

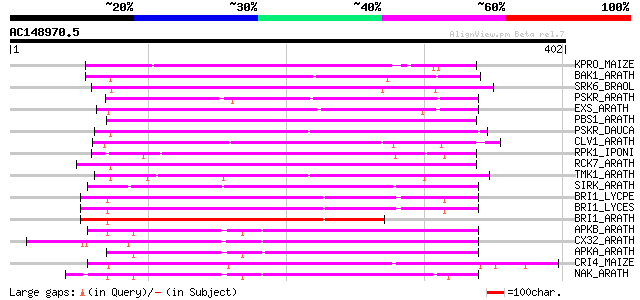
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148970.5 + phase: 0
(402 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
KPRO_MAIZE (P17801) Putative receptor protein kinase ZmPK1 precu... 188 2e-47
BAK1_ARATH (Q94F62) BRASSINOSTEROID INSENSITIVE 1-associated rec... 180 7e-45
SRK6_BRAOL (Q09092) Putative serine/threonine-protein kinase rec... 177 6e-44
PSKR_ARATH (Q9ZVR7) Putative phytosulfokine receptor precursor (... 177 6e-44
EXS_ARATH (Q9LYN8) Leucine-rich repeat receptor protein kinase E... 169 1e-41
PBS1_ARATH (Q9FE20) Serine/threonine-protein kinase PBS1 (EC 2.7... 166 8e-41
PSKR_DAUCA (Q8LPB4) Phytosulfokine receptor precursor (EC 2.7.1.... 164 3e-40
CLV1_ARATH (Q9SYQ8) Receptor protein kinase CLAVATA1 precursor (... 162 1e-39
RPK1_IPONI (P93194) Receptor-like protein kinase precursor (EC 2... 161 3e-39
RCK7_ARATH (Q9LQQ8) Putative serine/threonine-protein kinase RLC... 161 3e-39
TMK1_ARATH (P43298) Putative receptor protein kinase TMK1 precur... 160 4e-39
SIRK_ARATH (O64483) Senescence-induced receptor-like serine/thre... 160 4e-39
BRI1_LYCPE (Q8L899) Systemin receptor SR160 precursor (EC 2.7.1.... 155 2e-37
BRI1_LYCES (Q8GUQ5) Brassinosteroid LRR receptor kinase precurso... 154 3e-37
BRI1_ARATH (O22476) BRASSINOSTEROID INSENSITIVE 1 precursor (EC ... 154 3e-37
APKB_ARATH (P46573) Protein kinase APK1B, chloroplast precursor ... 154 3e-37
CX32_ARATH (P27450) Probable serine/threonine-protein kinase Cx3... 152 2e-36
APKA_ARATH (Q06548) Protein kinase APK1A, chloroplast precursor ... 150 4e-36
CRI4_MAIZE (O24585) Putative receptor protein kinase CRINKLY4 pr... 149 2e-35
NAK_ARATH (P43293) Probable serine/threonine-protein kinase NAK ... 148 3e-35
>KPRO_MAIZE (P17801) Putative receptor protein kinase ZmPK1
precursor (EC 2.7.1.37)
Length = 817
Score = 188 bits (477), Expect = 2e-47
Identities = 118/299 (39%), Positives = 166/299 (55%), Gaps = 25/299 (8%)
Query: 56 RFTPEKLDEITEKYSTILGSGAFGVVFKGELSNGENVAVKVLNCLDMGMEEQFKAEVITI 115
R++ +L + T K+ LG G G V+KG L + +VAVK L + G +E F+AE+ I
Sbjct: 523 RYSYRELVKATRKFKVELGRGESGTVYKGVLEDDRHVAVKKLENVRQG-KEVFQAELSVI 581
Query: 116 GRTYHINLVKLYGFCFHRDKRALVYEYVENGSLDKYIFGSKNRNDFDFQKLHKIAIGTAK 175
GR H+NLV+++GFC R LV EYVENGSL +F D++ IA+G AK
Sbjct: 582 GRINHMNLVRIWGFCSEGSHRLLVSEYVENGSLANILFSEGGNILLDWEGRFNIALGVAK 641
Query: 176 GIAYLHEECKHRIIHYDIKPENVLLDMKLEPKIADFGLAKLRSRESNIELNTHFRGTRGY 235
G+AYLH EC +IH D+KPEN+LLD EPKI DFGL KL +R + + +H RGT GY
Sbjct: 642 GLAYLHHECLEWVIHCDVKPENILLDQAFEPKITDFGLVKLLNRGGSTQNVSHVRGTLGY 701
Query: 236 AAPEMWKPYPVTYKCDVYSFGILLFEIVGRRRHFDSSYSESQQWFPRWTWEMFENNELVV 295
APE P+T K DVYS+G++L E++ R + + M +LV
Sbjct: 702 IAPEWVSSLPITAKVDVYSYGVVLLELLTGTRVSELVGGTDE------VHSML--RKLVR 753
Query: 296 ML-ALCEIEEK-------DSEI--------AERMLKVALWCVQYSPNDRPLMSTVVKML 338
ML A E EE+ DS++ A ++K+A+ C++ + RP M V+ L
Sbjct: 754 MLSAKLEGEEQSWIDGYLDSKLNRPVNYVQARTLIKLAVSCLEEDRSKRPTMEHAVQTL 812
>BAK1_ARATH (Q94F62) BRASSINOSTEROID INSENSITIVE 1-associated
receptor kinase 1 precursor (EC 2.7.1.37)
(BRI1-associated receptor kinase 1) (Somatic
embryogenesis receptor-like kinase 3)
Length = 615
Score = 180 bits (456), Expect = 7e-45
Identities = 111/293 (37%), Positives = 171/293 (57%), Gaps = 9/293 (3%)
Query: 56 RFTPEKLDEITEKYST--ILGSGAFGVVFKGELSNGENVAVKVLNC-LDMGMEEQFKAEV 112
RF+ +L ++ +S ILG G FG V+KG L++G VAVK L G E QF+ EV
