

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148969.8 - phase: 0
(219 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
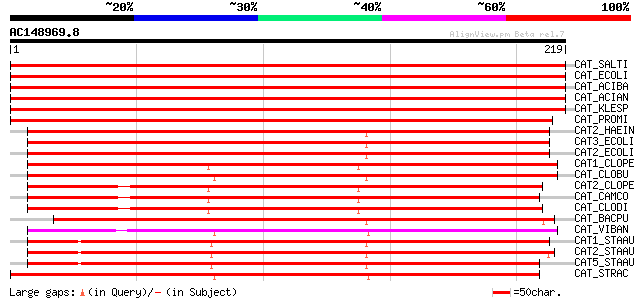
Score E
Sequences producing significant alignments: (bits) Value
CAT_SALTI (P62580) Chloramphenicol acetyltransferase (EC 2.3.1.2... 469 e-132
CAT_ECOLI (P62577) Chloramphenicol acetyltransferase (EC 2.3.1.2... 469 e-132
CAT_ACIBA (P62579) Chloramphenicol acetyltransferase (EC 2.3.1.2... 469 e-132
CAT_ACIAN (P62578) Chloramphenicol acetyltransferase (EC 2.3.1.2... 469 e-132
CAT_KLESP (P58777) Chloramphenicol acetyltransferase (EC 2.3.1.2... 466 e-131
CAT_PROMI (P07641) Chloramphenicol acetyltransferase (EC 2.3.1.2... 357 9e-99
CAT2_HAEIN (P22616) Chloramphenicol acetyltransferase II (EC 2.3... 220 2e-57
CAT3_ECOLI (P00484) Chloramphenicol acetyltransferase III (EC 2.... 215 6e-56
CAT2_ECOLI (P22615) Chloramphenicol acetyltransferase II (EC 2.3... 215 7e-56
CAT1_CLOPE (P26825) Chloramphenicol acetyltransferase (EC 2.3.1.... 206 3e-53
CAT_CLOBU (Q02736) Chloramphenicol acetyltransferase (EC 2.3.1.2... 204 2e-52
CAT2_CLOPE (P26826) Chloramphenicol acetyltransferase (EC 2.3.1.... 197 1e-50
CAT_CAMCO (P22782) Chloramphenicol acetyltransferase (EC 2.3.1.2... 193 3e-49
CAT_CLODI (P11504) Chloramphenicol acetyltransferase (EC 2.3.1.2... 191 1e-48
CAT_BACPU (P00487) Chloramphenicol acetyltransferase (EC 2.3.1.2... 189 6e-48
CAT_VIBAN (P49417) Chloramphenicol acetyltransferase (EC 2.3.1.2... 184 2e-46
CAT1_STAAU (P00485) Chloramphenicol acetyltransferase (EC 2.3.1.... 181 9e-46
CAT2_STAAU (P00486) Chloramphenicol acetyltransferase (EC 2.3.1.... 174 1e-43
CAT5_STAAU (P36883) Chloramphenicol acetyltransferase (EC 2.3.1.... 172 4e-43
CAT_STRAC (P20074) Chloramphenicol acetyltransferase (EC 2.3.1.2... 172 5e-43
>CAT_SALTI (P62580) Chloramphenicol acetyltransferase (EC 2.3.1.28)
(CAT)
Length = 219
Score = 469 bits (1206), Expect = e-132
Identities = 219/219 (100%), Positives = 219/219 (100%)
Query: 1 MEKKITGYTTVDISQWHRKEHFEAFQSVAQCTYNQTVQLDITAFLKTVKKNKHKFYPAFI 60
MEKKITGYTTVDISQWHRKEHFEAFQSVAQCTYNQTVQLDITAFLKTVKKNKHKFYPAFI
Sbjct: 1 MEKKITGYTTVDISQWHRKEHFEAFQSVAQCTYNQTVQLDITAFLKTVKKNKHKFYPAFI 60
Query: 61 HILARLMNAHPEFRMAMKDGELVIWDSVHPCYTVFHEQTETFSSLWSEYHDDFRQFLHIY 120
HILARLMNAHPEFRMAMKDGELVIWDSVHPCYTVFHEQTETFSSLWSEYHDDFRQFLHIY
