

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148970.7 - phase: 0
(548 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
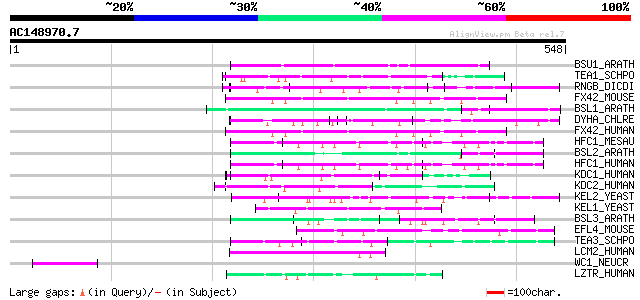
Score E
Sequences producing significant alignments: (bits) Value
BSU1_ARATH (Q9LR78) Serine/threonine protein phosphatase BSU1 (E... 82 5e-15
TEA1_SCHPO (P87061) Tip elongation aberrant protein 1 (Cell pola... 79 3e-14
RNGB_DICDI (Q7M3S9) Ring finger protein B (Protein rngB) 79 4e-14
FX42_MOUSE (Q6PDJ6) F-box only protein 42 76 3e-13
BSL1_ARATH (Q8L7U5) Serine/threonine protein phosphatase BSL1 (E... 76 3e-13
DYHA_CHLRE (Q39610) Dynein alpha chain, flagellar outer arm (DHC... 74 1e-12
FX42_HUMAN (Q6P3S6) F-box only protein 42 72 5e-12
HFC1_MESAU (P51611) Host cell factor C1 (HCF) (VP16 accessory pr... 68 7e-11
BSL2_ARATH (Q9SJF0) Serine/threonine protein phosphatase BSL2 (E... 67 1e-10
HFC1_HUMAN (P51610) Host cell factor C1 (HCF) (VP16 accessory pr... 67 2e-10
KDC1_HUMAN (Q8N7A1) Kelch domain containing protein 1 64 1e-09
KDC2_HUMAN (Q9Y2U9) Kelch domain containing protein 2 (Hepatocel... 62 3e-09
KEL2_YEAST (P50090) Kelch repeats protein 2 60 1e-08
KEL1_YEAST (P38853) Kelch repeats protein 1 58 7e-08
BSL3_ARATH (Q9SHS7) Serine/threonine protein phosphatase BSL3 (E... 57 2e-07
EFL4_MOUSE (P60882) Multiple EGF-like-domain protein 4 56 3e-07
TEA3_SCHPO (O14248) Tip elongation aberrant protein 3 (Cell pola... 54 1e-06
LCM2_HUMAN (O60294) Leucine carboxyl methyltransferase 2 (EC 2.1... 52 4e-06
WC1_NEUCR (Q01371) White collar 1 protein (WC1) 50 1e-05
LZTR_HUMAN (Q8N653) Leucine-zipper-like transcriptional regulato... 50 2e-05
>BSU1_ARATH (Q9LR78) Serine/threonine protein phosphatase BSU1 (EC
3.1.3.16) (Bri1 suppressor protein 1)
Length = 793
Score = 81.6 bits (200), Expect = 5e-15
Identities = 70/257 (27%), Positives = 108/257 (41%), Gaps = 9/257 (3%)
Query: 219 WRKLTVGGGVEPSRCNFSACAVGNRVVLFGGEGVNMQPMNDTFVLDLNSNNPEWQHVQVS 278
W +L G V R +A G +++ GG G + D ++LD+ +N +W V
Sbjct: 86 WTRLNPIGDVPSPRACHAAALYGTLILIQGGIGPSGPSDGDVYMLDMTNN--KWIKFLVG 143
Query: 279 SPPPG-RWGHTLSCVNGSRLVVFGGCGTQGLLNDVFVLDLDATPPTWREISGLA-PPLPR 336
P R+GH + LV+F G +L+D + LD P +W ++ P R
Sbjct: 144 GETPSPRYGHVMDIAAQRWLVIFSGNNGNEILDDTWALDTRG-PFSWDRLNPSGNQPSGR 202
Query: 337 SWHSSCTLDGTKLIVSGGCADSGVLLSDTFLLDMSMENPVWREIPVTWTPPSRLGHTLSV 396
+ S + + ++ GG SGV L DT+ L M +N VW +P P R HT +V
Sbjct: 203 MYASGSSREDGIFLLCGGIDHSGVTLGDTYGLKMDSDN-VWTPVPAV-APSPRYQHT-AV 259
Query: 397 YGGRKILMFGGLAKSGPLRFRSSDVFTMDLSEDEPCWRCVTGSGMPGAGNPEGIAPPPRL 456
+GG K+ + GG+ L + V +D E + GA R
Sbjct: 260 FGGSKLHVIGGILNRARLIDGEAVVAVLDTETGEWVDTNQPETSASGANRQNQYQLMRRC 319
Query: 457 DHVAVSLPGGRILIFGG 473
H A S G + + GG
Sbjct: 320 HHAAASF-GSHLYVHGG 335
Score = 43.9 bits (102), Expect = 0.001
Identities = 43/151 (28%), Positives = 63/151 (41%), Gaps = 9/151 (5%)
Query: 386 PPSRLGHTLS---VYGGRKILMFGGLAKSGPLRFRSSDVFTMD-LSEDEPCWRCVTGSGM 441
P R GHTL+ V ++++FGG + S ++D ++ + +T
Sbjct: 28 PGPRCGHTLTAVFVNNSHQLILFGGSTTAVANHNSSLPEISLDGVTNSVHSFDVLTRKWT 87
Query: 442 PGAGNPEGIAPPPRLDHVAVSLPGGRILIFGGSVAGLHSASQLYILDPTDEKPTWRILNV 501
NP G P PR H A +L G ILI GG S +Y+LD T+ K W V
Sbjct: 88 --RLNPIGDVPSPRACHAA-ALYGTLILIQGGIGPSGPSDGDVYMLDMTNNK--WIKFLV 142
Query: 502 PGRPPRFAWGHSTCVVGGTRAIVLGGQTGEE 532
G P +GH + ++ G G E
Sbjct: 143 GGETPSPRYGHVMDIAAQRWLVIFSGNNGNE 173
>TEA1_SCHPO (P87061) Tip elongation aberrant protein 1 (Cell
polarity protein tea1)
Length = 1147
Score = 79.0 bits (193), Expect = 3e-14
Identities = 63/222 (28%), Positives = 103/222 (46%), Gaps = 15/222 (6%)
Query: 211 LTTLEAAAWRKLTVGGGVEPSRCNFSACAVGNRVVLFGGEGVNMQPMNDTFVLDLNSNNP 270
L + W+K G R + +G+++ LFGG ++ ND DLN+ N
Sbjct: 167 LLNTSSLVWQKANASGARPSGRYGHTISCLGSKICLFGGRLLDYY-FNDLVCFDLNNLNT 225
Query: 271 E---WQHVQV-SSPPPGRWGHTLSCVNGSRLVVFGGCGTQGLLNDVFVLDLDATPPTWRE 326
W+ V + PPP R GH ++ +L +FGG ND++ + + E
Sbjct: 226 SDSRWELASVVNDPPPARAGH-VAFTFSDKLYIFGGTDGANFFNDLWCYHPKQSAWSKVE 284
Query: 327 ISGLAPPLPRSWHSSCTLDGTKLIVSGGCADSGVLLSDTFLLDMSMENPVWREI-PVTWT 385
G+AP PR+ H++ ++G L V GG A G L+D + +S ++ W ++ + +T
Sbjct: 285 TFGVAPN-PRAGHAASVVEGI-LYVFGGRASDGTFLNDLYAFRLSSKH--WYKLSDLPFT 340
Query: 386 PPSRLGHTLSVYGGRKILMFGGLAKSGPLRFRSSDVFTMDLS 427
P R HTLS G +L+ G K S+V+ +D S
Sbjct: 341 PSPRSSHTLSCSGLTLVLIGGKQGKGA----SDSNVYMLDTS 378
Score = 67.8 bits (164), Expect = 7e-11
Identities = 88/337 (26%), Positives = 131/337 (38%), Gaps = 83/337 (24%)
Query: 214 LEAAAWRKLTVGGG--VEP--SRCNFSACAVGNRVVLFGGEGVNMQPMNDTFVLD----- 264
+ A+ W KLTV G V P S + G + +FGG + QP ND +VL+
Sbjct: 61 ITASPWSKLTVRGSSNVLPRYSHASHLYAEGGQEIYIFGGVASDSQPKNDLWVLNLATSQ 120
Query: 265 -----------------------------------------------LNSNNPEWQHVQV 277
LN+++ WQ
Sbjct: 121 FTSLRSLGETPSPRLGHASILIGNAFIVFGGLTNHDVADRQDNSLYLLNTSSLVWQKANA 180
Query: 278 S-SPPPGRWGHTLSCVNGSRLVVFGGCGTQGLLNDVFVLD---LDATPPTWREISGL-AP 332
S + P GR+GHT+SC+ GS++ +FGG ND+ D L+ + W S + P
Sbjct: 181 SGARPSGRYGHTISCL-GSKICLFGGRLLDYYFNDLVCFDLNNLNTSDSRWELASVVNDP 239
Query: 333 PLPRSWHSSCTLDGTKLIVSGGCADSGVLLSDTFLLDMSMENPVWREIPVTWTPPS-RLG 391
P R+ H + T KL + GG D +D L + W ++ P+ R G
Sbjct: 240 PPARAGHVAFTF-SDKLYIFGG-TDGANFFND--LWCYHPKQSAWSKVETFGVAPNPRAG 295
Query: 392 HTLSVYGGRKILMFGGLAKSGPLRFRSSDVFTMDLSEDEPCWRCVTGSGMPGAGNPEGIA 451
H SV G + +FGG A G +D++ LS W + S +P
Sbjct: 296 HAASVVEG-ILYVFGGRASDGTF---LNDLYAFRLSSKH--WYKL--SDLP-------FT 340
Query: 452 PPPRLDHVAVSLPGGRILIFGGSVAGLHSASQLYILD 488
P PR H +S G +++ GG S S +Y+LD
Sbjct: 341 PSPRSSH-TLSCSGLTLVLIGGKQGKGASDSNVYMLD 376
>RNGB_DICDI (Q7M3S9) Ring finger protein B (Protein rngB)
Length = 943
Score = 78.6 bits (192), Expect = 4e-14
Identities = 64/217 (29%), Positives = 99/217 (45%), Gaps = 17/217 (7%)
Query: 219 WRKLTVGGGVEPSRCNFSACAVGNRVVLFGGEGVNMQPM-NDTFVLDLNSNNPEW-QHVQ 276
W+++T +E R +A +V+FGG + N + L SN EW Q V
Sbjct: 117 WKQVTTKS-IE-GRAGHTAVVYRQNLVVFGGHNNHKSKYYNSVLLFSLESN--EWRQQVC 172
Query: 277 VSSPPPGRWGHTLSCVNGSRLVVFGGCGTQGLLNDVFVLDLDATPPTWREISGLA-PPLP 335
P R H+ VN +++ +FGG + ND++ LDL+ W+++ PP P
Sbjct: 173 GGVIPSARATHSTFQVNNNKMFIFGGYDGKKYYNDIYYLDLETW--IWKKVEAKGTPPKP 230
Query: 336 RSWHSSCTLDGTKLIVSGGCADSGVLLSDTFLLDMSMENPVWREIPV---TWTPPSRLGH 392