Sbjct: 276 RFSLRELQVASDNFSNKNILGRGGFGKVYKGRLADGTLVAVKRLKEERTQGGELQFQTEV 335
Query: 113 ITIGRTYHINLVKLYGFCFHRDKRALVYEYVENGSLDKYIFGS-KNRNDFDFQKLHKIAI 171
I H NL++L GFC +R LVY Y+ NGS+ + +++ D+ K +IA+
Sbjct: 336 EMISMAVHRNLLRLRGFCMTPTERLLVYPYMANGSVASCLRERPESQPPLDWPKRQRIAL 395
Query: 172 GTAKGIAYLHEECKHRIIHYDIKPENVLLDMKLEPKIADFGLAKLRSRESNIELNTHFRG 231
G+A+G+AYLH+ C +IIH D+K N+LLD + E + DFGLAKL + + + T RG
Sbjct: 396 GSARGLAYLHDHCDPKIIHRDVKAANILLDEEFEAVVGDFGLAKLMDYK-DTHVTTAVRG 454
Query: 232 TRGYAAPEMWKPYPVTYKCDVYSFGILLFEIVGRRRHFDSS--YSESQQWFPRWTWEMFE 289
T G+ APE + K DV+ +G++L E++ +R FD + ++ W + +
Sbjct: 455 TIGHIAPEYLSTGKSSEKTDVFGYGVMLLELITGQRAFDLARLANDDDVMLLDWVKGLLK 514
Query: 290 NNELVVMLAL-CEIEEKDSEIAERMLKVALWCVQYSPNDRPLMSTVVKMLEGE 341
+L ++ + + KD E+ E++++VAL C Q SP +RP MS VV+MLEG+
Sbjct: 515 EKKLEALVDVDLQGNYKDEEV-EQLIQVALLCTQSSPMERPKMSEVVRMLEGD 566
>SRK6_BRAOL (Q09092) Putative serine/threonine-protein kinase
receptor precursor (EC 2.7.1.37) (S-receptor kinase)
(SRK)
Length = 849
Score = 177 bits (448), Expect = 6e-44
Identities = 103/300 (34%), Positives = 164/300 (54%), Gaps = 9/300 (3%)
Query: 60 EKLDEITEKYSTI--LGSGAFGVVFKGELSNGENVAVKVLNCLDMGMEEQFKAEVITIGR 117
E + + TE +S+ LG G FG+V+KG L +G+ +AVK L+ + ++F EV I R
Sbjct: 519 ETVVKATENFSSCNKLGQGGFGIVYKGRLLDGKEIAVKRLSKTSVQGTDEFMNEVTLIAR 578
Query: 118 TYHINLVKLYGFCFHRDKRALVYEYVENGSLDKYIFGSKNRNDFDFQKLHKIAIGTAKGI 177
HINLV++ G C D++ L+YEY+EN SLD Y+FG R+ ++ + I G A+G+
Sbjct: 579 LQHINLVQVLGCCIEGDEKMLIYEYLENLSLDSYLFGKTRRSKLNWNERFDITNGVARGL 638
Query: 178 AYLHEECKHRIIHYDIKPENVLLDMKLEPKIADFGLAKLRSRESNIELNTHFRGTRGYAA 237
YLH++ + RIIH D+K N+LLD + PKI+DFG+A++ R+ GT GY +
Sbjct: 639 LYLHQDSRFRIIHRDLKVSNILLDKNMIPKISDFGMARIFERDETEANTMKVVGTYGYMS 698
Query: 238 PEMWKPYPVTYKCDVYSFGILLFEIVGRRRH---FDSSYSESQQWFPRWTWEMFENNELV 294
PE + K DV+SFG+++ EIV +++ ++ Y + W+ E+V
Sbjct: 699 PEYAMYGIFSEKSDVFSFGVIVLEIVSGKKNRGFYNLDYENDLLSYVWSRWKEGRALEIV 758
Query: 295 VMLALCEIEEKDS----EIAERMLKVALWCVQYSPNDRPLMSTVVKMLEGEIDISSPPFP 350
+ + + + S + + +++ L CVQ RP MS+VV M E P P
Sbjct: 759 DPVIVDSLSSQPSIFQPQEVLKCIQIGLLCVQELAEHRPAMSSVVWMFGSEATEIPQPKP 818
>PSKR_ARATH (Q9ZVR7) Putative phytosulfokine receptor precursor (EC
2.7.1.37) (Phytosulfokine LRR receptor kinase)
Length = 1008
Score = 177 bits (448), Expect = 6e-44
Identities = 107/275 (38%), Positives = 155/275 (55%), Gaps = 10/275 (3%)
Query: 70 STILGSGAFGVVFKGELSNGENVAVKVLNCLDMGMEEQFKAEVITIGRTYHINLVKLYGF 129
+ I+G G FG+V+K L +G+ VA+K L+ +E +F+AEV T+ R H NLV L GF
Sbjct: 737 ANIIGCGGFGMVYKATLPDGKKVAIKKLSGDCGQIEREFEAEVETLSRAQHPNLVLLRGF 796
Query: 130 CFHRDKRALVYEYVENGSLDKYIFGSKNRND----FDFQKLHKIAIGTAKGIAYLHEECK 185
CF+++ R L+Y Y+ENGSLD ++ RND ++ +IA G AKG+ YLHE C
Sbjct: 797 CFYKNDRLLIYSYMENGSLDYWL---HERNDGPALLKWKTRLRIAQGAAKGLLYLHEGCD 853
Query: 186 HRIIHYDIKPENVLLDMKLEPKIADFGLAKLRSRESNIELNTHFRGTRGYAAPEMWKPYP 245
I+H DIK N+LLD +ADFGLA+L S ++T GT GY PE +
Sbjct: 854 PHILHRDIKSSNILLDENFNSHLADFGLARLMS-PYETHVSTDLVGTLGYIPPEYGQASV 912
Query: 246 VTYKCDVYSFGILLFEIVGRRRHFDSSYSESQQWFPRWTWEM-FENNELVVMLALCEIEE 304
TYK DVYSFG++L E++ +R D + + W +M E+ V L +E
Sbjct: 913 ATYKGDVYSFGVVLLELLTDKRPVDMCKPKGCRDLISWVVKMKHESRASEVFDPLIYSKE 972
Query: 305 KDSEIAERMLKVALWCVQYSPNDRPLMSTVVKMLE 339
D E+ R+L++A C+ +P RP +V L+
Sbjct: 973 NDKEMF-RVLEIACLCLSENPKQRPTTQQLVSWLD 1006
>EXS_ARATH (Q9LYN8) Leucine-rich repeat receptor protein kinase EXS
