGNAIDSLKKEWEGVKLMEDVVPFFQSVKLPQSATTAED 307
++CDHL GVK+GLPY+W ++ N +LKKE++G+ E+++PFFQ+ L + T+ +
Sbjct: 248 VICDHLGYGVKTGLPYIWHSKASNPFVNLKKEYKGIFWQEEIIPFFQAATLSKDCTSVQK 307
Query: 308 CVIEMAKSVKEQLGKVDPMFLKAADAMAEWVKLWKSVGS 346
C IE++K VKE+LG +DP F+K ADAM WV+ W + +
Sbjct: 308 CYIELSKQVKEKLGTIDPYFIKLADAMVTWVEAWDEINN 346
>UPTG_MAIZE (P80607) Alpha-1,4-glucan-protein synthase [UDP-forming]
(EC 2.4.1.112) (UDP-glucose:protein transglucosylase)
(UPTG) (Amylogenin) (Golgi associated protein se-wap41)
Length = 364
Score = 364 bits (935), Expect = e-100
Identities = 167/334 (50%), Positives = 239/334 (71%), Gaps = 6/334 (1%)
Query: 9 NEVDIVIGALHSNLTPFMNEWKSIFSRFHLIIVKDPALKEELQIPEGFSADVYTNSEIER 68
+E+DIVI + + F+ W++ F +HLIIV+D + +++PEGF ++Y ++I R
Sbjct: 20 DELDIVIPTIRN--LDFLEMWRAFFQPYHLIIVQDGDPTKTIKVPEGFDYELYNRNDINR 77
Query: 69 VVGSSTS-IRFSGYACRYFGFLVSKKKYVVCIDDDCVPAKDDAGNVVDAVAQHIVNLKTP 127
++G S I F ACR FG++VSKKKY+ IDDDC AKD +G ++A+ QHI NL +P
Sbjct: 78 ILGPKASCISFKDSACRCFGYMVSKKKYIYTIDDDCFVAKDPSGKDINALEQHIKNLLSP 137
Query: 128 ATPFFFNTLYDPFRKGADFVRGYPFSLRSGVDCALSCGLWLNLADLDAPTQALKPTQRNS 187
+TPFFFNTLYDP+R+GADFVRGYPFSLR G A+S GLWLN+ D DAPTQ +KP +RN
Sbjct: 138 STPFFFNTLYDPYREGADFVRGYPFSLREGAHTAVSHGLWLNIPDYDAPTQLVKPKERNE 197
Query: 188 RYVDAVLTVPTRAMLPVSGINIAFNRELVGPALVPALVLAGEGKLRWETVEDIWCGLCVK 247
RYVDAV+T+P + P+ G+N+AF+R+L+GPA+ L+ G+ R+ +D+W G CVK
Sbjct: 198 RYVDAVMTIPKGTLFPMCGMNLAFDRDLIGPAMYFGLMGDGQPIGRY---DDMWAGWCVK 254
Query: 248 IVCDHLSLGVKSGLPYVWRNERGNAIDSLKKEWEGVKLMEDVVPFFQSVKLPQSATTAED 307
++CDHLSLGVK+GLPY+W ++ N +LKKE++G+ ED++PFFQ+V +P+ T +
Sbjct: 255 VICDHLSLGVKTGLPYIWHSKASNPFVNLKKEYKGIFWQEDIIPFFQNVTIPKDCDTVQK 314
Query: 308 CVIEMAKSVKEQLGKVDPMFLKAADAMAEWVKLW 341
C I ++ VKE+LG +DP F+K DAM W++ W
Sbjct: 315 CYIYLSGQVKEKLGTIDPYFVKLGDAMVTWIEAW 348
>UPT2_SOLTU (Q8RU27) Alpha-1,4-glucan-protein synthase [UDP-forming]
2 (EC 2.4.1.112) (UDP-glucose:protein transglucosylase
2) (UPTG 2)
Length = 366
Score = 358 bits (919), Expect = 1e-98
Identities = 164/334 (49%), Positives = 235/334 (70%), Gaps = 6/334 (1%)
Query: 9 NEVDIVIGALHSNLTPFMNEWKSIFSRFHLIIVKDPALKEELQIPEGFSADVYTNSEIER 68
+E+DIVI + + F+ W+ F +HLIIV+D + + +PEGF ++Y ++I R
Sbjct: 14 DELDIVIPTIRN--LDFLEMWRPFFQPYHLIIVQDGDPSKIINVPEGFDYELYNRNDINR 71
Query: 69 VVGSSTS-IRFSGYACRYFGFLVSKKKYVVCIDDDCVPAKDDAGNVVDAVAQHIVNLKTP 127