Sbjct: 61 HILARLMNAHPEFRMAMKDGELVIWDSVHPCYTVFHEQTETFSSLWSEYHDDFRQFLHIY 120
Query: 121 SQDVACYGENLAYFPKGFIENMFFVSANPWVSFTSFDLNVANMDNFFAPVFTMGKYYTQG 180
SQDVACYGENLAYFPKGFIENMFFVSANPWVSFTSFDLNVANMDNFFAPVFTMGKYYTQG
Sbjct: 121 SQDVACYGENLAYFPKGFIENMFFVSANPWVSFTSFDLNVANMDNFFAPVFTMGKYYTQG 180
Query: 181 DKVLMPLAIQVHHAVCDGFHVGRMLNELQQYCDEWQGGA 219
DKVLMPLAIQVHHAVCDGFHVGRMLNELQQYCDEWQGGA
Sbjct: 181 DKVLMPLAIQVHHAVCDGFHVGRMLNELQQYCDEWQGGA 219
>CAT_ECOLI (P62577) Chloramphenicol acetyltransferase (EC 2.3.1.28)
(CAT)
Length = 219
Score = 469 bits (1206), Expect = e-132
Identities = 219/219 (100%), Positives = 219/219 (100%)
Query: 1 MEKKITGYTTVDISQWHRKEHFEAFQSVAQCTYNQTVQLDITAFLKTVKKNKHKFYPAFI 60
MEKKITGYTTVDISQWHRKEHFEAFQSVAQCTYNQTVQLDITAFLKTVKKNKHKFYPAFI
Sbjct: 1 MEKKITGYTTVDISQWHRKEHFEAFQSVAQCTYNQTVQLDITAFLKTVKKNKHKFYPAFI 60
Query: 61 HILARLMNAHPEFRMAMKDGELVIWDSVHPCYTVFHEQTETFSSLWSEYHDDFRQFLHIY 120
HILARLMNAHPEFRMAMKDGELVIWDSVHPCYTVFHEQTETFSSLWSEYHDDFRQFLHIY
Sbjct: 61 HILARLMNAHPEFRMAMKDGELVIWDSVHPCYTVFHEQTETFSSLWSEYHDDFRQFLHIY 120
Query: 121 SQDVACYGENLAYFPKGFIENMFFVSANPWVSFTSFDLNVANMDNFFAPVFTMGKYYTQG 180
SQDVACYGENLAYFPKGFIENMFFVSANPWVSFTSFDLNVANMDNFFAPVFTMGKYYTQG
Sbjct: 121 SQDVACYGENLAYFPKGFIENMFFVSANPWVSFTSFDLNVANMDNFFAPVFTMGKYYTQG 180
Query: 181 DKVLMPLAIQVHHAVCDGFHVGRMLNELQQYCDEWQGGA 219
DKVLMPLAIQVHHAVCDGFHVGRMLNELQQYCDEWQGGA
Sbjct: 181 DKVLMPLAIQVHHAVCDGFHVGRMLNELQQYCDEWQGGA 219
>CAT_ACIBA (P62579) Chloramphenicol acetyltransferase (EC 2.3.1.28)
(CAT)
Length = 219
Score = 469 bits (1206), Expect = e-132
Identities = 219/219 (100%), Positives = 219/219 (100%)
Query: 1 MEKKITGYTTVDISQWHRKEHFEAFQSVAQCTYNQTVQLDITAFLKTVKKNKHKFYPAFI 60
MEKKITGYTTVDISQWHRKEHFEAFQSVAQCTYNQTVQLDITAFLKTVKKNKHKFYPAFI
Sbjct: 1 MEKKITGYTTVDISQWHRKEHFEAFQSVAQCTYNQTVQLDITAFLKTVKKNKHKFYPAFI 60
Query: 61 HILARLMNAHPEFRMAMKDGELVIWDSVHPCYTVFHEQTETFSSLWSEYHDDFRQFLHIY 120
HILARLMNAHPEFRMAMKDGELVIWDSVHPCYTVFHEQTETFSSLWSEYHDDFRQFLHIY
Sbjct: 61 HILARLMNAHPEFRMAMKDGELVIWDSVHPCYTVFHEQTETFSSLWSEYHDDFRQFLHIY 120
Query: 121 SQDVACYGENLAYFPKGFIENMFFVSANPWVSFTSFDLNVANMDNFFAPVFTMGKYYTQG 180
SQDVACYGENLAYFPKGFIENMFFVSANPWVSFTSFDLNVANMDNFFAPVFTMGKYYTQG
Sbjct: 121 SQDVACYGENLAYFPKGFIENMFFVSANPWVSFTSFDLNVANMDNFFAPVFTMGKYYTQG 180
Query: 181 DKVLMPLAIQVHHAVCDGFHVGRMLNELQQYCDEWQGGA 219
DKVLMPLAIQVHHAVCDGFHVGRMLNELQQYCDEWQGGA