precursor (EC 2.7.1.37) (Extra sporogenous cells protein)
(EXCESS MICROSPOROCYTES1 protein)
Length = 1192
Score = 169 bits (428), Expect = 1e-41
Identities = 108/283 (38%), Positives = 160/283 (56%), Gaps = 11/283 (3%)
Query: 64 EITEKYS--TILGSGAFGVVFKGELSNGENVAVKVLNCLDMGMEEQFKAEVITIGRTYHI 121
E T+ +S I+G G FG V+K L + VAVK L+ +F AE+ T+G+ H
Sbjct: 912 EATDHFSKKNIIGDGGFGTVYKACLPGEKTVAVKKLSEAKTQGNREFMAEMETLGKVKHP 971
Query: 122 NLVKLYGFCFHRDKRALVYEYVENGSLDKYIFGSKNRND-FDFQKLHKIAIGTAKGIAYL 180
NLV L G+C +++ LVYEY+ NGSLD ++ + D+ K KIA+G A+G+A+L
Sbjct: 972 NLVSLLGYCSFSEEKLLVYEYMVNGSLDHWLRNQTGMLEVLDWSKRLKIAVGAARGLAFL 1031
Query: 181 HEECKHRIIHYDIKPENVLLDMKLEPKIADFGLAKLRSR-ESNIELNTHFRGTRGYAAPE 239
H IIH DIK N+LLD EPK+ADFGLA+L S ES++ +T GT GY PE
Sbjct: 1032 HHGFIPHIIHRDIKASNILLDGDFEPKVADFGLARLISACESHV--STVIAGTFGYIPPE 1089
Query: 240 MWKPYPVTYKCDVYSFGILLFEIVGRRRHFDSSYSESQQW-FPRWTWEMFENNELVVML- 297
+ T K DVYSFG++L E+V + + ES+ W + + V ++
Sbjct: 1090 YGQSARATTKGDVYSFGVILLELVTGKEPTGPDFKESEGGNLVGWAIQKINQGKAVDVID 1149
Query: 298 -ALCEIEEKDSEIAERMLKVALWCVQYSPNDRPLMSTVVKMLE 339
L + K+S++ R+L++A+ C+ +P RP M V+K L+
Sbjct: 1150 PLLVSVALKNSQL--RLLQIAMLCLAETPAKRPNMLDVLKALK 1190
>PBS1_ARATH (Q9FE20) Serine/threonine-protein kinase PBS1 (EC
2.7.1.37) (AvrPphB susceptible protein 1)
Length = 456
Score = 166 bits (421), Expect = 8e-41
Identities = 98/271 (36%), Positives = 145/271 (53%), Gaps = 3/271 (1%)
Query: 71 TILGSGAFGVVFKGEL-SNGENVAVKVLNCLDMGMEEQFKAEVITIGRTYHINLVKLYGF 129
T LG G FG V+KG L S G+ VAVK L+ + +F EV+ + +H NLV L G+
Sbjct: 90 TFLGEGGFGRVYKGRLDSTGQVVAVKQLDRNGLQGNREFLVEVLMLSLLHHPNLVNLIGY 149
Query: 130 CFHRDKRALVYEYVENGSLDKYIFG-SKNRNDFDFQKLHKIAIGTAKGIAYLHEECKHRI 188
C D+R LVYE++ GSL+ ++ ++ D+ KIA G AKG+ +LH++ +
Sbjct: 150 CADGDQRLLVYEFMPLGSLEDHLHDLPPDKEALDWNMRMKIAAGAAKGLEFLHDKANPPV 209
Query: 189 IHYDIKPENVLLDMKLEPKIADFGLAKLRSRESNIELNTHFRGTRGYAAPEMWKPYPVTY 248
I+ D K N+LLD PK++DFGLAKL ++T GT GY APE +T
Sbjct: 210 IYRDFKSSNILLDEGFHPKLSDFGLAKLGPTGDKSHVSTRVMGTYGYCAPEYAMTGQLTV 269
Query: 249 KCDVYSFGILLFEIVGRRRHFDSSYSESQQWFPRWTWEMFENNELVVMLALCEIEEK-DS 307
K DVYSFG++ E++ R+ DS +Q W +F + + LA ++ + +
Sbjct: 270 KSDVYSFGVVFLELITGRKAIDSEMPHGEQNLVAWARPLFNDRRKFIKLADPRLKGRFPT 329
Query: 308 EIAERMLKVALWCVQYSPNDRPLMSTVVKML 338
+ L VA C+Q RPL++ VV L
Sbjct: 330 RALYQALAVASMCIQEQAATRPLIADVVTAL 360
>PSKR_DAUCA (Q8LPB4) Phytosulfokine receptor precursor (EC 2.7.1.37)
(Phytosulfokine LRR receptor kinase)
Length = 1021
Score = 164 bits (416), Expect = 3e-40
Identities = 102/291 (35%), Positives = 156/291 (53%), Gaps = 8/291 (2%)
Query: 62 LDEITEKYST-----ILGSGAFGVVFKGELSNGENVAVKVLNCLDMGMEEQFKAEVITIG 116
LD+I + S+ I+G G FG+V+K L +G VA+K L+ M+ +F+AEV T+
Sbjct: 733 LDDILKSTSSFNQANIIGCGGFGLVYKATLPDGTKVAIKRLSGDTGQMDREFQAEVETLS 792
Query: 117 RTYHINLVKLYGFCFHRDKRALVYEYVENGSLDKYIFGS-KNRNDFDFQKLHKIAIGTAK 175
R H NLV L G+C +++ + L+Y Y++NGSLD ++ D++ +IA G A+
Sbjct: 793 RAQHPNLVHLLGYCNYKNDKLLIYSYMDNGSLDYWLHEKVDGPPSLDWKTRLRIARGAAE 852
Query: 176 GIAYLHEECKHRIIHYDIKPENVLLDMKLEPKIADFGLAKLRSRESNIELNTHFRGTRGY 235
G+AYLH+ C+ I+H DIK N+LL +ADFGLA+L + + T GT GY
Sbjct: 853 GLAYLHQSCEPHILHRDIKSSNILLSDTFVAHLADFGLARL-ILPYDTHVTTDLVGTLGY 911
Query: 236 AAPEMWKPYPVTYKCDVYSFGILLFEIVGRRRHFDSSYSESQQWFPRWTWEMFENNELVV 295
PE + TYK DVYSFG++L E++ RR D + W +M
Sbjct: 912 IPPEYGQASVATYKGDVYSFGVVLLELLTGRRPMDVCKPRGSRDLISWVLQMKTEKRESE 971
Query: 296 MLALCEIEEKDSEIAERMLKVALWCVQYSPNDRPLMSTVVKMLEGEIDISS 346