RS HS+ + KL++ GGC S L+D +L + N E P P +R H
Sbjct: 231 RSGHSATMIQNNKLMIFGGCGSSSNFLNDIHILHIEGANEYRWEQPSYLGLEIPQARFRH 290
Query: 393 TLSVYGGRKILMFGGLAKSGPLRFRSSDVFTMDLSED 429
T + GGR + G SG L D+ T++ +D
Sbjct: 291 TTNFIGGRVYIYAG--TGSGNL---MGDLHTLEFLDD 322
Score = 77.8 bits (190), Expect = 7e-14
Identities = 75/278 (26%), Positives = 121/278 (42%), Gaps = 34/278 (12%)
Query: 277 VSSPPPGRWGHT-LSCVNGSRLVVFGGCGTQGLLNDVFVLDLDATPPTWREISGLA-PPL 334
+ SP P RWGHT + NGS +VFGG + ND+ ++ +W +I + P
Sbjct: 11 IGSPEP-RWGHTGTTLPNGSGFIVFGG-NSNRAFNDIQYYNIFNN--SWSKIEAVGNAPS 66
Query: 335 PRSWHSSCTL--------DGTKLIVSGGCADSGVLLSDTFLLDMSMENPVWREIPVTWTP 386
R HS+ D ++I GG A S L S + +W+++ T +
Sbjct: 67 ERYGHSAVLYQSQSRPYSDSYQIIFFGGRATSKPFSDINILYVNSNRSFIWKQV-TTKSI 125
Query: 387 PSRLGHTLSVYGGRKILMFGGLAKSGPLRFRSSDVFTMDLSEDEPCWR-CVTGSGMPGAG 445
R GHT VY + +++FGG + S +F+++ +E WR V G
Sbjct: 126 EGRAGHTAVVY-RQNLVVFGGHNNHKSKYYNSVLLFSLESNE----WRQQVCG------- 173
Query: 446 NPEGIAPPPRLDHVAVSLPGGRILIFGGSVAGLHSASQLYILDPTDEKPTWRILNVPGRP 505
G+ P R H + ++ IFGG G + +Y LD E W+ + G P
Sbjct: 174 ---GVIPSARATHSTFQVNNNKMFIFGG-YDGKKYYNDIYYLDL--ETWIWKKVEAKGTP 227
Query: 506 PRFAWGHSTCVVGGTRAIVLGGQTGEEWMLSDLHELSL 543
P+ GHS ++ + ++ GG L+D+H L +
Sbjct: 228 PKPRSGHSATMIQNNKLMIFGGCGSSSNFLNDIHILHI 265
Score = 77.0 bits (188), Expect = 1e-13
Identities = 77/293 (26%), Positives = 130/293 (44%), Gaps = 36/293 (12%)
Query: 218 AWRKLTVGGGVEPSRCNFSACAVGNR---------VVLFGGEGVNMQPMNDTFVLDLNSN 268
+W K+ G R SA ++ ++ FGG + +P +D +L +NSN
Sbjct: 54 SWSKIEAVGNAPSERYGHSAVLYQSQSRPYSDSYQIIFFGGRATS-KPFSDINILYVNSN 112
Query: 269 NP-EWQHVQVSSPPPGRWGHTLSCVNGSRLVVFGGCGT--QGLLNDVFVLDLDATPPTWR 325
W+ V S GR GHT + V LVVFGG N V + L++ WR
Sbjct: 113 RSFIWKQVTTKS-IEGRAGHT-AVVYRQNLVVFGGHNNHKSKYYNSVLLFSLESN--EWR 168
Query: 326 -EISGLAPPLPRSWHSSCTLDGTKLIVSGGCADSGVLLSDTFLLDMSMENPVWREIPVTW 384
++ G P R+ HS+ ++ K+ + GG D +D + LD +E +W+++
Sbjct: 169 QQVCGGVIPSARATHSTFQVNNNKMFIFGG-YDGKKYYNDIYYLD--LETWIWKKVEAKG 225
Query: 385 TPPS-RLGHTLSVYGGRKILMFGGLAKSGPLRFRSSDVFTMDL-SEDEPCWRCVTGSGMP 442
TPP R GH+ ++ K+++FGG S +D+ + + +E W + G+
Sbjct: 226 TPPKPRSGHSATMIQNNKLMIFGGCGSSSNF---LNDIHILHIEGANEYRWEQPSYLGLE 282
Query: 443 GAGNPEGIAPPPRLDHVAVSLPGGRILIFGGSVAGLHSASQLYILDPTDEKPT 495
P R H + GGR+ I+ G+ +G + L+ L+ D+ T
Sbjct: 283 --------IPQARFRH-TTNFIGGRVYIYAGTGSG-NLMGDLHTLEFLDDNNT 325
Score = 74.7 bits (182), Expect = 6e-13
Identities = 62/208 (29%), Positives = 105/208 (49%), Gaps = 14/208 (6%)
Query: 211 LTTLEAAAWRKLTVGGGVEPSRCNFSACAV-GNRVVLFGGEGVNMQPMNDTFVLDLNSNN 269
L +LE+ WR+ GG + +R S V N++ +FGG + ND + LDL +
Sbjct: 159 LFSLESNEWRQQVCGGVIPSARATHSTFQVNNNKMFIFGGYD-GKKYYNDIYYLDLET-- 215
Query: 270 PEWQHVQV-SSPPPGRWGHTLSCVNGSRLVVFGGCG-TQGLLNDVFVLDLD-ATPPTWRE 326
W+ V+ +PP R GH+ + + ++L++FGGCG + LND+ +L ++ A W +
Sbjct: 216 WIWKKVEAKGTPPKPRSGHSATMIQNNKLMIFGGCGSSSNFLNDIHILHIEGANEYRWEQ 275
Query: 327 IS--GLAPPLPRSWHSSCTLDGTKLIVSGGCADSGVLLSDTFLLDMSMENPVWREIPVTW 384
S GL P R H++ + G I +G SG L+ D L+ +N IP+T
Sbjct: 276 PSYLGLEIPQARFRHTTNFIGGRVYIYAG--TGSGNLMGDLHTLEFLDDNNT-PLIPITI 332
Query: 385 TPPSRLGHTLSVYGGRKILMFGGLAKSG 412
+ P + ++ S+ G + G++ SG
Sbjct: 333 SIP--ITNSNSIVGSPNTSISCGVSNSG 358
>FX42_MOUSE (Q6PDJ6) F-box only protein 42
Length = 717
Score = 75.9 bits (185), Expect = 3e-13
Identities = 84/317 (26%), Positives = 138/317 (43%), Gaps = 46/317 (14%)
Query: 214 LEAAAWRKLTVGGGVEPSRCNFSACAVGNRVVLFGG-EGVNMQPMN--DTFVLDLNSNNP 270
L + W + G + + + +VLFGG + P++ + F ++++ +P
Sbjct: 156 LNSKEWIRPLASGSYPSPKAGATLVVYKDLLVLFGGWTRPSPYPLHQPERFFDEIHTYSP 215
Query: 271 E---WQHVQVSSPPPGRWGHTLSCVNGSRLVVFGGC-GTQGLLNDVFVLDLDATPPTWRE 326
W + + PP GH+ SCV G +++VFGG G++ + N+V+VLDL+ +
Sbjct: 216 SKNWWNCIVTTHGPPPMAGHS-SCVIGDKMIVFGGSLGSRQMSNEVWVLDLEQWAWSKPN 274
Query: 327 ISGLAPPLPRSWHSSCTLDGTKLIVSGGCADSGVLLSDTFLLDMSMENPVWREI------ 380
ISG +P PR S +D T L++ GGC L D +LL M W+ +
Sbjct: 275 ISGPSPH-PRGGQSQIVIDDTTLLILGGCGGPNALFKDAWLLHMHPGPWAWQPLKVENED 333
Query: 381 ---PVTWTPPS-RLGHTLSVY----GGRKILMFGGLAKSGPL------------RFRS-S 419
P W P+ R+G + V+ GR L ++ P+ +RS S
Sbjct: 334 HGAPELWCHPACRVGQCVVVFSQAPSGRAPLSPSLNSRPSPISATPPALVPETREYRSQS 393
Query: 420 DVFTMDLSEDEPCWRCVTGSGMPGA------GNPEGIAPPPRLDHVAVSLPGGRILIFGG 473
V +MD + PC G+ P A G+ EG P R D + L GG +
Sbjct: 394 PVRSMD---EAPCVNGRWGTLRPRAQRQTPSGSREGSLSPARGDGSPI-LNGGNLSPGTV 449
Query: 474 SVAGLHSASQLYILDPT 490
+V G S + ++ P+
Sbjct: 450 AVGGASLDSPVQVVSPS 466
Score = 39.3 bits (90), Expect = 0.027
Identities = 46/174 (26%), Positives = 71/174 (40%), Gaps = 23/174 (13%)
Query: 378 REIPVTWTP-PSRLGHTLSVY-GGRKILMFGGLAKSGPLRFRSSDVFTMDLSEDEPCWRC 435
R P TP R H+ Y + + +FGG +S +D++ +DL+ E
Sbjct: 107 RTYPYPGTPITQRFSHSACYYDANQSMYVFGGCTQSS-CNAAFNDLWRLDLNSKEWIRPL 165
Query: 436 VTGSGMPGAGNPEGIAPPPRLDHVAVSLPGGRILIFGG----SVAGLHSASQLY--ILDP 489
+GS P P+ V +++FGG S LH + + I
Sbjct: 166 ASGS-----------YPSPKAGATLVVYKD-LLVLFGGWTRPSPYPLHQPERFFDEIHTY 213
Query: 490 TDEKPTWRILNVPGRPPRFAWGHSTCVVGGTRAIVLGGQTGEEWMLSDLHELSL 543
+ K W + PP A GHS+CV+ G + IV GG G M +++ L L
Sbjct: 214 SPSKNWWNCIVTTHGPPPMA-GHSSCVI-GDKMIVFGGSLGSRQMSNEVWVLDL 265
>BSL1_ARATH (Q8L7U5) Serine/threonine protein phosphatase BSL1 (EC
3.1.3.16) (BSU1-like protein 1)
Length = 881
Score = 75.9 bits (185), Expect = 3e-13
Identities = 75/287 (26%), Positives = 112/287 (38%), Gaps = 13/287 (4%)
Query: 195 TVPGAKRLGWGRLARELTTLEAAAWRKLTVGGGVEPSRCNFSACAVGNRVVLFGGEGVNM 254
+VPG + G L W +L G R +A AVG VV GG G
Sbjct: 63 SVPGIRLAGVTNTVHSYDIL-TRKWTRLKPAGEPPSPRAAHAAAAVGTMVVFQGGIGPAG 121
Query: 255 QPMNDTFVLDLNSNNPEWQHVQVSSPPPG-RWGHTLSCVNGSRLVVFGGCGTQGLLNDVF 313
+D +VLD+ ++ +W V V PG R+GH + V+ LV G + L+D +
Sbjct: 122 HSTDDLYVLDMTNDKFKWHRVVVQGDGPGPRYGHVMDLVSQRYLVTVTGNDGKRALSDAW 181
Query: 314 VLDLDATPPTWREISGLAP-PLPRSWHSSCTLDGTKLIVSGGCADSGVLLSDTFLLDMSM 372
LD P W+ ++ P R + S ++ GG G L D + L M
Sbjct: 182 ALDTAQKPYVWQRLNPDGDRPSARMYASGSARSDGMFLLCGGRDTLGAPLGDAYGLLMH- 240
Query: 373 ENPVWREIPVTWTPPSRLGHTLSVYGGRKILMFGGLAKSGPLRFRSSDVFTMDLSEDEPC 432
N W PS +V+ G ++ + GG+ + G + + V +D +
Sbjct: 241 RNGQWEWTLAPGVAPSPRYQHAAVFVGARLHVSGGVLRGGRVIDAEASVAVLDTAAG--V 298
Query: 433 WRCVTGSGMPGAGNPEGIAPPP------RLDHVAVSLPGGRILIFGG 473
W G G+ I P R H A S+ G RI + GG