Sbjct: 181 DKVLMPLAIQVHHAVCDGFHVGRMLNELQQYCDEWQGGA 219
>CAT_ACIAN (P62578) Chloramphenicol acetyltransferase (EC 2.3.1.28)
(CAT)
Length = 219
Score = 469 bits (1206), Expect = e-132
Identities = 219/219 (100%), Positives = 219/219 (100%)
Query: 1 MEKKITGYTTVDISQWHRKEHFEAFQSVAQCTYNQTVQLDITAFLKTVKKNKHKFYPAFI 60
MEKKITGYTTVDISQWHRKEHFEAFQSVAQCTYNQTVQLDITAFLKTVKKNKHKFYPAFI
Sbjct: 1 MEKKITGYTTVDISQWHRKEHFEAFQSVAQCTYNQTVQLDITAFLKTVKKNKHKFYPAFI 60
Query: 61 HILARLMNAHPEFRMAMKDGELVIWDSVHPCYTVFHEQTETFSSLWSEYHDDFRQFLHIY 120
HILARLMNAHPEFRMAMKDGELVIWDSVHPCYTVFHEQTETFSSLWSEYHDDFRQFLHIY
Sbjct: 61 HILARLMNAHPEFRMAMKDGELVIWDSVHPCYTVFHEQTETFSSLWSEYHDDFRQFLHIY 120
Query: 121 SQDVACYGENLAYFPKGFIENMFFVSANPWVSFTSFDLNVANMDNFFAPVFTMGKYYTQG 180
SQDVACYGENLAYFPKGFIENMFFVSANPWVSFTSFDLNVANMDNFFAPVFTMGKYYTQG
Sbjct: 121 SQDVACYGENLAYFPKGFIENMFFVSANPWVSFTSFDLNVANMDNFFAPVFTMGKYYTQG 180
Query: 181 DKVLMPLAIQVHHAVCDGFHVGRMLNELQQYCDEWQGGA 219
DKVLMPLAIQVHHAVCDGFHVGRMLNELQQYCDEWQGGA
Sbjct: 181 DKVLMPLAIQVHHAVCDGFHVGRMLNELQQYCDEWQGGA 219
>CAT_KLESP (P58777) Chloramphenicol acetyltransferase (EC 2.3.1.28)
(CAT)
Length = 219
Score = 466 bits (1198), Expect = e-131
Identities = 218/219 (99%), Positives = 218/219 (99%)
Query: 1 MEKKITGYTTVDISQWHRKEHFEAFQSVAQCTYNQTVQLDITAFLKTVKKNKHKFYPAFI 60
MEKKITGYTTVDISQWHRKEHFEAFQSVAQCTYNQTVQLDITAFLKTVKKNKHKFYPAFI
Sbjct: 1 MEKKITGYTTVDISQWHRKEHFEAFQSVAQCTYNQTVQLDITAFLKTVKKNKHKFYPAFI 60
Query: 61 HILARLMNAHPEFRMAMKDGELVIWDSVHPCYTVFHEQTETFSSLWSEYHDDFRQFLHIY 120
HILARLMNAHPEFRMAMKDGELVIWDSVHPCYTVFHEQTETFSSLWSEYHDDFRQFLHIY
Sbjct: 61 HILARLMNAHPEFRMAMKDGELVIWDSVHPCYTVFHEQTETFSSLWSEYHDDFRQFLHIY 120
Query: 121 SQDVACYGENLAYFPKGFIENMFFVSANPWVSFTSFDLNVANMDNFFAPVFTMGKYYTQG 180
SQDVACYGENLAYFPKGFIENMFFVSANPWVSFTSFDLNVA MDNFFAPVFTMGKYYTQG
Sbjct: 121 SQDVACYGENLAYFPKGFIENMFFVSANPWVSFTSFDLNVAAMDNFFAPVFTMGKYYTQG 180
Query: 181 DKVLMPLAIQVHHAVCDGFHVGRMLNELQQYCDEWQGGA 219
DKVLMPLAIQVHHAVCDGFHVGRMLNELQQYCDEWQGGA
Sbjct: 181 DKVLMPLAIQVHHAVCDGFHVGRMLNELQQYCDEWQGGA 219
>CAT_PROMI (P07641) Chloramphenicol acetyltransferase (EC 2.3.1.28)
(CAT)
Length = 217
Score = 357 bits (917), Expect = 9e-99
Identities = 165/214 (77%), Positives = 190/214 (88%)
Query: 1 MEKKITGYTTVDISQWHRKEHFEAFQSVAQCTYNQTVQLDITAFLKTVKKNKHKFYPAFI 60
M+ K G VD+SQW RKEHFEAFQS AQCT++QTVQLDIT+ LKTVK+N +KFYP FI