+ ++ +E +L++A C+ +P RP +V LE ID+SS
Sbjct: 972 IFDPFIYDKDHAEEMLLVLEIACRCLGENPKTRPTTQQLVSWLE-NIDVSS 1021
>CLV1_ARATH (Q9SYQ8) Receptor protein kinase CLAVATA1 precursor (EC
2.7.1.-)
Length = 980
Score = 162 bits (410), Expect = 1e-39
Identities = 98/303 (32%), Positives = 161/303 (52%), Gaps = 16/303 (5%)
Query: 61 KLDEITE--KYSTILGSGAFGVVFKGELSNGENVAVKVLNCLDMGMEEQ-FKAEVITIGR 117
K +++ E K I+G G G+V++G + N +VA+K L G + F AE+ T+GR
Sbjct: 684 KSEDVLECLKEENIIGKGGAGIVYRGSMPNNVDVAIKRLVGRGTGRSDHGFTAEIQTLGR 743
Query: 118 TYHINLVKLYGFCFHRDKRALVYEYVENGSLDKYIFGSKNRNDFDFQKLHKIAIGTAKGI 177
H ++V+L G+ ++D L+YEY+ NGSL + + GSK + ++ H++A+ AKG+
Sbjct: 744 IRHRHIVRLLGYVANKDTNLLLYEYMPNGSLGELLHGSKGGH-LQWETRHRVAVEAAKGL 802
Query: 178 AYLHEECKHRIIHYDIKPENVLLDMKLEPKIADFGLAKLRSRESNIELNTHFRGTRGYAA 237
YLH +C I+H D+K N+LLD E +ADFGLAK + E + G+ GY A
Sbjct: 803 CYLHHDCSPLILHRDVKSNNILLDSDFEAHVADFGLAKFLVDGAASECMSSIAGSYGYIA 862
Query: 238 PEMWKPYPVTYKCDVYSFGILLFEIVGRRRHFDSSYSES---QQWFPRWTWEMFENNELV 294
PE V K DVYSFG++L E++ ++ + E +W E+ + ++
Sbjct: 863 PEYAYTLKVDEKSDVYSFGVVLLELIAGKKPV-GEFGEGVDIVRWVRNTEEEITQPSDAA 921
Query: 295 VMLALCEIEEKDSEIAE--RMLKVALWCVQYSPNDRPLMSTVVKMLEGEIDISSPPFPFH 352
+++A+ + + + K+A+ CV+ RP M VV ML ++PP
Sbjct: 922 IVVAIVDPRLTGYPLTSVIHVFKIAMMCVEEEAAARPTMREVVHML------TNPPKSVA 975
Query: 353 NLV 355
NL+
Sbjct: 976 NLI 978
>RPK1_IPONI (P93194) Receptor-like protein kinase precursor (EC
2.7.1.37)
Length = 1109
Score = 161 bits (407), Expect = 3e-39
Identities = 101/286 (35%), Positives = 155/286 (53%), Gaps = 11/286 (3%)
Query: 60 EKLDEITEKYSTILGSGAFGVVFKGELSNGENVAVK--VLNCLDMGMEEQFKAEVITIGR 117
E + + +KY ++G GA G ++K LS + AVK V + G + E+ TIG+
Sbjct: 811 EATENLNDKY--VIGKGAHGTIYKATLSPDKVYAVKKLVFTGIKNGSVSMVR-EIETIGK 867
Query: 118 TYHINLVKLYGFCFHRDKRALVYEYVENGSLDKYIFGSKNRNDFDFQKLHKIAIGTAKGI 177
H NL+KL F ++ ++Y Y+ENGSL + + D+ H IA+GTA G+
Sbjct: 868 VRHRNLIKLEEFWLRKEYGLILYTYMENGSLHDILHETNPPKPLDWSTRHNIAVGTAHGL 927
Query: 178 AYLHEECKHRIIHYDIKPENVLLDMKLEPKIADFGLAKLRSRESNIELNTHFRGTRGYAA 237
AYLH +C I+H DIKP N+LLD LEP I+DFG+AKL + + + +GT GY A
Sbjct: 928 AYLHFDCDPAIVHRDIKPMNILLDSDLEPHISDFGIAKLLDQSATSIPSNTVQGTIGYMA 987
Query: 238 PEMWKPYPVTYKCDVYSFGILLFEIVGRRRHFDSSYSESQQ--WFPRWTWEMFENNELVV 295
PE + + DVYS+G++L E++ R++ D S++ + R W + +V
Sbjct: 988 PENAFTTVKSRESDVYSYGVVLLELITRKKALDPSFNGETDIVGWVRSVWTQTGEIQKIV 1047
Query: 296 MLALCEIEEKDSEIAERM---LKVALWCVQYSPNDRPLMSTVVKML 338
+L + E DS + E++ L +AL C + + RP M VVK L
Sbjct: 1048 DPSLLD-ELIDSSVMEQVTEALSLALRCAEKEVDKRPTMRDVVKQL 1092
>RCK7_ARATH (Q9LQQ8) Putative serine/threonine-protein kinase
RLCKVII (EC 2.7.1.37)
Length = 423
Score = 161 bits (407), Expect = 3e-39
Identities = 98/295 (33%), Positives = 156/295 (52%), Gaps = 5/295 (1%)
Query: 49 INREKPVRFTPEKLDEITEKYST--ILGSGAFGVVFKGELSNGENV-AVKVLNCLDMGME 105
+ +K FT ++L E T + + LG G FG VFKG + + V A+K L+ +
Sbjct: 83 VTGKKAQTFTFQELAEATGNFRSDCFLGEGGFGKVFKGTIEKLDQVVAIKQLDRNGVQGI 142
Query: 106 EQFKAEVITIGRTYHINLVKLYGFCFHRDKRALVYEYVENGSLDKYIFG-SKNRNDFDFQ 164
+F EV+T+ H NLVKL GFC D+R LVYEY+ GSL+ ++ + D+
Sbjct: 143 REFVVEVLTLSLADHPNLVKLIGFCAEGDQRLLVYEYMPQGSLEDHLHVLPSGKKPLDWN 202
Query: 165 KLHKIAIGTAKGIAYLHEECKHRIIHYDIKPENVLLDMKLEPKIADFGLAKLRSRESNIE 224
KIA G A+G+ YLH+ +I+ D+K N+LL +PK++DFGLAK+
Sbjct: 203 TRMKIAAGAARGLEYLHDRMTPPVIYRDLKCSNILLGEDYQPKLSDFGLAKVGPSGDKTH 262
Query: 225 LNTHFRGTRGYAAPEMWKPYPVTYKCDVYSFGILLFEIVGRRRHFDSSYSESQQWFPRWT 284