Sbjct: 299 WLDRNGQVTSARGSKGQIDQDPSFELMRRCRHGAASV-GIRIYVHGG 344
Score = 47.8 bits (112), Expect = 8e-05
Identities = 32/99 (32%), Positives = 47/99 (47%), Gaps = 4/99 (4%)
Query: 447 PEGIAPPPRLDHVAVSLPGGRILIFGGSVAGL-HSASQLYILDPTDEKPTWRILNVPGRP 505
P G P PR H A ++ G +++F G + HS LY+LD T++K W + V G
Sbjct: 91 PAGEPPSPRAAHAAAAV--GTMVVFQGGIGPAGHSTDDLYVLDMTNDKFKWHRVVVQGDG 148
Query: 506 PRFAWGHSTCVVGGTRAIVLGGQTGEEWMLSDLHELSLA 544
P +GH +V + + G G+ LSD L A
Sbjct: 149 PGPRYGHVMDLVSQRYLVTVTGNDGKR-ALSDAWALDTA 186
>DYHA_CHLRE (Q39610) Dynein alpha chain, flagellar outer arm (DHC
alpha)
Length = 4499
Score = 73.6 bits (179), Expect = 1e-12
Identities = 53/186 (28%), Positives = 86/186 (45%), Gaps = 9/186 (4%)
Query: 219 WRKLTVGGGVEPSRCNFSACAVGNRVVLFGGEGVN---MQPMNDTFVLDLNSNNP-EWQH 274
W G R S +G R VLFGG G ND + LD + + +W+
Sbjct: 5 WEVPNAQGEAPCPRSGHSFTVLGERFVLFGGCGRKDGKAAAFNDLYELDTSDPDEYKWKE 64
Query: 275 VQVSSPPPGRWGHTLSCVNGSRLVVFGGCGTQGLLNDVFVLDLDATPPTWREISGLAPPL 334
+ V++ PP R H ++ RL+VFGG + NDV++ + D T E+ G AP
Sbjct: 65 LVVANAPPPRARHAAIALDDKRLLVFGGLNKRIRYNDVWLFNYDDKSWTCMEVEGAAPE- 123
Query: 335 PRSWHSSCTLDGTKLIVSGGCADSGVLLSDTFLLDMSMENPVWREIPVT---WTPPSRLG 391
PR+ H + T G+++ + GG SG + ++ ++L + W+ I + P R
Sbjct: 124 PRA-HFTATRFGSRVFIFGGYGGSGQVYNEMWVLHFGEDGFRWQNITESIEGTGPAPRFD 182
Query: 392 HTLSVY 397
H+ +Y
Sbjct: 183 HSAFIY 188
Score = 73.2 bits (178), Expect = 2e-12
Identities = 62/224 (27%), Positives = 100/224 (43%), Gaps = 31/224 (13%)
Query: 333 PLPRSWHSSCTLDGTKLIVSGGCA---DSGVLLSDTFLLDMSMENPV-WREIPVTWTPPS 388
P PRS HS L G + ++ GGC +D + LD S + W+E+ V PP
Sbjct: 15 PCPRSGHSFTVL-GERFVLFGGCGRKDGKAAAFNDLYELDTSDPDEYKWKELVVANAPPP 73
Query: 389 RLGHTLSVYGGRKILMFGGLAKSGPLRFRSSDVFTMDLSEDEPCWRCVTGSGMPGAGNPE 448
R H +++L+FGGL K R R +DV+ + D+ W C+ E
Sbjct: 74 RARHAAIALDDKRLLVFGGLNK----RIRYNDVWLFNY--DDKSWTCM---------EVE 118
Query: 449 GIAPPPRLDHVAVSLPGGRILIFGGSVAGLHSASQLYILDPTDEKPTWRIL--NVPGRPP 506
G AP PR H + G R+ IFGG +++++L ++ W+ + ++ G P
Sbjct: 119 GAAPEPRA-HFTATRFGSRVFIFGGYGGSGQVYNEMWVLHFGEDGFRWQNITESIEGTGP 177
Query: 507 RFAWGHSTCVVGGT-------RAIVLGGQTGEEWMLSDLHELSL 543
+ HS + T + +++GG+ + M D H L L
Sbjct: 178 APRFDHSAFIYPVTPNSDTYDKLLIMGGRDLSQ-MYQDSHMLDL 220
Score = 50.1 bits (118), Expect = 2e-05
Identities = 33/103 (32%), Positives = 48/103 (46%), Gaps = 13/103 (12%)
Query: 218 AWRKLTVGGGVEPSRCNFSACAVGNRVVLFGGEGVNMQPMNDTFVLDLNSNNPEWQHVQV 277
+W K V G R + C +GN +VL+GG+ + V LN+ EW+
Sbjct: 537 SWTKHRVMGAAPAKRKGATICTMGNELVLYGGD--------KSGVTVLNTEGAEWRWSPA 588
Query: 278 S---SPPPGRWGHTLSCVNGSRLVVFGG--CGTQGLLNDVFVL 315
+ S PP R H+ ++ LVVFGG Q LND++ L
Sbjct: 589 TVSGSTPPDRTAHSTVVLSDGELVVFGGINLADQNDLNDIYYL 631
Score = 48.1 bits (113), Expect = 6e-05
Identities = 34/123 (27%), Positives = 55/123 (44%), Gaps = 17/123 (13%)
Query: 218 AWRKLTVGGGVEPSRCNFSACAVGNRVVLFGGEGVNMQPMNDTFVLDLNSNNPEWQHVQV 277
+W + V G R +F+A G+RV +FGG G + Q N+ +VL + WQ++
Sbjct: 111 SWTCMEVEGAAPEPRAHFTATRFGSRVFIFGGYGGSGQVYNEMWVLHFGEDGFRWQNITE 170
Query: 278 S---SPPPGRWGH-------TLSCVNGSRLVVFGGCGTQGLLNDVFVLDL-------DAT 320
S + P R+ H T + +L++ GG + D +LDL +
Sbjct: 171 SIEGTGPAPRFDHSAFIYPVTPNSDTYDKLLIMGGRDLSQMYQDSHMLDLNKMAWENETQ 230
Query: 321 PPT 323
PPT
Sbjct: 231 PPT 233
Score = 40.0 bits (92), Expect = 0.016
Identities = 38/126 (30%), Positives = 58/126 (45%), Gaps = 19/126 (15%)
Query: 331 APPLPRSWHSSCTLDGTKLIVSGGCADSGVLLSDTFLLDMSMENPVWREIPVTW---TPP 387
A P R + CT+ G +L++ GG SGV + +T E WR P T TPP
Sbjct: 546 AAPAKRKGATICTM-GNELVLYGG-DKSGVTVLNT-------EGAEWRWSPATVSGSTPP 596
Query: 388 SRLGHTLSVYGGRKILMFGG--LAKSGPLRFRSSDVFTMDLSEDEPCWRCVTGSGMPGAG 445
R H+ V ++++FGG LA L +D++ + + W C + S P
Sbjct: 597 DRTAHSTVVLSDGELVVFGGINLADQNDL----NDIYYLRKQGEGWVWSCPSES-RPYIR 651
Query: 446 NPEGIA 451
+P+G A
Sbjct: 652 HPKGAA 657
Score = 38.5 bits (88), Expect = 0.046
Identities = 39/169 (23%), Positives = 69/169 (40%), Gaps = 17/169 (10%)
Query: 205 GRLARELTTLEA-----AAWRKLTVGGGVEPSRCNFSACAVGNRVVLFGGEGVNMQ---- 255
G L EL ++ A W + + G V R +A A GN +VLFGG+ N
Sbjct: 759 GDLMTELAMVDTSDRTCAHWLEPILKGDVPVPRKACAAAATGNTIVLFGGQTQNADGEAT 818
Query: 256 PMNDTFVLDLNSNNPEWQHVQVSSP-----PPGRWGHTLSCVNGSRLVVFGGCGTQGL-L 309
D ++++ N V ++P P R+G + + +L + GG L
Sbjct: 819 VTGDLVIMEVTGPNSIQCAVNPAAPGASGSPAARYGAVMQEFSNGKLFLHGGMDAASKPL 878
Query: 310 NDVFVLDLDATPPTWREISGLAPPLPRSWHSSCTLDGTKLIVSGGCADS 358
ND ++ D+ + TW+ + + + TL G ++++ S
Sbjct: 879 NDGWLFDVPS--KTWQCVYVGSSDVVLPTGQLATLSGNRIVLVSAAVGS 925
Score = 33.5 bits (75), Expect = 1.5
Identities = 31/112 (27%), Positives = 47/112 (41%), Gaps = 20/112 (17%)
Query: 386 PPSRLGHTLSVYGGRKILMFGGLAKSGPLRFRSSDVFTMDLSEDEPCWRCVTGSGMPGAG 445
P R G T+ G +++++GG KSG V ++ E W T SG
Sbjct: 548 PAKRKGATICTMGN-ELVLYGG-DKSG--------VTVLNTEGAEWRWSPATVSGS---- 593
Query: 446 NPEGIAPPPRLDHVAVSLPGGRILIFGG-SVAGLHSASQLYILDPTDEKPTW 496
PP R H V L G +++FGG ++A + + +Y L E W
Sbjct: 594 -----TPPDRTAHSTVVLSDGELVVFGGINLADQNDLNDIYYLRKQGEGWVW 640
>FX42_HUMAN (Q6P3S6) F-box only protein 42
Length = 717
Score = 71.6 bits (174), Expect = 5e-12
Identities = 82/317 (25%), Positives = 135/317 (41%), Gaps = 46/317 (14%)
Query: 214 LEAAAWRKLTVGGGVEPSRCNFSACAVGNRVVLFGG-EGVNMQPMN--DTFVLDLNSNNP 270
L + W + G + + + +VLFGG + P++ + F ++++ +P
Sbjct: 156 LNSKEWIRPLASGSYPSPKAGATLVVYKDLLVLFGGWTRPSPYPLHQPERFFDEIHTYSP 215
Query: 271 E---WQHVQVSSPPPGRWGHTLSCVNGSRLVVFGGC-GTQGLLNDVFVLDLDATPPTWRE 326
W + + PP GH+ SCV +++VFGG G++ + NDV+VLDL+ +
Sbjct: 216 SKNWWNCIVTTHGPPPMAGHS-SCVIDDKMIVFGGSLGSRQMSNDVWVLDLEQWAWSKPN 274
Query: 327 ISGLAPPLPRSWHSSCTLDGTKLIVSGGCADSGVLLSDTFLLDMSMENPVWREI------ 380
ISG +P PR S +D +++ GGC L D +LL M W+ +
Sbjct: 275 ISGPSPH-PRGGQSQIVIDDATILILGGCGGPNALFKDAWLLHMHSGPWAWQPLKVENEE 333
Query: 381 ---PVTWTPPS-RLGHTLSVY----GGRKILMFGGLAKSGPL------------RFRS-S 419
P W P+ R+G + V+ GR L ++ P+ +RS S
Sbjct: 334 HGAPELWCHPACRVGQCVVVFSQAPSGRAPLSPSLNSRPSPISATPPALVPETREYRSQS 393
Query: 420 DVFTMDLSEDEPCWRCVTGSGMPGA------GNPEGIAPPPRLDHVAVSLPGGRILIFGG 473
V +MD + PC G+ P A G+ EG P R D + L GG +
Sbjct: 394 PVRSMD---EAPCVNGRWGTLRPRAQRQTPSGSREGSLSPARGDGSPI-LNGGSLSPGTA 449
Query: 474 SVAGLHSASQLYILDPT 490
+V G S + + P+
Sbjct: 450 AVGGSSLDSPVQAISPS 466
>HFC1_MESAU (P51611) Host cell factor C1 (HCF) (VP16 accessory