Sbjct: 1 MDTKRVGILVVDLSQWGRKEHFEAFQSFAQCTFSQTVQLDITSLLKTVKQNGYKFYPTFI 60
Query: 61 HILARLMNAHPEFRMAMKDGELVIWDSVHPCYTVFHEQTETFSSLWSEYHDDFRQFLHIY 120
+I++ L+N H EFRMAMKDGELVIWDSV+P Y +FHEQTETFSSLWS YH D +FL Y
Sbjct: 61 YIISLLVNKHAEFRMAMKDGELVIWDSVNPGYNIFHEQTETFSSLWSYYHKDINRFLKTY 120
Query: 121 SQDVACYGENLAYFPKGFIENMFFVSANPWVSFTSFDLNVANMDNFFAPVFTMGKYYTQG 180
S+D+A YG++LAYFPK FIENMFFVSANPWVSFTSF+LN+AN++NFFAPVFT+GKYYTQG
Sbjct: 121 SEDIAQYGDDLAYFPKEFIENMFFVSANPWVSFTSFNLNMANINNFFAPVFTIGKYYTQG 180
Query: 181 DKVLMPLAIQVHHAVCDGFHVGRMLNELQQYCDE 214
DKVLMPLAIQVHHAVCDGFHVGR+LNE+QQYCDE
Sbjct: 181 DKVLMPLAIQVHHAVCDGFHVGRLLNEIQQYCDE 214
>CAT2_HAEIN (P22616) Chloramphenicol acetyltransferase II (EC
2.3.1.28) (CAT-II)
Length = 213
Score = 220 bits (560), Expect = 2e-57
Identities = 96/207 (46%), Positives = 144/207 (69%), Gaps = 1/207 (0%)
Query: 8 YTTVDISQWHRKEHFEAFQSVAQCTYNQTVQLDITAFLKTVKKNKHKFYPAFIHILARLM 67
+T +D++ W+R+EHF ++ +C ++ T +LDITAF + + +KFYP I++++R++
Sbjct: 3 FTRIDLNTWNRREHFALYRQQIKCGFSLTTKLDITAFRTALAETDYKFYPVMIYLISRVV 62
Query: 68 NAHPEFRMAMKDGELVIWDSVHPCYTVFHEQTETFSSLWSEYHDDFRQFLHIYSQDVACY 127
N PEFRMAMKD L+ WD P +TVFH++TETFS+L+ Y D +F+ Y+ +A Y
Sbjct: 63 NQFPEFRMAMKDNALIYWDQTDPVFTVFHKETETFSALFCRYCPDISEFMAGYNAVMAEY 122
Query: 128 GENLAYFPKGFI-ENMFFVSANPWVSFTSFDLNVANMDNFFAPVFTMGKYYTQGDKVLMP 186
N A FP+G + EN +S+ PWVSF F+LN+ D++FAPVFTM K+ + ++VL+P
Sbjct: 123 QHNTALFPQGALPENHLNISSLPWVSFDGFNLNITGNDDYFAPVFTMAKFQQEDNRVLLP 182
Query: 187 LAIQVHHAVCDGFHVGRMLNELQQYCD 213
+++QVHHAVCDGFH R +N LQ CD
Sbjct: 183 VSVQVHHAVCDGFHAARFINTLQMMCD 209
>CAT3_ECOLI (P00484) Chloramphenicol acetyltransferase III (EC
2.3.1.28) (catIII)
Length = 213
Score = 215 bits (548), Expect = 6e-56
Identities = 98/207 (47%), Positives = 141/207 (67%), Gaps = 1/207 (0%)
Query: 8 YTTVDISQWHRKEHFEAFQSVAQCTYNQTVQLDITAFLKTVKKNKHKFYPAFIHILARLM 67
YT D+ W R+EHFE ++ C ++ T ++DIT K++ + +KFYP I+++A+ +
Sbjct: 3 YTKFDVKNWVRREHFEFYRHRLPCGFSLTSKIDITTLKKSLDDSAYKFYPVMIYLIAQAV 62
Query: 68 NAHPEFRMAMKDGELVIWDSVHPCYTVFHEQTETFSSLWSEYHDDFRQFLHIYSQDVACY 127
N E RMA+KD EL++WDSV P +TVFH++TETFS+L Y D QF+ Y + Y
Sbjct: 63 NQFDELRMAIKDDELIVWDSVDPQFTVFHQETETFSALSCPYSSDIDQFMVNYLSVMERY 122
Query: 128 GENLAYFPKGFI-ENMFFVSANPWVSFTSFDLNVANMDNFFAPVFTMGKYYTQGDKVLMP 186
+ FP+G EN +SA PWV+F SF+LNVAN ++FAP+ TM KY +GD++L+P