++T GT GY AP+ +T+K D+YSFG++L E++ R+ D++ + Q W
Sbjct: 263 VSTRVMGTYGYCAPDYAMTGQLTFKSDIYSFGVVLLELITGRKAIDNTKTRKDQNLVGWA 322
Query: 285 WEMFENNELVVMLALCEIE-EKDSEIAERMLKVALWCVQYSPNDRPLMSTVVKML 338
+F++ + ++ + + L ++ CVQ P RP++S VV L
Sbjct: 323 RPLFKDRRNFPKMVDPLLQGQYPVRGLYQALAISAMCVQEQPTMRPVVSDVVLAL 377
>TMK1_ARATH (P43298) Putative receptor protein kinase TMK1 precursor
(EC 2.7.1.-)
Length = 942
Score = 160 bits (406), Expect = 4e-39
Identities = 99/296 (33%), Positives = 153/296 (51%), Gaps = 12/296 (4%)
Query: 62 LDEITEKYST--ILGSGAFGVVFKGELSNGENVAVKVLN---CLDMGMEEQFKAEVITIG 116
L +T +S+ ILGSG FGVV+KGEL +G +AVK + G E FK+E+ +
Sbjct: 581 LRSVTNNFSSDNILGSGGFGVVYKGELHDGTKIAVKRMENGVIAGKGFAE-FKSEIAVLT 639
Query: 117 RTYHINLVKLYGFCFHRDKRALVYEYVENGSLDKYIF--GSKNRNDFDFQKLHKIAIGTA 174
+ H +LV L G+C +++ LVYEY+ G+L +++F + +++ +A+ A
Sbjct: 640 KVRHRHLVTLLGYCLDGNEKLLVYEYMPQGTLSRHLFEWSEEGLKPLLWKQRLTLALDVA 699
Query: 175 KGIAYLHEECKHRIIHYDIKPENVLLDMKLEPKIADFGLAKLRSRESNIELNTHFRGTRG 234
+G+ YLH IH D+KP N+LL + K+ADFGL +L + E + T GT G
Sbjct: 700 RGVEYLHGLAHQSFIHRDLKPSNILLGDDMRAKVADFGLVRL-APEGKGSIETRIAGTFG 758
Query: 235 YAAPEMWKPYPVTYKCDVYSFGILLFEIVGRRRHFDSSYSESQQWFPRWTWEMFENNELV 294
Y APE VT K DVYSFG++L E++ R+ D S E W M+ N E
Sbjct: 759 YLAPEYAVTGRVTTKVDVYSFGVILMELITGRKSLDESQPEESIHLVSWFKRMYINKEAS 818
Query: 295 VMLAL---CEIEEKDSEIAERMLKVALWCVQYSPNDRPLMSTVVKMLEGEIDISSP 347
A+ +++E+ + ++A C P RP M V +L +++ P
Sbjct: 819 FKKAIDTTIDLDEETLASVHTVAELAGHCCAREPYQRPDMGHAVNILSSLVELWKP 874
>SIRK_ARATH (O64483) Senescence-induced receptor-like
serine/threonine kinase precursor (FLG22-induced
receptor-like kinase 1)
Length = 876
Score = 160 bits (406), Expect = 4e-39
Identities = 97/283 (34%), Positives = 156/283 (54%), Gaps = 3/283 (1%)
Query: 57 FTPEKLDEITEKYSTILGSGAFGVVFKGELSNGENVAVKVLNCLDMGMEEQFKAEVITIG 116
F ++ IT + ++G G FG V+ G + NGE VAVKVL+ ++F+AEV +
Sbjct: 564 FKYSEVVNITNNFERVIGKGGFGKVYHGVI-NGEQVAVKVLSEESAQGYKEFRAEVDLLM 622
Query: 117 RTYHINLVKLYGFCFHRDKRALVYEYVENGSLDKYIFGSKNRNDFDFQKLHKIAIGTAKG 176
R +H NL L G+C + L+YEY+ N +L Y+ G K +++ KI++ A+G
Sbjct: 623 RVHHTNLTSLVGYCNEINHMVLIYEYMANENLGDYLAG-KRSFILSWEERLKISLDAAQG 681
Query: 177 IAYLHEECKHRIIHYDIKPENVLLDMKLEPKIADFGLAKLRSRESNIELNTHFRGTRGYA 236
+ YLH CK I+H D+KP N+LL+ KL+ K+ADFGL++ S E + +++T G+ GY
Sbjct: 682 LEYLHNGCKPPIVHRDVKPTNILLNEKLQAKMADFGLSRSFSVEGSGQISTVVAGSIGYL 741
Query: 237 APEMWKPYPVTYKCDVYSFGILLFEIVGRRRHFDSSYSESQQWFPRWTWEMFENNELVVM 296
PE + + K DVYS G++L E++ + SS +E + N ++ +
Sbjct: 742 DPEYYSTRQMNEKSDVYSLGVVLLEVITGQPAIASSKTEKVH-ISDHVRSILANGDIRGI 800
Query: 297 LALCEIEEKDSEIAERMLKVALWCVQYSPNDRPLMSTVVKMLE 339
+ E D A +M ++AL C +++ RP MS VV L+
Sbjct: 801 VDQRLRERYDVGSAWKMSEIALACTEHTSAQRPTMSQVVMELK 843
>BRI1_LYCPE (Q8L899) Systemin receptor SR160 precursor (EC 2.7.1.37)
(Brassinosteroid LRR receptor kinase)
Length = 1207
Score = 155 bits (392), Expect = 2e-37
Identities = 104/296 (35%), Positives = 164/296 (55%), Gaps = 12/296 (4%)
Query: 52 EKPVR-FTPEKLDEITEKY--STILGSGAFGVVFKGELSNGENVAVKVLNCLDMGMEEQF 108
EKP+R T L E T + +++GSG FG V+K +L +G VA+K L + + +F
Sbjct: 870 EKPLRKLTFADLLEATNGFHNDSLVGSGGFGDVYKAQLKDGSVVAIKKLIHVSGQGDREF 929
Query: 109 KAEVITIGRTYHINLVKLYGFCFHRDKRALVYEYVENGSLDKYIFGSKNRN-DFDFQKLH 167
AE+ TIG+ H NLV L G+C ++R LVYEY++ GSL+ + K ++
Sbjct: 930 TAEMETIGKIKHRNLVPLLGYCKVGEERLLVYEYMKYGSLEDVLHDRKKTGIKLNWPARR 989