protein) (HFC1) (VCAF) (CFF)
Length = 2090
Score = 67.8 bits (164), Expect = 7e-11
Identities = 79/276 (28%), Positives = 121/276 (43%), Gaps = 38/276 (13%)
Query: 270 PEWQHVQVSSPPPGRWGHTLSCVNGSRLVVFGGCGTQGLLNDVFVLDLDATPPTWREISG 329
P W+ V S P R H V L+V G G +G+++++ V + + G
Sbjct: 17 PRWKRVVGWSGPVPRPRHGHRAVAIKELIVVFGGGNEGIVDELHVYNTATNQWFIPAVRG 76
Query: 330 LAPPLPRSWHSSCTLDGTKLIVSGGCADSGVLLSDTFLLDMSMENPVWREI----PVTWT 385
PP ++ C DGT+L+V GG + G +D + L S W+ + P
Sbjct: 77 DIPPGCAAYGFVC--DGTRLLVFGGMVEYGKYSNDLYELQASRWE--WKRLKAKTPKNGP 132
Query: 386 PP-SRLGHTLSVYGGRKILMFGGLA------KSGPLRFRSSDVFTMDLSEDEPCWRCVTG 438
PP RLGH+ S+ G K +FGGLA K+ R+ +D++ ++L G
Sbjct: 133 PPCPRLGHSFSLV-GNKCYLFGGLANDSEDPKNNIPRY-LNDLYILELR---------PG 181
Query: 439 SGMPGAGNP--EGIAPPPRLDHVAV-----SLPGGRILIFGGSVAGLHSASQLYILDPTD 491
SG+ P G+ PPPR H AV +++I+GG ++G L+ LD
Sbjct: 182 SGVVAWDIPITYGVLPPPRESHTAVVYTEKDNKKSKLVIYGG-MSGCR-LGDLWTLD--I 237
Query: 492 EKPTWRILNVPGRPPRFAWGHSTCVVGGTRAIVLGG 527
E TW ++ G P HS + G + V GG
Sbjct: 238 ETLTWNKPSLSGVAPLPRSLHSATTI-GNKMYVFGG 272
Score = 54.3 bits (129), Expect = 8e-07
Identities = 56/216 (25%), Positives = 93/216 (42%), Gaps = 39/216 (18%)
Query: 219 WRKLTVGGGVEPSRCNFSACAVGNRVVLFGGEGVNMQPMNDTFVLDLNSNNPEWQHVQVS 278
W V G + P + G R+++FGG + ND + +L ++ EW+ ++
Sbjct: 69 WFIPAVRGDIPPGCAAYGFVCDGTRLLVFGGMVEYGKYSNDLY--ELQASRWEWKRLKAK 126
Query: 279 SPPPG-----RWGHTLSCVNGSRLVVFGGCGTQG---------LLNDVFVLDLDA----- 319
+P G R GH+ S V G++ +FGG LND+++L+L
Sbjct: 127 TPKNGPPPCPRLGHSFSLV-GNKCYLFGGLANDSEDPKNNIPRYLNDLYILELRPGSGVV 185
Query: 320 ---TPPTWREISGLAPPLPRSWHSSCTL-----DGTKLIVSGGCADSGVLLSDTFLLDMS 371
P T+ G+ PP PR H++ +KL++ GG SG L D + LD
Sbjct: 186 AWDIPITY----GVLPP-PRESHTAVVYTEKDNKKSKLVIYGGM--SGCRLGDLWTLD-- 236
Query: 372 MENPVWREIPVTWTPPSRLGHTLSVYGGRKILMFGG 407
+E W + ++ P + G K+ +FGG
Sbjct: 237 IETLTWNKPSLSGVAPLPRSLHSATTIGNKMYVFGG 272
Score = 34.3 bits (77), Expect = 0.86
Identities = 35/113 (30%), Positives = 50/113 (43%), Gaps = 16/113 (14%)
Query: 430 EPCWRCVTGSGMPGAGNPEGIAPPPRLDHVAVSLPGGRILIFGGSVAGLHSASQLYILDP 489
+P W+ V G P P PR H AV++ I++FGG G+ +L++ +
Sbjct: 16 QPRWKRVVGWSGP--------VPRPRHGHRAVAIKE-LIVVFGGGNEGI--VDELHVYNT 64
Query: 490 TDEKPTWRILNVPGR-PPRFAWGHSTCVVGGTRAIVLGGQTGEEWMLSDLHEL 541
+ W I V G PP A C GTR +V GG +DL+EL
Sbjct: 65 ATNQ--WFIPAVRGDIPPGCAAYGFVC--DGTRLLVFGGMVEYGKYSNDLYEL 113
Score = 32.3 bits (72), Expect = 3.3
Identities = 35/156 (22%), Positives = 61/156 (38%), Gaps = 25/156 (16%)
Query: 214 LEAAAWRKLTVGGGVEPSRCNFSACAVGNRVVLFGG------EGVNMQPMNDTF-----V 262
+E W K ++ G R SA +GN++ +FGG + V + + +
Sbjct: 237 IETLTWNKPSLSGVAPLPRSLHSATTIGNKMYVFGGWVPLVMDDVKVATHEKEWKCTNTL 296
Query: 263 LDLNSNNPEWQHVQV----SSPPPGRWGHTLSCVNGSRLVVFGG-------CGTQGLLND 311
LN + W+ + + + P R GH +N +RL ++ G Q D
Sbjct: 297 ACLNLDTMAWETILMDTLEDNIPRARAGHCAVAIN-TRLYIWSGRDGYRKAWNNQVCCKD 355
Query: 312 VFVLDLDATPPTWRE--ISGLAPPLPRSWHSSCTLD 345
++ L+ + PP R + L SW + T D
Sbjct: 356 LWYLETEKPPPPARVQLVRANTNSLEVSWGAVATAD 391
>BSL2_ARATH (Q9SJF0) Serine/threonine protein phosphatase BSL2 (EC
3.1.3.16) (BSU1-like protein 2)
Length = 1088
Score = 67.0 bits (162), Expect = 1e-10
Identities = 77/268 (28%), Positives = 101/268 (36%), Gaps = 34/268 (12%)
Query: 219 WRKLTVGGGVEPSRCNFSACAVGNRVVLFGGEGVNMQPMNDTFVLDLNSNNPEWQHVQVS 278
W +LT G R A AVG VV+ GG G D VLDL P W V V
Sbjct: 192 WTRLTPFGEPPTPRAAHVATAVGTMVVIQGGIGPAGLSAEDLHVLDLTQQRPRWHRVVVQ 251
Query: 279 SPPPG-RWGHTLSCVNGSRLVVFGGCGTQGLLNDVFVLDLDATPPTWREISGLAPPLPRS 337
P PG R+GH ++ V L+ GG ND G PP
Sbjct: 252 GPGPGPRYGHVMALVGQRYLMAIGG-------ND----------------EGEGPPPCMY 288
Query: 338 WHSSCTLDGTKLIVSGGCADSGVLLSDTFLLDMSMENPVWREIPVTWTPPSRLGHTLSVY 397
+S DG L+ G A+S L S L W P +P SR H +V+
Sbjct: 289 ATASARSDGLLLLCGGRDANSVPLASAYGLAKHRDGRWEWAIAPGV-SPSSRYQHA-AVF 346
Query: 398 GGRKILMFGGLAKSGPLRFRSSDVFTMDLSEDEPC-WRCVTGSGMPG------AGNPEGI 450
++ + GG G + SS V +D + C + V S G AG +
Sbjct: 347 VNARLHVSGGALGGGRMVEDSSSVAVLDTAAGVWCDTKSVVTSPRTGRYSADAAGGDASV 406
Query: 451 APPPRLDHVAVSLPGGRILIFGGSVAGL 478
R H A ++ G I I+GG G+
Sbjct: 407 ELTRRCRHAAAAV-GDLIFIYGGLRGGV 433
Score = 50.1 bits (118), Expect = 2e-05
Identities = 32/82 (39%), Positives = 45/82 (54%), Gaps = 3/82 (3%)
Query: 447 PEGIAPPPRLDHVAVSLPGGRILIFGG-SVAGLHSASQLYILDPTDEKPTWRILNVPGRP 505
P G P PR HVA ++ G ++I GG AGL SA L++LD T ++P W + V G
Sbjct: 197 PFGEPPTPRAAHVATAV-GTMVVIQGGIGPAGL-SAEDLHVLDLTQQRPRWHRVVVQGPG 254
Query: 506 PRFAWGHSTCVVGGTRAIVLGG 527
P +GH +VG + +GG
Sbjct: 255 PGPRYGHVMALVGQRYLMAIGG 276
>HFC1_HUMAN (P51610) Host cell factor C1 (HCF) (VP16 accessory
protein) (HFC1) (VCAF) (CFF)
Length = 2035
Score = 66.6 bits (161), Expect = 2e-10
Identities = 78/276 (28%), Positives = 121/276 (43%), Gaps = 38/276 (13%)
Query: 270 PEWQHVQVSSPPPGRWGHTLSCVNGSRLVVFGGCGTQGLLNDVFVLDLDATPPTWREISG 329
P W+ V S P R H V L+V G G +G+++++ V + + G
Sbjct: 17 PRWKRVVGWSGPVPRPRHGHRAVAIKELIVVFGGGNEGIVDELHVYNTATNQWFIPAVRG 76
Query: 330 LAPPLPRSWHSSCTLDGTKLIVSGGCADSGVLLSDTFLLDMSMENPVWREI----PVTWT 385
PP ++ C DGT+L+V GG + G +D + L S W+ + P
Sbjct: 77 DIPPGCAAYGFVC--DGTRLLVFGGMVEYGKYSNDLYELQASRWE--WKRLKAKTPKNGP 132
Query: 386 PP-SRLGHTLSVYGGRKILMFGGLA------KSGPLRFRSSDVFTMDLSEDEPCWRCVTG 438
PP RLGH+ S+ G K +FGGLA K+ R+ +D++ ++L G
Sbjct: 133 PPCPRLGHSFSLV-GNKCYLFGGLANDSEDPKNNIPRY-LNDLYILELR---------PG 181
Query: 439 SGMPGAGNP--EGIAPPPRLDHVAV-----SLPGGRILIFGGSVAGLHSASQLYILDPTD 491
SG+ P G+ PPPR H AV +++I+GG ++G L+ LD
Sbjct: 182 SGVVAWDIPITYGVLPPPRESHTAVVYTEKDNKKSKLVIYGG-MSGCR-LGDLWTLD--I 237
Query: 492 EKPTWRILNVPGRPPRFAWGHSTCVVGGTRAIVLGG 527
+ TW ++ G P HS + G + V GG
Sbjct: 238 DTLTWNKPSLSGVAPLPRSLHSATTI-GNKMYVFGG 272
Score = 54.3 bits (129), Expect = 8e-07
Identities = 59/224 (26%), Positives = 95/224 (42%), Gaps = 55/224 (24%)
Query: 219 WRKLTVGGGVEPSRCNFSACAVGNRVVLFGGEGVNMQPMNDTFVLDLNSNNPEWQHVQVS 278
W V G + P + G R+++FGG + ND + +L ++ EW+ ++
Sbjct: 69 WFIPAVRGDIPPGCAAYGFVCDGTRLLVFGGMVEYGKYSNDLY--ELQASRWEWKRLKAK 126
Query: 279 SPPPG-----RWGHTLSCVNGSRLVVFGGCGTQG---------LLNDVFVLDLDA----- 319
+P G R GH+ S V G++ +FGG LND+++L+L
Sbjct: 127 TPKNGPPPCPRLGHSFSLV-GNKCYLFGGLANDSEDPKNNIPRYLNDLYILELRPGSGVV 185