Sbjct: 123 KSDTKLFPQGVTPENHLNISALPWVNFDSFNLNVANFTDYFAPIITMAKYQQEGDRLLLP 182
Query: 187 LAIQVHHAVCDGFHVGRMLNELQQYCD 213
L++QVHHAVCDGFHV R +N LQ+ C+
Sbjct: 183 LSVQVHHAVCDGFHVARFINRLQELCN 209
>CAT2_ECOLI (P22615) Chloramphenicol acetyltransferase II (EC
2.3.1.28) (CAT-II)
Length = 213
Score = 215 bits (547), Expect = 7e-56
Identities = 95/207 (45%), Positives = 141/207 (67%), Gaps = 1/207 (0%)
Query: 8 YTTVDISQWHRKEHFEAFQSVAQCTYNQTVQLDITAFLKTVKKNKHKFYPAFIHILARLM 67
+T +D++ W+R+EHF ++ +C ++ T +LDITA + + +KFYP I++++R +
Sbjct: 3 FTRIDLNTWNRREHFALYRQQIKCGFSLTTKLDITALRTALAETGYKFYPLMIYLISRAV 62
Query: 68 NAHPEFRMAMKDGELVIWDSVHPCYTVFHEQTETFSSLWSEYHDDFRQFLHIYSQDVACY 127
N PEFRMA+KD EL+ WD P +TVFH++TETFS+L Y D +F+ Y+ A Y
Sbjct: 63 NQFPEFRMALKDNELIYWDQSDPVFTVFHKETETFSALSCRYFPDLSEFMAGYNAVTAEY 122
Query: 128 GENLAYFPKGFI-ENMFFVSANPWVSFTSFDLNVANMDNFFAPVFTMGKYYTQGDKVLMP 186
+ FP+G + EN +S+ PWVSF F+LN+ D++FAPVFTM K+ +GD+VL+P
Sbjct: 123 QHDTRLFPQGNLPENHLNISSLPWVSFDGFNLNITGNDDYFAPVFTMAKFQQEGDRVLLP 182
Query: 187 LAIQVHHAVCDGFHVGRMLNELQQYCD 213
+++QVHHAVCDGFH R +N LQ CD
Sbjct: 183 VSVQVHHAVCDGFHAARFINTLQLMCD 209
>CAT1_CLOPE (P26825) Chloramphenicol acetyltransferase (EC 2.3.1.28)
(CAT)
Length = 219
Score = 206 bits (524), Expect = 3e-53
Identities = 89/211 (42%), Positives = 139/211 (65%), Gaps = 2/211 (0%)
Query: 8 YTTVDISQWHRKEHFEAFQSVAQCTYNQTVQLDITAFLKTVKKNKHKFYPAFIHILARLM 67
+ +DI W+RK +FE + + +CTY+ T ++IT L+ +K K YP I+I+ ++
Sbjct: 3 FNLIDIEDWNRKPYFEHYLNAVRCTYSMTANIEITGLLREIKLKGLKLYPTLIYIITTVV 62
Query: 68 NAHPEFRMAM-KDGELVIWDSVHPCYTVFHEQTETFSSLWSEYHDDFRQFLHIYSQDVAC 126
N H EFR + G+L WDS++P YTVFH+ ETFSS+W+EY ++F +F + Y +D+
Sbjct: 63 NRHKEFRTCFDQKGKLGYWDSMNPSYTVFHKDNETFSSIWTEYDENFPRFYYNYLEDIRN 122
Query: 127 YGENLAYFPK-GFIENMFFVSANPWVSFTSFDLNVANMDNFFAPVFTMGKYYTQGDKVLM 185
Y + L + PK G N VS+ PWV+FT F+LN+ N + P+FT+GKY+ Q +K+L+
Sbjct: 123 YSDVLNFMPKTGEPANTINVSSIPWVNFTGFNLNIYNDATYLIPIFTLGKYFQQDNKILL 182
Query: 186 PLAIQVHHAVCDGFHVGRMLNELQQYCDEWQ 216
P+++QVHHAVCDG+H+ R NE Q+ ++
Sbjct: 183 PMSVQVHHAVCDGYHISRFFNEAQELASNYE 213
>CAT_CLOBU (Q02736) Chloramphenicol acetyltransferase (EC 2.3.1.28)
(CAT)
Length = 219
Score = 204 bits (518), Expect = 2e-52
Identities = 91/211 (43%), Positives = 137/211 (64%), Gaps = 2/211 (0%)
Query: 8 YTTVDISQWHRKEHFEAFQSVAQCTYNQTVQLDITAFLKTVKKNKHKFYPAFIHILARLM 67
+ +DI+ W RK +FE + + +CTY+ T ++IT L +K KFYP I+++A ++