Query: 168 KIAIGTAKGIAYLHEECKHRIIHYDIKPENVLLDMKLEPKIADFGLAKLRS-RESNIELN 226
KIAIG A+G+A+LH C IIH D+K NVLLD LE +++DFG+A+L S ++++ ++
Sbjct: 990 KIAIGAARGLAFLHHNCIPHIIHRDMKSSNVLLDENLEARVSDFGMARLMSAMDTHLSVS 1049
Query: 227 THFRGTRGYAAPEMWKPYPVTYKCDVYSFGILLFEIVGRRRHFDSSYSESQQWFPRWTWE 286
T GT GY PE ++ + + K DVYS+G++L E++ ++ DS+ W
Sbjct: 1050 T-LAGTPGYVPPEYYQSFRCSTKGDVYSYGVVLLELLTGKQPTDSADFGDNNLV---GWV 1105
Query: 287 MFENNELVVMLALCEIEEKDSEIAERM---LKVALWCVQYSPNDRPLMSTVVKMLE 339
+ + E+ ++D+ I + LKVA C+ RP M V+ M +
Sbjct: 1106 KLHAKGKITDVFDRELLKEDASIEIELLQHLKVACACLDDRHWKRPTMIQVMAMFK 1161
>BRI1_LYCES (Q8GUQ5) Brassinosteroid LRR receptor kinase precursor (EC
2.7.1.37) (tBRI1) (Altered brassinolide sensitivity 1)
(Systemin receptor SR160)
Length = 1207
Score = 154 bits (390), Expect = 3e-37
Identities = 104/296 (35%), Positives = 164/296 (55%), Gaps = 12/296 (4%)
Query: 52 EKPVR-FTPEKLDEITEKY--STILGSGAFGVVFKGELSNGENVAVKVLNCLDMGMEEQF 108
EKP+R T L E T + +++GSG FG V+K +L +G VA+K L + + +F
Sbjct: 870 EKPLRKLTFADLLEATNGFHNDSLVGSGGFGDVYKAQLKDGSVVAIKKLIHVSGQGDREF 929
Query: 109 KAEVITIGRTYHINLVKLYGFCFHRDKRALVYEYVENGSLDKYIFGSKNRN-DFDFQKLH 167
AE+ TIG+ H NLV L G+C ++R LVYEY++ GSL+ + K ++
Sbjct: 930 TAEMETIGKIKHRNLVPLLGYCKVGEERLLVYEYMKYGSLEDVLHDRKKIGIKLNWPARR 989
Query: 168 KIAIGTAKGIAYLHEECKHRIIHYDIKPENVLLDMKLEPKIADFGLAKLRS-RESNIELN 226
KIAIG A+G+A+LH C IIH D+K NVLLD LE +++DFG+A+L S ++++ ++
Sbjct: 990 KIAIGAARGLAFLHHNCIPHIIHRDMKSSNVLLDENLEARVSDFGMARLMSAMDTHLSVS 1049
Query: 227 THFRGTRGYAAPEMWKPYPVTYKCDVYSFGILLFEIVGRRRHFDSSYSESQQWFPRWTWE 286
T GT GY PE ++ + + K DVYS+G++L E++ ++ DS+ W
Sbjct: 1050 T-LAGTPGYVPPEYYQSFRCSTKGDVYSYGVVLLELLTGKQPTDSADFGDNNLV---GWV 1105
Query: 287 MFENNELVVMLALCEIEEKDSEIAERM---LKVALWCVQYSPNDRPLMSTVVKMLE 339
+ + E+ ++D+ I + LKVA C+ RP M V+ M +
Sbjct: 1106 KLHAKGKITDVFDRELLKEDASIEIELLQHLKVACACLDDRHWKRPTMIQVMAMFK 1161
>BRI1_ARATH (O22476) BRASSINOSTEROID INSENSITIVE 1 precursor (EC
2.7.1.37) (AtBRI1) (Brassinosteroid LRR receptor kinase)
Length = 1196
Score = 154 bits (390), Expect = 3e-37
Identities = 90/225 (40%), Positives = 139/225 (61%), Gaps = 6/225 (2%)
Query: 52 EKPVR-FTPEKLDEITEKY--STILGSGAFGVVFKGELSNGENVAVKVLNCLDMGMEEQF 108
EKP+R T L + T + +++GSG FG V+K L +G VA+K L + + +F
Sbjct: 865 EKPLRKLTFADLLQATNGFHNDSLIGSGGFGDVYKAILKDGSAVAIKKLIHVSGQGDREF 924
Query: 109 KAEVITIGRTYHINLVKLYGFCFHRDKRALVYEYVENGSLDKYIFGSKNRN-DFDFQKLH 167
AE+ TIG+ H NLV L G+C D+R LVYE+++ GSL+ + K ++
Sbjct: 925 MAEMETIGKIKHRNLVPLLGYCKVGDERLLVYEFMKYGSLEDVLHDPKKAGVKLNWSTRR 984
Query: 168 KIAIGTAKGIAYLHEECKHRIIHYDIKPENVLLDMKLEPKIADFGLAKLRS-RESNIELN 226
KIAIG+A+G+A+LH C IIH D+K NVLLD LE +++DFG+A+L S ++++ ++
Sbjct: 985 KIAIGSARGLAFLHHNCSPHIIHRDMKSSNVLLDENLEARVSDFGMARLMSAMDTHLSVS 1044
Query: 227 THFRGTRGYAAPEMWKPYPVTYKCDVYSFGILLFEIVGRRRHFDS 271
T GT GY PE ++ + + K DVYS+G++L E++ +R DS
Sbjct: 1045 T-LAGTPGYVPPEYYQSFRCSTKGDVYSYGVVLLELLTGKRPTDS 1088
>APKB_ARATH (P46573) Protein kinase APK1B, chloroplast precursor (EC
2.7.1.-)
Length = 412
Score = 154 bits (390), Expect = 3e-37
Identities = 104/301 (34%), Positives = 156/301 (51%), Gaps = 23/301 (7%)
Query: 57 FTPEKLDEITEKY--STILGSGAFGVVFKGELSN----------GENVAVKVLNCLDMGM 104
FT +L T + ++LG G FG VFKG + G +AVK LN
Sbjct: 57 FTFAELKAATRNFRPDSVLGEGGFGSVFKGWIDEQTLTASKPGTGVVIAVKKLNQDGWQG 116
Query: 105 EEQFKAEVITIGRTYHINLVKLYGFCFHRDKRALVYEYVENGSLDKYIFGSKNRNDFDFQ 164
+++ AEV +G+ H NLVKL G+C + R LVYE++ GSL+ ++F R FQ