Query: 320 ---TPPTWREISGLAPPLPRSWHSSCTL-----DGTKLIVSGGCADSGVLLSDTFLLDMS 371
P T+ G+ PP PR H++ +KL++ GG SG L D + LD+
Sbjct: 186 AWDIPITY----GVLPP-PRESHTAVVYTEKDNKKSKLVIYGGM--SGCRLGDLWTLDID 238
Query: 372 MENPVWREIPVTWTPPSRLG--------HTLSVYGGRKILMFGG 407
+TW PS G H+ + G K+ +FGG
Sbjct: 239 ---------TLTWNKPSLSGVAPLPRSLHSATTI-GNKMYVFGG 272
Score = 34.3 bits (77), Expect = 0.86
Identities = 35/113 (30%), Positives = 50/113 (43%), Gaps = 16/113 (14%)
Query: 430 EPCWRCVTGSGMPGAGNPEGIAPPPRLDHVAVSLPGGRILIFGGSVAGLHSASQLYILDP 489
+P W+ V G P P PR H AV++ I++FGG G+ +L++ +
Sbjct: 16 QPRWKRVVGWSGP--------VPRPRHGHRAVAIKE-LIVVFGGGNEGI--VDELHVYNT 64
Query: 490 TDEKPTWRILNVPGR-PPRFAWGHSTCVVGGTRAIVLGGQTGEEWMLSDLHEL 541
+ W I V G PP A C GTR +V GG +DL+EL
Sbjct: 65 ATNQ--WFIPAVRGDIPPGCAAYGFVC--DGTRLLVFGGMVEYGKYSNDLYEL 113
Score = 31.2 bits (69), Expect = 7.3
Identities = 34/156 (21%), Positives = 61/156 (38%), Gaps = 25/156 (16%)
Query: 214 LEAAAWRKLTVGGGVEPSRCNFSACAVGNRVVLFGG------EGVNMQPMNDTF-----V 262
++ W K ++ G R SA +GN++ +FGG + V + + +
Sbjct: 237 IDTLTWNKPSLSGVAPLPRSLHSATTIGNKMYVFGGWVPLVMDDVKVATHEKEWKCTNTL 296
Query: 263 LDLNSNNPEWQHVQV----SSPPPGRWGHTLSCVNGSRLVVFGG-------CGTQGLLND 311
LN + W+ + + + P R GH +N +RL ++ G Q D
Sbjct: 297 ACLNLDTMAWETILMDTLEDNIPRARAGHCAVAIN-TRLYIWSGRDGYRKAWNNQVCCKD 355
Query: 312 VFVLDLDATPPTWRE--ISGLAPPLPRSWHSSCTLD 345
++ L+ + PP R + L SW + T D
Sbjct: 356 LWYLETEKPPPPARVQLVRANTNSLEVSWGAVATAD 391
>KDC1_HUMAN (Q8N7A1) Kelch domain containing protein 1
Length = 406
Score = 63.5 bits (153), Expect = 1e-09
Identities = 54/215 (25%), Positives = 88/215 (40%), Gaps = 25/215 (11%)
Query: 215 EAAAWRKLTVGGGVEPS-RCNFSACAVGNRVVLFGGEGVNMQPM---------------- 257
E W K+T G P+ R S +R++ FGG G
Sbjct: 106 ETYIWEKITDFEGQPPTPRDKLSCWVYKDRLIYFGGYGCRRHSELQDCFDVHDASWEEQI 165
Query: 258 -----NDTFVLDLNSNNPEWQHVQVSSPPPGRWGHTLSCVNGSRLVVFGGCGTQGLLNDV 312
ND + D + ++ PP R HT + V G++ +FGG Q +ND+
Sbjct: 166 FWGWHNDVHIFDTKTQTWFQPEIKGGVPPQPRAAHTCA-VLGNKGYIFGGRVLQTRMNDL 224
Query: 313 FVLDLDATPPTWREISGLAPPLPRSWHSSCTLDGTKLIVSGGCADSGVLLSDTFLLDMSM 372
L+LD + R P RSWH+ + KL + GG + + LSD ++ +++
Sbjct: 225 HYLNLDTWTWSGRITINGESPKHRSWHTLTPIADDKLFLCGGLSADNIPLSDGWIHNVTT 284
Query: 373 ENPVWREIPVTWTPPSRLGHTLSVYGGRKILMFGG 407
W+++ RL HT + +I++FGG
Sbjct: 285 N--CWKQLTHLPKTRPRLWHTACLGKENEIMVFGG 317
Score = 57.8 bits (138), Expect = 7e-08
Identities = 43/149 (28%), Positives = 73/149 (48%), Gaps = 6/149 (4%)
Query: 219 WRKLTVGGGVEPSRCNFSACAV-GNRVVLFGGEGVNMQPMNDTFVLDLNSNNPEWQHVQV 277
W + + GGV P CAV GN+ +FGG + + MND L+L++ +
Sbjct: 183 WFQPEIKGGVPPQPRAAHTCAVLGNKGYIFGGRVLQTR-MNDLHYLNLDTWTWSGRITIN 241
Query: 278 SSPPPGRWGHTLSCVNGSRLVVFGGCGTQGL-LNDVFVLDLDATPPTWREISGLAPPLPR 336
P R HTL+ + +L + GG + L+D ++ ++ T W++++ L PR
Sbjct: 242 GESPKHRSWHTLTPIADDKLFLCGGLSADNIPLSDGWIHNV--TTNCWKQLTHLPKTRPR 299
Query: 337 SWHSSCTLDGTKLIVSGGCADSGVLLSDT 365
WH++C +++V GG D +L DT
Sbjct: 300 LWHTACLGKENEIMVFGGSKDD-LLALDT 327
Score = 53.5 bits (127), Expect = 1e-06
Identities = 70/295 (23%), Positives = 113/295 (37%), Gaps = 61/295 (20%)
Query: 214 LEAAAWRKLTVGGGVEPSRCNFSACAVGNRVVLFGGEG----------VNMQPMNDTFVL 263
+++ WR + G + S + ++ +FGG VN++ ++T++
Sbjct: 51 IDSGLWRMHLMEGELPASMSGSCGACINGKLYIFGGYDDKGYSNRLYFVNLRTRDETYI- 109
Query: 264 DLNSNNPEWQHVQVSSPPPGRWGHTLSC-VNGSRLVVFGGCGTQ---------------- 306
W+ + P LSC V RL+ FGG G +
Sbjct: 110 --------WEKITDFEGQPPTPRDKLSCWVYKDRLIYFGGYGCRRHSELQDCFDVHDASW 161
Query: 307 ------GLLNDVFVLDLDATPPTWREISGLAPPLPRSWHSSCTLDGTKLIVSGGCADSGV 360
G NDV + D EI G PP PR+ H+ C + G K + GG
Sbjct: 162 EEQIFWGWHNDVHIFDTKTQTWFQPEIKGGVPPQPRAAHT-CAVLGNKGYIFGGRVLQ-T 219
Query: 361 LLSDTFLLDMSMENPVWREIPVTWTPPSRLGHTLSVYGGRKILMFGGLAKSG-PLRFRSS 419
++D L++ R +P R HTL+ K+ + GGL+ PL S
Sbjct: 220 RMNDLHYLNLDTWTWSGRITINGESPKHRSWHTLTPIADDKLFLCGGLSADNIPL----S 275
Query: 420 DVFTMDLSEDEPCWRCVTGSGMPGAGNPEGIAPPPRLDHVAVSLPGGRILIFGGS 474
D + +++ + CW+ +T +P PRL H A I++FGGS
Sbjct: 276 DGWIHNVTTN--CWKQLTH--LPKTR--------PRLWHTACLGKENEIMVFGGS 318
Score = 40.4 bits (93), Expect = 0.012
Identities = 58/234 (24%), Positives = 95/234 (39%), Gaps = 43/234 (18%)
Query: 336 RSWHSSCTLDGTKLIVSGGCA---DSGVLLSDTFLLDMSMENPVWREIPVTWTPPSRLGH 392
RS H + +DG L V GG A D+ V L + + +++ +WR + P+ +
Sbjct: 13 RSGHCA-VVDGNFLYVWGGYASIEDNEVYLPNDEIWTYDIDSGLWRMHLMEGELPASMSG 71
Query: 393 TLSVYGGRKILMFGGLAKSGPLRFRSSDVFTMDL-SEDEP-CWRCVTGSGMPGAGNPEGI 450
+ K+ +FGG G S+ ++ ++L + DE W +T EG
Sbjct: 72 SCGACINGKLYIFGGYDDKG----YSNRLYFVNLRTRDETYIWEKITDF--------EGQ 119
Query: 451 APPPRLDHVAVSLPGGRILIFGGSVAGLHSASQ---------------------LYILDP 489
P PR D ++ + R++ FGG HS Q ++I D
Sbjct: 120 PPTPR-DKLSCWVYKDRLIYFGGYGCRRHSELQDCFDVHDASWEEQIFWGWHNDVHIFDT 178
Query: 490 TDEKPTWRILNVPGRPPRFAWGHSTCVVGGTRAIVLGGQTGEEWMLSDLHELSL 543
+ TW + G P TC V G + + GG+ + M +DLH L+L
Sbjct: 179 KTQ--TWFQPEIKGGVPPQPRAAHTCAVLGNKGYIFGGRVLQTRM-NDLHYLNL 229
>KDC2_HUMAN (Q9Y2U9) Kelch domain containing protein 2
(Hepatocellular carcinoma-associated antigen 33) (Host
cell factor homolog LCP)
Length = 406
Score = 62.4 bits (150), Expect = 3e-09
Identities = 69/293 (23%), Positives = 111/293 (37%), Gaps = 51/293 (17%)
Query: 214 LEAAAWRKLTVGGGVEPSRCNFSACAVGNRVVLFGGEGVNMQPMNDTFVLDLNSNNP--E 271
+E W+K+ G V PS A V + LFGG + N ++LD S + +
Sbjct: 73 METGRWKKINTEGDVPPSMSGSCAVCVDRVLYLFGGHH-SRGNTNKFYMLDSRSTDRVLQ 131
Query: 272 WQHVQVSSPPPGRWGHTLSCVNGSRLVVFGGCG-----------------------TQGL 308
W+ + PP V ++L+ FGG G +G
Sbjct: 132 WERIDCQGIPPSSKDKLGVWVYKNKLIFFGGYGYLPEDKVLGTFEFDETSFWNSSHPRGW 191
Query: 309 LNDVFVLDLDATPPTWRE-ISGLAPPLPRSWHSSCTLDGTKLIVSGGCADSGVLLSDTFL 367
+ V +LD + TW + I+ P PR+ H+ T+ + G D+ ++D
Sbjct: 192 NDHVHILDTETF--TWSQPITTGKAPSPRAAHACATVGNRGFVFGGRYRDA--RMNDLHY 247
Query: 368 LDMSMENPVWRE-IPVTWTPPSRLGHTLSVYGGRKILMFGGLAKSGPLRFRSSDVFTMDL 426
L +++ W E IP P R H+L+ + +FGG FT D
Sbjct: 248 L--NLDTWEWNELIPQGICPVGRSWHSLTPVSSDHLFLFGG--------------FTTDK 291
Query: 427 SEDEPCWR-CVTGSGMPGAGNPEGIAPPPRLDHVAVSLPGGRILIFGGSVAGL 478
W C++ + +P PRL H A + G +++FGG L
Sbjct: 292 QPLSDAWTYCISKNEWIQFNHP--YTEKPRLWHTACASDEGEVIVFGGCANNL 342
Score = 61.2 bits (147), Expect = 7e-09
Identities = 48/158 (30%), Positives = 72/158 (45%), Gaps = 8/158 (5%)