Sbjct: 3 FNLIDINHWSRKPYFEHYLNNVKCTYSMTANIEITDLLYEIKLKNIKFYPTLIYMIATVV 62
Query: 68 NAHPEFRMAMKD-GELVIWDSVHPCYTVFHEQTETFSSLWSEYHDDFRQFLHIYSQDVAC 126
N H EFR+ G L WDS++P YT+FH++ ETFSS+W+EY+ F +F Y D+
Sbjct: 63 NNHKEFRICFDHKGSLGYWDSMNPSYTIFHKENETFSSIWTEYNKSFLRFYSDYLDDIKN 122
Query: 127 YGENLAYFPKGFI-ENMFFVSANPWVSFTSFDLNVANMDNFFAPVFTMGKYYTQGDKVLM 185
YG + + PK +N F VS+ PWVSFT F+LNV N + P+FT GKY+ Q +K+ +
Sbjct: 123 YGNIMKFTPKSNEPDNTFSVSSIPWVSFTGFNLNVYNEGTYLIPIFTAGKYFKQENKIFI 182
Query: 186 PLAIQVHHAVCDGFHVGRMLNELQQYCDEWQ 216
P++IQVHHA+CDG+H R +NE+Q+ +Q
Sbjct: 183 PISIQVHHAICDGYHASRFINEMQELAFSFQ 213
>CAT2_CLOPE (P26826) Chloramphenicol acetyltransferase (EC 2.3.1.28)
(CAT)
Length = 207
Score = 197 bits (502), Expect = 1e-50
Identities = 96/205 (46%), Positives = 136/205 (65%), Gaps = 6/205 (2%)
Query: 8 YTTVDISQWHRKEHFEAFQSVAQCTYNQTVQLDITAFLKTVKKNKHKFYPAFIHILARLM 67
+ +D + W+RKE+F+ + + CTY+ TV++DIT +K+ K YPA ++ +A ++
Sbjct: 3 FEKIDKNSWNRKEYFDHYFASVPCTYSMTVKVDITQ----IKEKGMKLYPAMLYYIAMIV 58
Query: 68 NAHPEFRMAM-KDGELVIWDSVHPCYTVFHEQTETFSSLWSEYHDDFRQFLHIYSQDVAC 126
N H EFR A+ +DGEL I+D + P YT+FH TETFSSLW+E DF+ FL Y D
Sbjct: 59 NRHSEFRTAINQDGELGIYDEMIPSYTIFHNDTETFSSLWTECKSDFKSFLADYESDTQR 118
Query: 127 YGENLAYFPK-GFIENMFFVSANPWVSFTSFDLNVANMDNFFAPVFTMGKYYTQGDKVLM 185
YG N K EN+F VS PW +F F+LN+ ++ P+FTMGKYY + +K+++
Sbjct: 119 YGNNHRMEGKPNAPENIFNVSMIPWSTFDGFNLNLQKGYDYLIPIFTMGKYYKEDNKIIL 178
Query: 186 PLAIQVHHAVCDGFHVGRMLNELQQ 210
PLAIQVHHAVCDGFH+ R +NELQ+
Sbjct: 179 PLAIQVHHAVCDGFHICRFVNELQE 203
>CAT_CAMCO (P22782) Chloramphenicol acetyltransferase (EC 2.3.1.28)
(CAT)
Length = 207
Score = 193 bits (490), Expect = 3e-49
Identities = 92/204 (45%), Positives = 136/204 (66%), Gaps = 6/204 (2%)
Query: 8 YTTVDISQWHRKEHFEAFQSVAQCTYNQTVQLDITAFLKTVKKNKHKFYPAFIHILARLM 67
+T +DI+ W RKE+F+ + CTY+ TV+LDI+ +KK+ K YP ++ + ++
Sbjct: 3 FTKIDINNWTRKEYFDHYFGNTPCTYSMTVKLDISK----LKKDGKKLYPTLLYGVTTII 58
Query: 68 NAHPEFRMAM-KDGELVIWDSVHPCYTVFHEQTETFSSLWSEYHDDFRQFLHIYSQDVAC 126
N H EFR A+ ++G++ ++ + PCYTVFH++TETFSS+W+E+ D+ +FL Y +D+
Sbjct: 59 NRHEEFRTALDENGQVGVFSEMLPCYTVFHKETETFSSIWTEFTADYTEFLQNYQKDIDA 118
Query: 127 YGENLAYFPK-GFIENMFFVSANPWVSFTSFDLNVANMDNFFAPVFTMGKYYTQGDKVLM 185
+GE + K EN F VS PW SF F+LN+ ++ P+FT GKYY +G K +
Sbjct: 119 FGERMGMSAKPNPPENTFPVSMIPWTSFEGFNLNLKKGYDYLLPIFTFGKYYEEGGKYYI 178