Sbjct: 117 HQEWLAEVNYLGQFSHPNLVKLIGYCLEDEHRLLVYEFMPRGSLENHLF----RRGSYFQ 172
Query: 165 KLH-----KIAIGTAKGIAYLHEECKHRIIHYDIKPENVLLDMKLEPKIADFGLAKLRSR 219
L K+A+G AKG+A+LH + +I+ D K N+LLD + K++DFGLAK
Sbjct: 173 PLSWTLRLKVALGAAKGLAFLHN-AETSVIYRDFKTSNILLDSEYNAKLSDFGLAKDGPT 231
Query: 220 ESNIELNTHFRGTRGYAAPEMWKPYPVTYKCDVYSFGILLFEIVGRRRHFDSSYSESQQW 279
++T GT GYAAPE +T K DVYS+G++L E++ RR D + +Q
Sbjct: 232 GDKSHVSTRIMGTYGYAAPEYLATGHLTTKSDVYSYGVVLLEVLSGRRAVDKNRPPGEQK 291
Query: 280 FPRWTWEMFENNELVVMLALCEIEEKDS-EIAERMLKVALWCVQYSPNDRPLMSTVVKML 338
W + N + + ++++ S E A ++ +AL C+ + RP M+ VV L
Sbjct: 292 LVEWARPLLANKRKLFRVIDNRLQDQYSMEEACKVATLALRCLTFEIKLRPNMNEVVSHL 351
Query: 339 E 339
E
Sbjct: 352 E 352
>CX32_ARATH (P27450) Probable serine/threonine-protein kinase Cx32,
chloroplast precursor (EC 2.7.1.37)
Length = 419
Score = 152 bits (383), Expect = 2e-36
Identities = 107/346 (30%), Positives = 170/346 (48%), Gaps = 23/346 (6%)
Query: 13 DNQISSNSFSSTNNIEVVVQTDSKHEFATMER-IFSNINR--EKP----VRFTPEKLDEI 65
+N + FSST + +F+ + I S+ + E P F K
Sbjct: 25 NNHSNGTEFSSTTGATTNSSVGQQSQFSDISTGIISDSGKLLESPNLKVYNFLDLKTATK 84
Query: 66 TEKYSTILGSGAFGVVFKG----------ELSNGENVAVKVLNCLDMGMEEQFKAEVITI 115
K ++LG G FG V++G + +G VA+K LN + ++++EV +
Sbjct: 85 NFKPDSMLGQGGFGKVYRGWVDATTLAPSRVGSGMIVAIKRLNSESVQGFAEWRSEVNFL 144
Query: 116 GRTYHINLVKLYGFCFHRDKRALVYEYVENGSLDKYIFGSKNRND-FDFQKLHKIAIGTA 174
G H NLVKL G+C + LVYE++ GSL+ ++F RND F + KI IG A
Sbjct: 145 GMLSHRNLVKLLGYCREDKELLLVYEFMPKGSLESHLF---RRNDPFPWDLRIKIVIGAA 201
Query: 175 KGIAYLHEECKHRIIHYDIKPENVLLDMKLEPKIADFGLAKLRSRESNIELNTHFRGTRG 234
+G+A+LH + +I+ D K N+LLD + K++DFGLAKL + + T GT G
Sbjct: 202 RGLAFLHS-LQREVIYRDFKASNILLDSNYDAKLSDFGLAKLGPADEKSHVTTRIMGTYG 260
Query: 235 YAAPEMWKPYPVTYKCDVYSFGILLFEIVGRRRHFDSSYSESQQWFPRWTWEMFENNELV 294
YAAPE + K DV++FG++L EI+ ++ Q+ W N V
Sbjct: 261 YAAPEYMATGHLYVKSDVFAFGVVLLEIMTGLTAHNTKRPRGQESLVDWLRPELSNKHRV 320
Query: 295 VMLALCEIE-EKDSEIAERMLKVALWCVQYSPNDRPLMSTVVKMLE 339
+ I+ + +++A M ++ L C++ P +RP M VV++LE
Sbjct: 321 KQIMDKGIKGQYTTKVATEMARITLSCIEPDPKNRPHMKEVVEVLE 366
>APKA_ARATH (Q06548) Protein kinase APK1A, chloroplast precursor (EC
2.7.1.-)
Length = 410
Score = 150 bits (380), Expect = 4e-36
Identities = 100/285 (35%), Positives = 149/285 (52%), Gaps = 21/285 (7%)
Query: 71 TILGSGAFGVVFKGELSN----------GENVAVKVLNCLDMGMEEQFKAEVITIGRTYH 120
++LG G FG VFKG + G +AVK LN +++ AEV +G+ H
Sbjct: 72 SVLGEGGFGCVFKGWIDEKSLTASRPGTGLVIAVKKLNQDGWQGHQEWLAEVNYLGQFSH 131
Query: 121 INLVKLYGFCFHRDKRALVYEYVENGSLDKYIFGSKNRNDFDFQKLH-----KIAIGTAK 175
+LVKL G+C + R LVYE++ GSL+ ++F R FQ L K+A+G AK
Sbjct: 132 RHLVKLIGYCLEDEHRLLVYEFMPRGSLENHLF----RRGLYFQPLSWKLRLKVALGAAK 187
Query: 176 GIAYLHEECKHRIIHYDIKPENVLLDMKLEPKIADFGLAKLRSRESNIELNTHFRGTRGY 235
G+A+LH + R+I+ D K N+LLD + K++DFGLAK ++T GT GY
Sbjct: 188 GLAFLHSS-ETRVIYRDFKTSNILLDSEYNAKLSDFGLAKDGPIGDKSHVSTRVMGTHGY 246
Query: 236 AAPEMWKPYPVTYKCDVYSFGILLFEIVGRRRHFDSSYSESQQWFPRWTWEMFENNELVV 295
AAPE +T K DVYSFG++L E++ RR D + ++ W N +
Sbjct: 247 AAPEYLATGHLTTKSDVYSFGVVLLELLSGRRAVDKNRPSGERNLVEWAKPYLVNKRKIF 306
Query: 296 MLALCEIEEKDS-EIAERMLKVALWCVQYSPNDRPLMSTVVKMLE 339
+ ++++ S E A ++ ++L C+ RP MS VV LE
Sbjct: 307 RVIDNRLQDQYSMEEACKVATLSLRCLTTEIKLRPNMSEVVSHLE 351
>CRI4_MAIZE (O24585) Putative receptor protein kinase CRINKLY4
precursor (EC 2.7.1.-)
Length = 901
Score = 149 bits (375), Expect = 2e-35