Query: 203 GWGRLARELTTLEAAAWRKLTVGGGVEPSRCNFSACAVGNRVVLFGGEGVNMQPMNDTFV 262
GW L T E W + G R + VGNR +FGG + + MND
Sbjct: 190 GWNDHVHILDT-ETFTWSQPITTGKAPSPRAAHACATVGNRGFVFGGRYRDAR-MNDLHY 247
Query: 263 LDLNSNNPEWQH-VQVSSPPPGRWGHTLSCVNGSRLVVFGGCGTQGL-LNDVFVLDLDAT 320
L+L++ EW + P GR H+L+ V+ L +FGG T L+D + +
Sbjct: 248 LNLDTW--EWNELIPQGICPVGRSWHSLTPVSSDHLFLFGGFTTDKQPLSDAWTYCISKN 305
Query: 321 PPTWREISGLAPPLPRSWHSSCTLDGTKLIVSGGCADS 358
W + + PR WH++C D ++IV GGCA++
Sbjct: 306 E--WIQFNHPYTEKPRLWHTACASDEGEVIVFGGCANN 341
Score = 38.5 bits (88), Expect = 0.046
Identities = 68/289 (23%), Positives = 94/289 (31%), Gaps = 50/289 (17%)
Query: 266 NSNNPEWQHVQVSSP-PPGRWGHTLSCVNGSRLVVFGGCGTQGLLNDVFVLDLDATPPT- 323
N W+ + PP G CV+ L +FGG ++G N ++LD +T
Sbjct: 72 NMETGRWKKINTEGDVPPSMSGSCAVCVDRV-LYLFGGHHSRGNTNKFYMLDSRSTDRVL 130
Query: 324 -WREISGLAPPLPRSWHSSCTLDGTKLIVSGGCADSGVLLSD----TFLLD--------- 369
W I P + KLI GG G L D TF D
Sbjct: 131 QWERIDCQGIPPSSKDKLGVWVYKNKLIFFGG---YGYLPEDKVLGTFEFDETSFWNSSH 187
Query: 370 ----------MSMENPVWREIPVTWTPPS-RLGHTLSVYGGRKILMFGGLAKSGPLRFRS 418
+ E W + T PS R H + G R +FGG + R
Sbjct: 188 PRGWNDHVHILDTETFTWSQPITTGKAPSPRAAHACATVGNRG-FVFGGRYRDA----RM 242
Query: 419 SDVFTMDLSEDEPCWRCVTGSGMPGAGNPEGIAPPPRLDHVAVSLPGGRILIFGGSVAGL 478
+D+ ++L E W + P+GI P R H + + +FGG
Sbjct: 243 NDLHYLNLDTWE--WNELI---------PQGICPVGRSWHSLTPVSSDHLFLFGGFTTDK 291
Query: 479 HSASQLYILDPTDEKPTWRILNVPGRPPRFAWGHSTCVVGGTRAIVLGG 527
S + K W N P W H+ C IV GG
Sbjct: 292 QPLSDAWTY--CISKNEWIQFNHPYTEKPRLW-HTACASDEGEVIVFGG 337
Score = 37.0 bits (84), Expect = 0.13
Identities = 29/101 (28%), Positives = 46/101 (44%), Gaps = 9/101 (8%)
Query: 214 LEAAAWRKLTVGGGVEPSRCNFSACAVGN-RVVLFGGEGVNMQPMNDTFVLDLNSNNPEW 272
L+ W +L G R S V + + LFGG + QP++D + ++ N EW
Sbjct: 250 LDTWEWNELIPQGICPVGRSWHSLTPVSSDHLFLFGGFTTDKQPLSDAWTYCISKN--EW 307
Query: 273 QHVQVSSP---PPGRWGHTLSCVNGSRLVVFGGCGTQGLLN 310
+Q + P P W HT + ++VFGGC L++
Sbjct: 308 --IQFNHPYTEKPRLW-HTACASDEGEVIVFGGCANNLLVH 345
>KEL2_YEAST (P50090) Kelch repeats protein 2
Length = 882
Score = 60.1 bits (144), Expect = 1e-08
Identities = 76/295 (25%), Positives = 122/295 (40%), Gaps = 38/295 (12%)
Query: 266 NSNNPEWQHVQVSSPPPGRWGHTLSCV--NGSRLVVFGGCGTQGLLNDVFVL--DLDATP 321
N+ W V++ + P R+ H+ S + N +R+ V GG Q + DV+ + + D T
Sbjct: 67 NTGKYIWNRVKLKNSPFPRYRHSSSFIVTNDNRIFVTGGLHDQSVYGDVWQIAANADGTS 126
Query: 322 PTWREIS-GLAPPLPRSWHSSCTLDGTKLIVSGG----CADSGVLLSDTFLLDMSMENPV 376
T + I P PR H+S T+ G +V GG +G+L D +L +++
Sbjct: 127 FTSKRIDIDQNTPPPRVGHAS-TICGNAYVVFGGDTHKLNKNGLLDDDLYLFNINSYK-- 183
Query: 377 WR-EIPVTWTPPSRLGHTLSVYGGR----KILMFGGLAKSGPLRFRSSDVFTMDLS---E 428
W P+ P R GH +S+ K+ +FGG +D+ DLS
Sbjct: 184 WTIPQPIGRRPLGRYGHKISIIASNPMQTKLYLFGGQVDETYF----NDLVVFDLSSFRR 239
Query: 429 DEPCWRCVTGSGMPGAGNPEGIAPPPRLDHVAVSLPGGRILIFGGSVAGLHSASQLYILD 488
W + P G PPP +H V+ ++ +FGG S + Y D
Sbjct: 240 PNSHWEFL---------EPVGDLPPPLTNHTMVAY-DNKLWVFGGETPKTIS-NDTYRYD 288
Query: 489 PTDEKPTWRILNVPGRPPRFAWGHSTCVVGGTRAIVLGGQTGEEWMLSDLHELSL 543
P + W + G P H++ VV VLGG+ +D++ L+L
Sbjct: 289 PAQSE--WSKVKTTGEKPPPIQEHAS-VVYKHLMCVLGGKDTHNAYSNDVYFLNL 340
Score = 54.3 bits (129), Expect = 8e-07
Identities = 67/267 (25%), Positives = 111/267 (41%), Gaps = 32/267 (11%)
Query: 220 RKLTVGGGVEPSRCNFSACAVGNRVVLFGGE--GVNMQPMNDTFVLDLNSNNPEWQHVQ- 276
+++ + P R ++ GN V+FGG+ +N + D + N N+ +W Q
Sbjct: 130 KRIDIDQNTPPPRVGHASTICGNAYVVFGGDTHKLNKNGLLDDDLYLFNINSYKWTIPQP 189
Query: 277 VSSPPPGRWGHTLSCVNG----SRLVVFGGCGTQGLLNDVFVLDLDA--TPPTWRE---- 326
+ P GR+GH +S + ++L +FGG + ND+ V DL + P + E
Sbjct: 190 IGRRPLGRYGHKISIIASNPMQTKLYLFGGQVDETYFNDLVVFDLSSFRRPNSHWEFLEP 249
Query: 327 ISGLAPPLPRSWHSSCTLDGTKLIVSGGCADSGVLLSDTFLLDMSMENPVWREIPVTWTP 386
+ L PPL H+ D KL V GG + +DT+ D + W ++ T
Sbjct: 250 VGDLPPPLTN--HTMVAYD-NKLWVFGG-ETPKTISNDTYRYDPAQSE--WSKVKTTGEK 303
Query: 387 PSRLGHTLSVYGGRKILMFGGLAKSGPLRFRSSDVFTMDLSEDEPCWRCVTGSGMPGAGN 446
P + SV + + GG S+DV+ ++L + W +
Sbjct: 304 PPPIQEHASVVYKHLMCVLGGKDTHNAY---SNDVYFLNLLSLK--WYKLPRM------- 351
Query: 447 PEGIAPPPRLDHVAVSLPGGRILIFGG 473
EGI P R H + ++LI GG
Sbjct: 352 KEGI-PQERSGHSLTLMKNEKLLIMGG 377
>KEL1_YEAST (P38853) Kelch repeats protein 1
Length = 1164
Score = 57.8 bits (138), Expect = 7e-08
Identities = 57/190 (30%), Positives = 87/190 (45%), Gaps = 13/190 (6%)
Query: 243 RVVLFGGEGVNMQPMNDTFVLDLNSNNPEWQHVQVSSP----PPGRWGHTLSCVNGSRLV 298
++ +FGG+ + ND V DL+S H + P PP T+ + S+L
Sbjct: 253 KLYVFGGQFDDTY-FNDLAVYDLSSFRRPDSHWEFLKPRTFTPPPITNFTMISYD-SKLW 310
Query: 299 VFGGCGTQGLLNDVFVLDLDATPPTWREISGLAPPLPRSWHSSCTLDGTKLIVSGGCADS 358
VFGG QGL+NDVF+ D + +G PP P H++ + +V G
Sbjct: 311 VFGGDTLQGLVNDVFMYDPAINDWFIIDTTGEKPP-PVQEHATVVYNDLMCVVGGKDEHD 369
Query: 359 GVLLSDTFLLDMSMENPVWREIPV--TWTPPSRLGHTLSVYGGRKILMFGGLAKSGPLRF 416
L S FL ++++ W ++PV P R GH+L++ KIL+ GG K R
Sbjct: 370 AYLNSVYFL---NLKSRKWFKLPVFTAGIPQGRSGHSLTLLKNDKILIMGG-DKFDYARV 425
Query: 417 RSSDVFTMDL 426
D+ T D+
Sbjct: 426 EEYDLHTSDI 435
Score = 38.5 bits (88), Expect = 0.046
Identities = 46/184 (25%), Positives = 67/184 (36%), Gaps = 50/184 (27%)
Query: 368 LDMSMENPVWREIPVTWTPPSRLGHTLSVY--GGRKILMFGGL-------------AKSG 412
L E VW I + +P R H S Y +I + GGL A
Sbjct: 104 LSKQREYTVWNRIKLQNSPFPRYRHVASAYVTDKNQIYVIGGLHDQSVYGDTWILTAFDN 163
Query: 413 PLRFRSSDVFTMDLSEDEPCWRCVTGSGMPGAGNPEGIAPPPRLDHVAVSLPGGRILIFG 472
RF ++ T+D+SE PPPR+ H AV L G ++FG
Sbjct: 164 ATRFSTT---TIDISE---------------------ATPPPRVGHAAV-LCGNAFVVFG 198
Query: 473 GSVAGLHSA----SQLYILDPTDEKPTWRILNVPGRPPRFAWGHSTCVVG----GTRAIV 524
G ++ +Y+L+ K W + G P +GH ++ T+ V
Sbjct: 199 GDTHKVNKEGLMDDDIYLLNINSYK--WTVPAPVGPRPLGRYGHKISIIATTQMKTKLYV 256
Query: 525 LGGQ 528
GGQ
Sbjct: 257 FGGQ 260
Score = 32.0 bits (71), Expect = 4.3
Identities = 36/159 (22%), Positives = 59/159 (36%), Gaps = 21/159 (13%)
Query: 324 WREISGLAPPLPRSWH--SSCTLDGTKLIVSGGCADSGVLLSDTFLLDMSMENPVWREIP 381
W I P PR H S+ D ++ V GG D V DT++L +
Sbjct: 113 WNRIKLQNSPFPRYRHVASAYVTDKNQIYVIGGLHDQSVY-GDTWILTAFDNATRFSTTT 171
Query: 382 VTW---TPPSRLGHTLSVYGGRKILMFGGLAKSGPLRFRSSDVFTMDLSEDEPCWRCVTG 438
+ TPP R+GH + G ++ G K D++ ++++ + W
Sbjct: 172 IDISEATPPPRVGHAAVLCGNAFVVFGGDTHKVNKEGLMDDDIYLLNINSYK--WTVPA- 228
Query: 439 SGMPGAGNPEGIAPPPRLDH----VAVSLPGGRILIFGG 473