++G S I F ACR FG++VSKKKY+ IDDDC AKD +G ++A+ QHI NL P
Sbjct: 72 ILGPKASCISFKDSACRCFGYMVSKKKYIYTIDDDCFVAKDPSGKDINALEQHIKNLLCP 131
Query: 128 ATPFFFNTLYDPFRKGADFVRGYPFSLRSGVDCALSCGLWLNLADLDAPTQALKPTQRNS 187
+TP FFNTLYDP+R+GADFVRGYPFS+R G A+S GLWLN+ D DAPTQ +KP +RN+
Sbjct: 132 STPHFFNTLYDPYREGADFVRGYPFSMREGAATAVSHGLWLNIPDYDAPTQLVKPRERNT 191
Query: 188 RYVDAVLTVPTRAMLPVSGINIAFNRELVGPALVPALVLAGEGKLRWETVEDIWCGLCVK 247
RYVDAV+T+P + P+ G+N+AF+REL+GPA+ L+ G+ R+ +D+W G C+K
Sbjct: 192 RYVDAVMTIPKGTLFPMCGMNLAFDRELIGPAMYFGLMGDGQPIGRY---DDMWAGWCIK 248
Query: 248 IVCDHLSLGVKSGLPYVWRNERGNAIDSLKKEWEGVKLMEDVVPFFQSVKLPQSATTAED 307
++CDHL LGVK+GLPY+W ++ N +LKKE++G+ E+++PF QS LP+ T+ +
Sbjct: 249 VICDHLGLGVKTGLPYIWHSKASNPFVNLKKEYKGIYWQEEIIPFSQSATLPKDCTSVQQ 308
Query: 308 CVIEMAKSVKEQLGKVDPMFLKAADAMAEWVKLW 341
C +E++K VKE+L +DP F K ADAM W++ W
Sbjct: 309 CYLELSKQVKEKLSTIDPYFTKLADAMVTWIEAW 342
>UPT1_SOLTU (Q9SC19) Alpha-1,4-glucan-protein synthase [UDP-forming]
1 (EC 2.4.1.112) (UDP-glucose:protein transglucosylase
1) (UPTG 1)
Length = 365
Score = 353 bits (907), Expect = 3e-97
Identities = 160/334 (47%), Positives = 233/334 (68%), Gaps = 6/334 (1%)
Query: 9 NEVDIVIGALHSNLTPFMNEWKSIFSRFHLIIVKDPALKEELQIPEGFSADVYTNSEIER 68
+E+DIVI + + F+ W+ F +HLIIV+D + +++PEGF ++Y ++I R
Sbjct: 10 DELDIVIPTIRN--LDFLEMWRPFFQPYHLIIVQDGDPSKIIKVPEGFDYELYNRNDINR 67
Query: 69 VVGSSTS-IRFSGYACRYFGFLVSKKKYVVCIDDDCVPAKDDAGNVVDAVAQHIVNLKTP 127
++G S I F ACR FG++VSKKKY+ IDDDC AKD +G ++A+ QHI NL P
Sbjct: 68 ILGPKASCISFKDSACRCFGYMVSKKKYIYTIDDDCFVAKDPSGKDINALEQHIKNLLCP 127
Query: 128 ATPFFFNTLYDPFRKGADFVRGYPFSLRSGVDCALSCGLWLNLADLDAPTQALKPTQRNS 187
+TP FFNTLYDP+R GADFVRGYPFS+R G A+S GLWLN+ D DAPTQ +KP +RN+
Sbjct: 128 STPHFFNTLYDPYRDGADFVRGYPFSMREGAPTAVSHGLWLNIPDYDAPTQLVKPHERNT 187
Query: 188 RYVDAVLTVPTRAMLPVSGINIAFNRELVGPALVPALVLAGEGKLRWETVEDIWCGLCVK 247
RYVDAV+T+P + P+ G+N+AF+R+L+GPA+ L+ G+ R+ +D+W G C K
Sbjct: 188 RYVDAVMTIPKGTLFPMCGMNLAFDRDLIGPAMYFGLMGDGQPIGRY---DDMWAGWCTK 244
Query: 248 IVCDHLSLGVKSGLPYVWRNERGNAIDSLKKEWEGVKLMEDVVPFFQSVKLPQSATTAED 307
++CDHL LG+K+GLPY+W ++ N +LKKE+ G+ E+++PFFQ+ LP+ TT +
Sbjct: 245 VICDHLGLGIKTGLPYIWHSKASNPFVNLKKEYNGIFWQEEIIPFFQAATLPKECTTVQQ 304
Query: 308 CVIEMAKSVKEQLGKVDPMFLKAADAMAEWVKLW 341
C +E++K VK++L +DP F K +AM W++ W
Sbjct: 305 CYLELSKQVKKKLSSIDPYFTKLGEAMVTWIEAW 338