Query: 186 PLAIQVHHAVCDGFHVGRMLNELQ 209
PL+IQVHHAVCDGFHV R L+ELQ
Sbjct: 179 PLSIQVHHAVCDGFHVCRFLDELQ 202
>CAT_CLODI (P11504) Chloramphenicol acetyltransferase (EC 2.3.1.28)
(CAT)
Length = 212
Score = 191 bits (485), Expect = 1e-48
Identities = 94/205 (45%), Positives = 134/205 (64%), Gaps = 6/205 (2%)
Query: 8 YTTVDISQWHRKEHFEAFQSVAQCTYNQTVQLDITAFLKTVKKNKHKFYPAFIHILARLM 67
+ +D + W+RKE+F+ + + CTY+ TV++DIT +K+ K YPA ++ +A ++
Sbjct: 3 FEKIDKNSWNRKEYFDHYFASVPCTYSMTVKVDITQ----IKEKGMKLYPAMLYYIAMIV 58
Query: 68 NAHPEFRMAM-KDGELVIWDSVHPCYTVFHEQTETFSSLWSEYHDDFRQFLHIYSQDVAC 126
N H EFR A+ +DGEL I+D + P YT+FH TETFSSLW+E DF+ FL Y D
Sbjct: 59 NRHSEFRTAINQDGELGIYDEMIPSYTIFHNDTETFSSLWTECKSDFKSFLADYESDTQR 118
Query: 127 YGENLAYFPK-GFIENMFFVSANPWVSFTSFDLNVANMDNFFAPVFTMGKYYTQGDKVLM 185
YG N K EN+F VS PW +F F+LN+ ++ P+FTMGK + +K+++
Sbjct: 119 YGNNHRMEGKPNAPENIFNVSMIPWSTFDGFNLNLQKGYDYLIPIFTMGKIIKKDNKIIL 178
Query: 186 PLAIQVHHAVCDGFHVGRMLNELQQ 210
PLAIQVHHAVCDGFH+ R +NELQ+
Sbjct: 179 PLAIQVHHAVCDGFHICRFVNELQE 203
>CAT_BACPU (P00487) Chloramphenicol acetyltransferase (EC 2.3.1.28)
(CAT) (Cat-86)
Length = 220
Score = 189 bits (479), Expect = 6e-48
Identities = 84/202 (41%), Positives = 135/202 (66%), Gaps = 4/202 (1%)
Query: 18 RKEHFEAFQSVAQCTYNQTVQLDITAFLKTVKKNKHKFYPAFIHILARLMNAHPEFRMAM 77
RKEHF + ++ +C+Y+ + LDIT +K+ K K YP I++LAR + PEFRM
Sbjct: 11 RKEHFHHYMTLTRCSYSLVINLDITKLHAILKEKKLKVYPVQIYLLARAVQKIPEFRMDQ 70
Query: 78 KDGELVIWDSVHPCYTVFHEQTETFSSLWSEYHDDFRQFLHIYSQDVACYGENLAYFPKG 137
+ EL W+ +HP YT+ +++T+TFSS+W+ + ++F QF D+ + ++ FPK
Sbjct: 71 VNDELGYWEILHPSYTILNKETKTFSSIWTPFDENFAQFYKSCVADIETFSKSSNLFPKP 130
Query: 138 FI-ENMFFVSANPWVSFTSFDLNVANMDNFFAPVFTMGKYYTQGDKVLMPLAIQVHHAVC 196
+ ENMF +S+ PW+ FTSF+LNV+ + + P+FT+GK+ + K+++P+AIQVHHAVC
Sbjct: 131 HMPENMFNISSLPWIDFTSFNLNVSTDEAYLLPIFTIGKFKVEEGKIILPVAIQVHHAVC 190
Query: 197 DGFHVGRMLNELQ---QYCDEW 215
DG+H G+ + L+ ++CDEW
Sbjct: 191 DGYHAGQYVEYLRWLIEHCDEW 212
>CAT_VIBAN (P49417) Chloramphenicol acetyltransferase (EC 2.3.1.28)
(CAT)
Length = 216
Score = 184 bits (466), Expect = 2e-46
Identities = 90/211 (42%), Positives = 127/211 (59%), Gaps = 6/211 (2%)
Query: 8 YTTVDISQWHRKEHFEAFQSVAQCTYNQTVQLDITAFLKTVKKNKHKFYPAFIHILARLM 67
+ VD+ W RKE+F + CTY+ TV+LDIT T+K K K YPA ++ ++ ++
Sbjct: 3 FRLVDLKTWKRKEYFTHYFESVPCTYSMTVKLDIT----TIKTGKAKLYPALLYAVSTVV 58