P G P R H +A + ++ +FGG
Sbjct: 229 --------PVGPRPLGRYGHKISIIATTQMKTKLYVFGG 259
Score = 30.8 bits (68), Expect = 9.5
Identities = 20/77 (25%), Positives = 37/77 (47%), Gaps = 1/77 (1%)
Query: 227 GVEPSRCNFSACAVGNRVV-LFGGEGVNMQPMNDTFVLDLNSNNPEWQHVQVSSPPPGRW 285
G +P A V N ++ + GG+ + +N + L+L S V + P GR
Sbjct: 341 GEKPPPVQEHATVVYNDLMCVVGGKDEHDAYLNSVYFLNLKSRKWFKLPVFTAGIPQGRS 400
Query: 286 GHTLSCVNGSRLVVFGG 302
GH+L+ + ++++ GG
Sbjct: 401 GHSLTLLKNDKILIMGG 417
>BSL3_ARATH (Q9SHS7) Serine/threonine protein phosphatase BSL3 (EC
3.1.3.16) (BSU1-like protein 3)
Length = 715
Score = 56.6 bits (135), Expect = 2e-07
Identities = 62/246 (25%), Positives = 93/246 (37%), Gaps = 77/246 (31%)
Query: 281 PPGRWGHTLSCVN-----------GSRLVVFGGC---------------------GTQGL 308
P R GHTL+ V G RL++FGG G G
Sbjct: 67 PGPRCGHTLTAVPAVGEEGTSSYIGPRLILFGGATALEGNSGGTGTPTSSNVSYAGLAGA 126
Query: 309 LNDVFVLDLDATPPTWREISGLA-PPLPRSWHSSCTLDGTKLIVSGGCADSGVLLSDTFL 367
DV D+ + W ++ PP PR+ H + T GT +++ GG +G+ D +
Sbjct: 127 TADVHCYDVLSNK--WSRLTPYGEPPSPRAAHVA-TAVGTMVVIQGGIGPAGLSAEDLHV 183
Query: 368 LDMSMENPVWREIPVTWTPPS-RLGHTLSVYGGRKILMFG---GLAKSGPLRFRSSDVFT 423
LD++ + P W + V P R GH +++ G R ++ GLAK R+
Sbjct: 184 LDLTQQRPRWHRVVVQGPGPGPRYGHVMALVGQRYLMPLASAYGLAKHRDGRWE------ 237
Query: 424 MDLSEDEPCWRCVTGSGMPGAGNPEGIAPPPRLDHVAV-----------SLPGGRILIFG 472
W G++P R H AV +L GGR++
Sbjct: 238 ---------WAIA-----------PGVSPSARYQHAAVFVNARLHVSGGALGGGRMVEDS 277
Query: 473 GSVAGL 478
SVAG+
Sbjct: 278 SSVAGV 283
Score = 52.0 bits (123), Expect = 4e-06
Identities = 44/148 (29%), Positives = 60/148 (39%), Gaps = 11/148 (7%)
Query: 219 WRKLTVGGGVEPSRCNFSACAVGNRVVLFGGEGVNMQPMNDTFVLDLNSNNPEWQHVQVS 278
W +LT G R A AVG VV+ GG G D VLDL P W V V
Sbjct: 140 WSRLTPYGEPPSPRAAHVATAVGTMVVIQGGIGPAGLSAEDLHVLDLTQQRPRWHRVVVQ 199
Query: 279 SPPPG-RWGHTLSCVNGSRLVVFGGCGTQGLLNDVFVLDLDATPPTWREISGLAPPLPRS 337
P PG R+GH ++ V L+ D W G++P R
Sbjct: 200 GPGPGPRYGHVMALVGQRYLMPLASAYGLAKHRD--------GRWEWAIAPGVSPS-ARY 250
Query: 338 WHSSCTLDGTKLIVSGGCADSGVLLSDT 365
H++ ++ +L VSGG G ++ D+
Sbjct: 251 QHAAVFVN-ARLHVSGGALGGGRMVEDS 277
Score = 51.2 bits (121), Expect = 7e-06
Identities = 49/153 (32%), Positives = 69/153 (45%), Gaps = 24/153 (15%)
Query: 386 PPSRLGHTL-----------SVYGGRKILMFGGLA-----KSGPLRFRSSDVFTMDLS-- 427
P R GHTL S Y G ++++FGG G SS+V L+
Sbjct: 67 PGPRCGHTLTAVPAVGEEGTSSYIGPRLILFGGATALEGNSGGTGTPTSSNVSYAGLAGA 126
Query: 428 -EDEPCWRCVTGSGMPGAGNPEGIAPPPRLDHVAVSLPGGRILIFGG-SVAGLHSASQLY 485
D C+ ++ P G P PR HVA ++ G ++I GG AGL SA L+
Sbjct: 127 TADVHCYDVLSNKW--SRLTPYGEPPSPRAAHVATAV-GTMVVIQGGIGPAGL-SAEDLH 182
Query: 486 ILDPTDEKPTWRILNVPGRPPRFAWGHSTCVVG 518
+LD T ++P W + V G P +GH +VG
Sbjct: 183 VLDLTQQRPRWHRVVVQGPGPGPRYGHVMALVG 215
Score = 38.1 bits (87), Expect = 0.060
Identities = 37/123 (30%), Positives = 50/123 (40%), Gaps = 26/123 (21%)
Query: 240 VGNRVVLFGG-------EGVNMQPMN-------------DTFVLDLNSNNPEWQHVQ-VS 278
+G R++LFGG G P + D D+ SN +W +
Sbjct: 90 IGPRLILFGGATALEGNSGGTGTPTSSNVSYAGLAGATADVHCYDVLSN--KWSRLTPYG 147
Query: 279 SPPPGRWGHTLSCVNGSRLVVFGGCGTQGL-LNDVFVLDLDATPPTWREISGLAP-PLPR 336
PP R H + V G+ +V+ GG G GL D+ VLDL P W + P P PR
Sbjct: 148 EPPSPRAAHVATAV-GTMVVIQGGIGPAGLSAEDLHVLDLTQQRPRWHRVVVQGPGPGPR 206
Query: 337 SWH 339
H
Sbjct: 207 YGH 209
>EFL4_MOUSE (P60882) Multiple EGF-like-domain protein 4
Length = 2330
Score = 55.8 bits (133), Expect = 3e-07
Identities = 71/268 (26%), Positives = 118/268 (43%), Gaps = 33/268 (12%)
Query: 284 RWGHTLSCVNGSRLVVFGGCGT-QGLLNDVFVLDLDATPPTWREI-----SGLAPPLPRS 337
R GHT+ + L +FGG G QGLL +++ + W ++ G P PRS
Sbjct: 1051 RLGHTMVEGPDATLWMFGGLGLPQGLLGNLYRYSVSER--RWTQMLAGAEDGGPGPSPRS 1108
Query: 338 WHSSCTLDGTK--LIVSGGCADSGVLLSDTFLLDMSMENPVWREIPVTWTPPSRLGHTLS 395
+H++ + + + + GG GV D ++L+++ + P P+ GHTL+
Sbjct: 1109 FHAAAYVPAGRGAMYLLGGLTAGGVT-RDFWVLNLTTLQWRQEKPPQNMELPAVAGHTLT 1167
Query: 396 VYGGRKILMFGGLAKSGPLRFRSSDVFTMDLSEDEPCWRCVTGSGMPGAGNPEGIAPPPR 455
G +L+ GG + + F L E ++ TG+ + GA G P
Sbjct: 1168 ARRGLSLLLVGG--------YSPENGFNQQLLE----YQLATGTWVSGA--QSGTPPTGL 1213
Query: 456 LDHVAVSLPG-GRILIFGGSVAGLHSAS---QLYILDPTDEKPTWRIL-NVPGRPPRFAW 510
H AV + +FGG + A+ +LY L D TW +L G PR
Sbjct: 1214 YGHSAVYHEATDSLYVFGGFRFHVELAAPSPELYSLHCPDR--TWSLLAPSQGAKPRPRL 1271
Query: 511 GHSTCVVGGTRAIVLGGQTGEEWMLSDL 538
H++ ++G T +VLGG++ + SD+
Sbjct: 1272 FHASALLGDT-MVVLGGRSDPDEFSSDV 1298
Score = 39.3 bits (90), Expect = 0.027
Identities = 80/320 (25%), Positives = 123/320 (38%), Gaps = 42/320 (13%)
Query: 219 WRKLTVG---GGVEPSRCNFSACAV----GNRVVLFGG---EGVNMQPMNDTFVLDLNSN 268
W ++ G GG PS +F A A + L GG GV D +VL+L +
Sbjct: 1090 WTQMLAGAEDGGPGPSPRSFHAAAYVPAGRGAMYLLGGLTAGGVT----RDFWVLNLTTL 1145
Query: 269 NPEWQHVQVSSPPPGRWGHTLSCVNGSRLVVFGGCGTQGLLNDVFVLDLDATPPTWREIS 328
+ + P GHTL+ G L++ GG + N +L+ TW +S
Sbjct: 1146 QWRQEKPPQNMELPAVAGHTLTARRGLSLLLVGGYSPENGFNQQ-LLEYQLATGTW--VS 1202
Query: 329 GL---APPLPRSWHSSCTLDGT-KLIVSGGCADSGVLLSDT-FLLDMSMENPVWREIPVT 383
G PP HS+ + T L V GG L + + L + + W + +
Sbjct: 1203 GAQSGTPPTGLYGHSAVYHEATDSLYVFGGFRFHVELAAPSPELYSLHCPDRTWSLLAPS 1262
Query: 384 W--TPPSRLGHTLSVYGGRKILMFGGLAKSGPLRFRSSDVFTMDLSEDEPCWRCVTGSGM 441
P RL H ++ G +++ G +S P F SSDV ++ + +T
Sbjct: 1263 QGAKPRPRLFHASALLGDTMVVLGG---RSDPDEF-SSDVLLYQVNCNTWLLPALTRPAF 1318
Query: 442 PGAGNPEGIAPPPRLDHVAVSLPGGRILIFGGSVAGLHSASQLYILDPTDEKPTWRILNV 501
G+ E +A AV+ G R+ I GG G+ L + P D R+L
Sbjct: 1319 VGSPMEESVAH-------AVAAVGSRLYISGG-FGGVALGRLLALTLPPD---PCRLLPS 1367
Query: 502 P---GRPPRFAWGHSTCVVG 518
P + W H C+ G
Sbjct: 1368 PEACNQSGACTWCHGACLSG 1387
Score = 37.0 bits (84), Expect = 0.13
Identities = 22/60 (36%), Positives = 29/60 (47%), Gaps = 2/60 (3%)
Query: 432 CWRCVTGSGMPGAGNPEGIAPPP--RLDHVAVSLPGGRILIFGGSVAGLHSASQLYILDP 489
C + V+G+ + G PEG A PP R HVA L G +L+ GG S Y + P
Sbjct: 29 CHQWVSGAELAPPGTPEGRAAPPSGRYSHVAAVLGGSVLLVAGGYSGRPRGDSMAYKVPP 88
>TEA3_SCHPO (O14248) Tip elongation aberrant protein 3 (Cell
polarity protein tea3)
Length = 1125
Score = 53.9 bits (128), Expect = 1e-06
Identities = 45/184 (24%), Positives = 84/184 (45%), Gaps = 34/184 (18%)
Query: 219 WRKLTVGGGVEPSRCNFSACAVGNRVVLFGGEGVNMQPMNDTFVLDL------------- 265
W ++ + R S V +++ +FGGE + +ND + D