>CFI_ARATH (P41088) Chalcone--flavonone isomerase (EC 5.5.1.6)
(Chalcone isomerase) (TRANSPARENT TESTA 5 protein)
Length = 246
Score = 30.4 bits (67), Expect = 6.9
Identities = 22/85 (25%), Positives = 37/85 (42%), Gaps = 15/85 (17%)
Query: 270 GNAIDSLKKEWEG--VKLMEDVVPFFQSV-------------KLPQSATTAEDCVIEMAK 314
GNA+ SL +W+G + + + +PFF+ + KLP + + V E
Sbjct: 67 GNAVPSLSVKWKGKTTEELTESIPFFREIVTGAFEKFIKVTMKLPLTGQQYSEKVTENCV 126
Query: 315 SVKEQLGKVDPMFLKAADAMAEWVK 339
++ +QLG KA + E K
Sbjct: 127 AIWKQLGLYTDCEAKAVEKFLEIFK 151
>YNEF_ECOLI (P76147) Hypothetical protein yneF
Length = 315
Score = 30.0 bits (66), Expect = 9.0
Identities = 17/65 (26%), Positives = 30/65 (46%), Gaps = 6/65 (9%)
Query: 186 NSRYVDAVLTVPTRAMLPVSGINIAFNRELVGPAL------VPALVLAGEGKLRWETVED 239
+ ++ VL VP L + G+ F E + PA+ + ++V+ G G L +
Sbjct: 7 SEQFSTGVLIVPCMLTLAIPGVLPRFKAEQMMPAIALIVSVIASVVIGGAGSLAFPLPAL 66
Query: 240 IWCGL 244
IWC +
Sbjct: 67 IWCAV 71
>YNEF_ECO57 (Q8XAZ4) Hypothetical protein yneF
Length = 315
Score = 30.0 bits (66), Expect = 9.0
Identities = 17/65 (26%), Positives = 30/65 (46%), Gaps = 6/65 (9%)
Query: 186 NSRYVDAVLTVPTRAMLPVSGINIAFNRELVGPAL------VPALVLAGEGKLRWETVED 239
+ ++ VL VP L + G+ F E + PA+ + ++V+ G G L +
Sbjct: 7 SEQFSTGVLIVPCMLTLAIPGVLPRFKAEQMMPAIALIVSVIASVVIGGAGSLAFPLPAL 66
Query: 240 IWCGL 244
IWC +
Sbjct: 67 IWCAV 71
>TAL_CAMJE (Q9PIL5) Transaldolase (EC 2.2.1.2)
Length = 325
Score = 30.0 bits (66), Expect = 9.0
Identities = 24/100 (24%), Positives = 45/100 (45%), Gaps = 8/100 (8%)
Query: 99 IDDDCVPAKDDAGNVVDAVAQHIVNLKTPATPFFFNTLYDPFRKGADFVRGYPFSLRSGV 158
+ D+ + +A + A+ + + +K PAT + +Y+ + G FSL
Sbjct: 99 LHDNTTLSLGEAKRLYSAIGKENIMIKIPATKASYEVMYELMKNGISVNATLIFSLEQSQ 158
Query: 159 DC--ALSCGLWLNLADLDAPTQALKPTQRNSRYVDAVLTV 196
C AL+ G L + ALK ++N+R AV+++
Sbjct: 159 KCFEALNAG----LVEFRKNNIALK--EQNTRTPQAVISI 192
>N192_YEAST (P47054) Nucleoporin NUP192 (Nuclear pore protein NUP192)
Length = 1683
Score = 30.0 bits (66), Expect = 9.0
Identities = 16/76 (21%), Positives = 35/76 (46%)
Query: 39 IIVKDPALKEELQIPEGFSADVYTNSEIERVVGSSTSIRFSGYACRYFGFLVSKKKYVVC 98
I P E++ + + S ++Y ++E ++ +R F VSK++++
Sbjct: 1215 IFTNVPLNLEQVTLNKYCSGNIYDFHKMENLMRLIKRVRAESLHSNSFSLTVSKEQFLKD 1274
Query: 99 IDDDCVPAKDDAGNVV 114
D +C+ AK N++
Sbjct: 1275 ADVECIKAKSHFTNII 1290