Identities = 113/369 (30%), Positives = 175/369 (46%), Gaps = 30/369 (8%)
Query: 57 FTPEKLDEITEKYS--TILGSGAFGVVFKGELSNGENVAVK-VLNCLDMGME-EQFKAEV 112
F+ E+L++ T +S + +G G+F VFKG L +G VAVK + D+ ++F E+
Sbjct: 493 FSYEELEQATGGFSEDSQVGKGSFSCVFKGILRDGTVVAVKRAIKASDVKKSSKEFHNEL 552
Query: 113 ITIGRTYHINLVKLYGFCFHRDKRALVYEYVENGSLDKYIFGSKN--RNDFDFQKLHKIA 170
+ R H +L+ L G+C +R LVYE++ +GSL +++ G + ++ + IA
Sbjct: 553 DLLSRLNHAHLLNLLGYCEDGSERLLVYEFMAHGSLYQHLHGKDPNLKKRLNWARRVTIA 612
Query: 171 IGTAKGIAYLHEECKHRIIHYDIKPENVLLDMKLEPKIADFGLAKLRSRESNIELNTHFR 230
+ A+GI YLH +IH DIK N+L+D ++ADFGL+ L +S L+
Sbjct: 613 VQAARGIEYLHGYACPPVIHRDIKSSNILIDEDHNARVADFGLSILGPADSGTPLSELPA 672
Query: 231 GTRGYAAPEMWKPYPVTYKCDVYSFGILLFEIVGRRRHFDSSYSESQQWFPRWTWEMFEN 290
GT GY PE ++ + +T K DVYSFG++L EI+ R+ D + E W + +
Sbjct: 673 GTLGYLDPEYYRLHYLTTKSDVYSFGVVLLEILSGRKAIDMQFEEGN--IVEWAVPLIKA 730
Query: 291 NELVVMLALCEIEEKDSEIAERMLKVALWCVQYSPNDRPLMSTVVKMLE-------GEID 343
++ +L D E +++ VA CV+ DRP M V LE G
Sbjct: 731 GDIFAILDPVLSPPSDLEALKKIASVACKCVRMRGKDRPSMDKVTTALEHALALLMGSPC 790
Query: 344 ISSPPFP---------FHNLVPAKEN-STQEGSTADSDT-----TTSSWRTESSRESGFK 388
I P P H + N S E AD + SW T S S +
Sbjct: 791 IEQPILPTEVVLGSSRMHKVSQMSSNHSCSENELADGEDQGIGYRAPSWITFPSVTSSQR 850
Query: 389 TKHNAFEIE 397
K +A E +
Sbjct: 851 RKSSASEAD 859
>NAK_ARATH (P43293) Probable serine/threonine-protein kinase NAK (EC
2.7.1.37)
Length = 389
Score = 148 bits (373), Expect = 3e-35
Identities = 101/319 (31%), Positives = 159/319 (49%), Gaps = 30/319 (9%)
Query: 41 TMERIFSNINREKPVRFTPEKLDEITEKY--STILGSGAFGVVFKGELSN---------- 88
T I N N + F+ +L T + +++G G FG VFKG +
Sbjct: 43 TEGEILQNANLKN---FSLSELKSATRNFRPDSVVGEGGFGCVFKGWIDESSLAPSKPGT 99
Query: 89 GENVAVKVLNCLDMGMEEQFKAEVITIGRTYHINLVKLYGFCFHRDKRALVYEYVENGSL 148
G +AVK LN ++ AE+ +G+ H NLVKL G+C + R LVYE++ GSL
Sbjct: 100 GIVIAVKRLNQEGFQGHREWLAEINYLGQLDHPNLVKLIGYCLEEEHRLLVYEFMTRGSL 159
Query: 149 DKYIFGSKNRNDFDFQKLH-----KIAIGTAKGIAYLHEECKHRIIHYDIKPENVLLDMK 203
+ ++F R +Q L ++A+G A+G+A+LH + ++I+ D K N+LLD
Sbjct: 160 ENHLF----RRGTFYQPLSWNTRVRMALGAARGLAFLHN-AQPQVIYRDFKASNILLDSN 214
Query: 204 LEPKIADFGLAKLRSRESNIELNTHFRGTRGYAAPEMWKPYPVTYKCDVYSFGILLFEIV 263
K++DFGLA+ N ++T GT+GYAAPE ++ K DVYSFG++L E++
Sbjct: 215 YNAKLSDFGLARDGPMGDNSHVSTRVMGTQGYAAPEYLATGHLSVKSDVYSFGVVLLELL 274
Query: 264 GRRRHFDSSYSESQQWFPRWTWEMFENNELVVMLALCEIEEKDSEIAERMLKV---ALWC 320
RR D + + W N ++ + ++ + S R LK+ AL C
Sbjct: 275 SGRRAIDKNQPVGEHNLVDWARPYLTNKRRLLRVMDPRLQGQYS--LTRALKIAVLALDC 332
Query: 321 VQYSPNDRPLMSTVVKMLE 339
+ RP M+ +VK +E
Sbjct: 333 ISIDAKSRPTMNEIVKTME 351
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.318 0.134 0.394
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 47,915,839
Number of Sequences: 164201
Number of extensions: 2091270
Number of successful extensions: 9208
Number of sequences better than 10.0: 1650
Number of HSP's better than 10.0 without gapping: 987
Number of HSP's successfully gapped in prelim test: 663
Number of HSP's that attempted gapping in prelim test: 5890
Number of HSP's gapped (non-prelim): 1872
length of query: 402
length of database: 59,974,054
effective HSP length: 112
effective length of query: 290
effective length of database: 41,583,542
effective search space: 12059227180
effective search space used: 12059227180
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 67 (30.4 bits)
Medicago: description of AC148970.5