Sbjct: 156 WNLVSTQSPLPSPRTGHSMLLVDSKLWIFGGE-CQGKYLNDIHLFDTKGVDRRTQSELKQ 214
Query: 266 --NSNNPEWQHVQV-------------SSPPPGRWGHTLSCVNGSRLVVFGGCGTQGLLN 310
N+NN E +++ SS PP R H+++ V G ++ V GG G L+
Sbjct: 215 KANANNVEKANMEFDETDWSWETPFLHSSSPPPRSNHSVTLVQG-KIFVHGGHNDTGPLS 273
Query: 311 DVFVLDLDATPPTWREISGLAP-PLPRSWHSSCTLDGTKLIVSGGCADSGVLLSDTFLLD 369
D+++ DL+ +W E+ + P PR H + T+D T + + GG + G++L++ + +
Sbjct: 274 DLWLFDLETL--SWTEVRSIGRFPGPREGHQATTIDDT-VYIYGGRDNKGLILNELWAFN 330
Query: 370 MSME 373
S +
Sbjct: 331 YSQQ 334
Score = 45.1 bits (105), Expect = 5e-04
Identities = 63/275 (22%), Positives = 105/275 (37%), Gaps = 39/275 (14%)
Query: 289 LSCVNGSRLVVFGGCGTQGLLNDVFVLDLDATPPTWREISGLAPPLPRSWHSSCTLDGTK 348
L N + + GG G+ N +F LDLD+ + G P S
Sbjct: 66 LDAANEKFMYLHGGREKSGISNSLFKLDLDSC-TVYSHNRGEDNDSPARVGHSIVCSADT 124
Query: 349 LIVSGGC---ADSGVLLSDTFLLDMSMENPVWREIPVTWTPPS-RLGHTLSVYGGRKILM 404
+ + GGC DS + D L + ++ W + PS R GH++ + K+ +
Sbjct: 125 IYLFGGCDSETDSTFEVGDNSLYAYNFKSNQWNLVSTQSPLPSPRTGHSMLLVDS-KLWI 183
Query: 405 FGGLAKSGPL---------------------RFRSSDVFTMDLSEDEPCWRCVTGSGMPG 443
FGG + L + +++V ++ DE W T
Sbjct: 184 FGGECQGKYLNDIHLFDTKGVDRRTQSELKQKANANNVEKANMEFDETDWSWETPF---- 239
Query: 444 AGNPEGIAPPPRLDHVAVSLPGGRILIFGGSVAGLHSASQLYILDPTDEKPTWRILNVPG 503
+PPPR +H +V+L G+I + GG S L++ D E +W + G
Sbjct: 240 ---LHSSSPPPRSNH-SVTLVQGKIFVHGGH-NDTGPLSDLWLFDL--ETLSWTEVRSIG 292
Query: 504 RPPRFAWGHSTCVVGGTRAIVLGGQTGEEWMLSDL 538
R P GH + T + GG+ + +L++L
Sbjct: 293 RFPGPREGHQATTIDDT-VYIYGGRDNKGLILNEL 326
>LCM2_HUMAN (O60294) Leucine carboxyl methyltransferase 2 (EC
2.1.1.-) (p21WAF1/CIP1 promoter-interacting protein)
Length = 686
Score = 52.0 bits (123), Expect = 4e-06
Identities = 43/161 (26%), Positives = 70/161 (42%), Gaps = 7/161 (4%)
Query: 218 AWRKLTVGGGVEPSRCNFSACAVGNRVVLFGGEGVNMQPMNDTFVLDLNSNNPEWQHVQV 277
AW ++ V G V +R + SAC ++ GG G + +P+N L S W+ V +
Sbjct: 520 AWVRIPVEGEVPEARHSHSACTWQGGALIAGGLGASEEPLNSVLFLRPISCGFLWESVDI 579
Query: 278 SSPPPGRWGHTLSCVNGSRLVVFGGCGTQGLLNDVFVLDLDATPPTWREISGLAPPLPRS 337
P R+ HT +NG L+V G V V++L + +I P P
Sbjct: 580 QPPITPRYSHTAHVLNGKLLLVGGIWIHSSSFPGVTVINLTTGLSSEYQIDTTYVPWPLM 639
Query: 338 WHSSCTL---DGTKLIVSGG---CADSGVLLS-DTFLLDMS 371
H+ ++ + +L++ GG C G + T LD+S
Sbjct: 640 LHNHTSILLPEEQQLLLLGGGGNCFSFGTYFNPHTVTLDLS 680
Score = 43.9 bits (102), Expect = 0.001
Identities = 49/180 (27%), Positives = 74/180 (40%), Gaps = 16/180 (8%)
Query: 242 NRVVLFGGEGVNMQPMNDTFVLDL-NSNNPEWQHVQVSSPPPGR--------WGHT---L 289
+RV++ GG + P L S + + ++V+ GR W H+ +
Sbjct: 428 SRVLVLGGRLSPVSPALGVLQLHFFKSEDNNTEDLKVTITKAGRKDDSTLCCWRHSTTEV 487
Query: 290 SCVNGSRLVVFGGCGT-QGLLNDVFVLDLDATPPTWREISGLAPPLPRSWHSSCTLDGTK 348
SC N L V+GG + +L+D L + + G P S HS+CT G
Sbjct: 488 SCQNQEYLFVYGGRSVVEPVLSDWHFLHVGTMAWVRIPVEGEVPEARHS-HSACTWQGGA 546
Query: 349 LIVSGGCADSGVLLSDTFLLDMSMENPVWREIPVTWTPPSRLGHTLSVYGGRKILMFGGL 408
LI G A L S FL +S +W + + R HT V G K+L+ GG+
Sbjct: 547 LIAGGLGASEEPLNSVLFLRPISC-GFLWESVDIQPPITPRYSHTAHVLNG-KLLLVGGI 604
>WC1_NEUCR (Q01371) White collar 1 protein (WC1)
Length = 1167
Score = 50.4 bits (119), Expect = 1e-05
Identities = 23/64 (35%), Positives = 38/64 (58%)
Query: 23 AKRRHPLVDSSVISEIRKCIDEGVEFQGELLNFRKDGSPLMNRLRLTPIYGEDDEITHVI 82
A + V+++ + ++K I EG E Q L+N+RK G P +N L + PI + +EI + I
Sbjct: 440 AGTKREFVENNAVYTLKKTIAEGQEIQQSLINYRKGGKPFLNLLTMIPIPWDTEEIRYFI 499
Query: 83 GIQL 86
G Q+
Sbjct: 500 GFQI 503
>LZTR_HUMAN (Q8N653) Leucine-zipper-like transcriptional regulator 1
(LZTR-1)
Length = 840
Score = 50.1 bits (118), Expect = 2e-05
Identities = 55/232 (23%), Positives = 86/232 (36%), Gaps = 39/232 (16%)
Query: 215 EAAAWRKLTVGGGVEPSRCNFSACAVGNRVVLFGGEGVNMQPMNDTFVLDLNSNNPEW-- 272
E W ++ G + PS CNF +++ +F G+ + N+ F + W
Sbjct: 211 ELTCWEEVAQSGEIPPSCCNFPVAVCRDKMFVFSGQS-GAKITNNLFQFEFKDKT--WTR 267
Query: 273 ---QHVQVSSPPP--GRWGHTLSCVNGSRLVVFGGCGTQGLLNDVFVLDLDATPPTWREI 327
+H+ SPPP R+GHT+ + L VFGG L N++ D+D TW +
Sbjct: 268 IPTEHLLRGSPPPPQRRYGHTMVAFD-RHLYVFGGAADNTLPNELHCYDVDF--QTWEVV 324
Query: 328 SGLAPPLPRSWHSSCTLDGTKLIVSGGCADSGV------------LLSDTFLLDMSMENP 375
SS + G + CA V D F LD +
Sbjct: 325 Q----------PSSDSEVGGAEVPERACASEEVPTLTYEERVGFKKSRDVFGLDFGTTSA 374
Query: 376 VWREIPVTWTPPSRLGHTLSVYGGRKILMFGGLAKSGPLRFRSSDVFTMDLS 427
P + P RL H +V + +FGG + RS +++ S
Sbjct: 375 KQPTQPASELPSGRLFHAAAVISD-AMYIFGGTVDN---NIRSGEMYRFQFS 422
Score = 37.4 bits (85), Expect = 0.10
Identities = 66/268 (24%), Positives = 105/268 (38%), Gaps = 31/268 (11%)
Query: 219 WRKLT-VGGGVEPSRCNFSACAVGNRVVLFGGEGVNMQPMNDTFVLDLNSNNPEWQHVQV 277
WR+L V R + A + + +FGG+ +ND D+ + W
Sbjct: 54 WRRLPPCDEFVGARRSKHTVVAYKDAIYVFGGDNGKTM-LNDLLRFDVKDCS--WCRAFT 110
Query: 278 SSPPPGRWGHTLSCVNGSRLVVFGG-----CGTQGLLNDVFVLDLDATPPTWRE--ISGL 330
+ PP H + V GS + VFGG L N + + W E I G
Sbjct: 111 TGTPPAPRYHHSAVVYGSSMFVFGGYTGDIYSNSNLKNKNDLFEYKFATGQWTEWKIEGR 170
Query: 331 APPLPRSWHSSCTLDGTKLIVSGGCADSGVLLSDTFLLDM-SMENPVWREIPVTW-TPPS 388
P+ RS H + T+ KL + G D L+D + + + E W E+ + PPS
Sbjct: 171 L-PVARSAHGA-TVYSDKLWIFAG-YDGNARLNDMWTIGLQDRELTCWEEVAQSGEIPPS 227
Query: 389 RLGHTLSVYGGRKILMFGGLAKSGPLRFRSSDVFTMDLSEDEPCWRCVTGSGMPGAGNPE 448
++V K+ +F G +S T +L + E ++ T + +P
Sbjct: 228 CCNFPVAVCRD-KMFVFSG---------QSGAKITNNLFQFE--FKDKTWTRIPTEHLLR 275
Query: 449 GIAPPP--RLDHVAVSLPGGRILIFGGS 474
G PPP R H V+ + +FGG+
Sbjct: 276 GSPPPPQRRYGHTMVAF-DRHLYVFGGA 302
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.320 0.139 0.443
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 72,670,658
Number of Sequences: 164201
Number of extensions: 3358779
Number of successful extensions: 7415
Number of sequences better than 10.0: 68
Number of HSP's better than 10.0 without gapping: 30
Number of HSP's successfully gapped in prelim test: 38
Number of HSP's that attempted gapping in prelim test: 7168
Number of HSP's gapped (non-prelim): 166
length of query: 548
length of database: 59,974,054
effective HSP length: 115
effective length of query: 433
effective length of database: 41,090,939
effective search space: 17792376587
effective search space used: 17792376587
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 68 (30.8 bits)
Medicago: description of AC148970.7