>DIVL_CAUCR (Q9RQQ9) Sensor protein divL (EC 2.7.3.-)
Length = 769
Score = 30.0 bits (66), Expect = 9.0
Identities = 38/120 (31%), Positives = 48/120 (39%), Gaps = 7/120 (5%)
Query: 115 DAVAQHIVNLKTPATPFFFNTLYDPFRKGADFV---RGYPFSLRSGVDCALSCGLWLNLA 171
DA +V + A P + L F +G V RG P L S A WL LA
Sbjct: 90 DAEVSAVVAALSDADPNYAQKLTALFERGEPCVFEARG-PHGLVSVEGRAAGALAWLRLA 148
Query: 172 DLDAPTQALKPTQRNSRYVDAVLTVPTRAMLPVSGI--NIAFNRELVGPALVPALVLAGE 229
+D L R + +VD+V+ A I N AF R VG A A LAG+
Sbjct: 149 PIDRADSGLPTAARFAAFVDSVVEPCWIAGADGQAIWGNAAFVR-AVGAASAQAPALAGK 207
>CLPB_HELPY (P71404) Chaperone clpB
Length = 856
Score = 30.0 bits (66), Expect = 9.0
Identities = 24/99 (24%), Positives = 40/99 (40%), Gaps = 9/99 (9%)
Query: 16 GALHSNLTPFMNEWKSIFS-------RFHLIIVKDPALKEELQIPEGFSADVYTNSEIER 68
G LH+ + E++ F RF I++ +P++ E LQI G + T+ I
Sbjct: 303 GELHTIGATTLKEYRKYFEKDMALQRRFQPILLNEPSINEALQILRGLKETLETHHNI-- 360
Query: 69 VVGSSTSIRFSGYACRYFGFLVSKKKYVVCIDDDCVPAK 107
+ S I + + RY K + ID+ K
Sbjct: 361 TINDSALIASAKLSSRYITDRFLPDKAIDLIDEGAAQLK 399
>CLPB_HELPJ (Q9ZMH1) Chaperone clpB
Length = 856
Score = 30.0 bits (66), Expect = 9.0
Identities = 24/99 (24%), Positives = 40/99 (40%), Gaps = 9/99 (9%)
Query: 16 GALHSNLTPFMNEWKSIFS-------RFHLIIVKDPALKEELQIPEGFSADVYTNSEIER 68
G LH+ + E++ F RF I++ +P++ E LQI G + T+ I
Sbjct: 303 GELHTIGATTLKEYRKYFEKDMALQRRFQPILLNEPSINEALQILRGLKETLETHHNI-- 360
Query: 69 VVGSSTSIRFSGYACRYFGFLVSKKKYVVCIDDDCVPAK 107
+ S I + + RY K + ID+ K
Sbjct: 361 TINDSALIASAKLSSRYITDRFLPDKAIDLIDEGAAQLK 399
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.321 0.137 0.421
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 42,576,883
Number of Sequences: 164201
Number of extensions: 1805003
Number of successful extensions: 3697
Number of sequences better than 10.0: 12
Number of HSP's better than 10.0 without gapping: 5
Number of HSP's successfully gapped in prelim test: 7
Number of HSP's that attempted gapping in prelim test: 3681
Number of HSP's gapped (non-prelim): 15
length of query: 347
length of database: 59,974,054
effective HSP length: 111
effective length of query: 236
effective length of database: 41,747,743
effective search space: 9852467348
effective search space used: 9852467348
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 66 (30.0 bits)
Medicago: description of AC148994.1