Query: 68 NAHPEFRMAMKD-GELVIWDSVHPCYTVFHEQTETFSSLWSEYHDDFRQFLHIYSQDVAC 126
N H EFRM + D G++ I+ + PCYT+F + TE FS++W+EY D+ +F Y +D+
Sbjct: 59 NRHEEFRMTVDDEGQIGIFSEMMPCYTIFQKDTEMFSNIWTEYIGDYTEFCKQYEKDMQQ 118
Query: 127 YGENLAYFPKGFIE-NMFFVSANPWVSFTSFDLNVANMDNFFAPVFTMGKYYTQGDKVLM 185
YGEN K N F VS PW +F F+LN+ + P+FT G+YY + K +
Sbjct: 119 YGENKGMMAKPNPPVNTFPVSMIPWTTFEGFNLNLQKGYGYLLPIFTFGRYYEENGKYWI 178
Query: 186 PLAIQVHHAVCDGFHVGRMLNELQQYCDEWQ 216
PL+IQVHHAVCDGFH R +NELQ Q
Sbjct: 179 PLSIQVHHAVCDGFHTCRFINELQDVIQSLQ 209
>CAT1_STAAU (P00485) Chloramphenicol acetyltransferase (EC 2.3.1.28)
(CAT)
Length = 216
Score = 181 bits (460), Expect = 9e-46
Identities = 82/208 (39%), Positives = 130/208 (62%), Gaps = 3/208 (1%)
Query: 8 YTTVDISQWHRKEHFEAFQSVAQCTYNQTVQLDITAFLKTVKKNKHKFYPAFIHILARLM 67
+ +D+ W RKE F + + Q T++ T ++DI+ + +K+ +KFYPAFI ++ R++
Sbjct: 3 FNKIDLDNWKRKEIFNHYLN-QQTTFSITTEIDISVLYRNIKQEGYKFYPAFIFLVTRVI 61
Query: 68 NAHPEFRMAMK-DGELVIWDSVHPCYTVFHEQTETFSSLWSEYHDDFRQFLHIYSQDVAC 126
N++ FR DGEL WD + P YT+F ++TFS +W+ +DF++F +Y DV
Sbjct: 62 NSNTAFRTGYNSDGELGYWDKLEPLYTIFDGVSKTFSGIWTPVKNDFKEFYDLYLSDVEK 121
Query: 127 YGENLAYFPKGFI-ENMFFVSANPWVSFTSFDLNVANMDNFFAPVFTMGKYYTQGDKVLM 185
Y + FPK I EN F +S PW SFT F+LN+ N N+ P+ T GK+ +G+ + +
Sbjct: 122 YNGSGKLFPKTPIPENAFSLSIIPWTSFTGFNLNINNNSNYLLPIITAGKFINKGNSIYL 181
Query: 186 PLAIQVHHAVCDGFHVGRMLNELQQYCD 213
PL++QVHH+VCDG+H G +N +Q+ D
Sbjct: 182 PLSLQVHHSVCDGYHAGLFMNSIQELSD 209
>CAT2_STAAU (P00486) Chloramphenicol acetyltransferase (EC 2.3.1.28)
(CAT)
Length = 215
Score = 174 bits (441), Expect = 1e-43
Identities = 80/213 (37%), Positives = 133/213 (61%), Gaps = 6/213 (2%)
Query: 8 YTTVDISQWHRKEHFEAFQSVAQCTYNQTVQLDITAFLKTVKKNKHKFYPAFIHILARLM 67
+ + + W RKE+FE + + Q TY+ T ++DIT F +KK ++ YP+ I+ + ++
Sbjct: 3 FNIIKLENWDRKEYFEHYFN-QQTTYSITKEIDITLFKDMIKKKGYEIYPSLIYAIMEVV 61
Query: 68 NAHPEFRMAMK-DGELVIWDSVHPCYTVFHEQTETFSSLWSEYHDDFRQFLHIYSQDVAC 126
N + FR + + +L WD ++P YTVF++QTE F+++W+E ++F F + Y D+
Sbjct: 62 NKNKVFRTGINSENKLGYWDKLNPLYTVFNKQTEKFTNIWTESDNNFTSFYNNYKNDLLE 121
Query: 127 YGENLAYFPKGFI-ENMFFVSANPWVSFTSFDLNVANMDNFFAPVFTMGKYYTQGDKVLM 185
Y + FPK I EN +S PW+ F+SF+LN+ N NF P+ T+GK+Y++ +K+ +
Sbjct: 122 YKDKEEMFPKKPIPENTIPISMIPWIDFSSFNLNIGNNSNFLLPIITIGKFYSENNKIYI 181
Query: 186 PLAIQVHHAVCDGFHVGRMLNELQQY---CDEW 215
P+A+Q+HHAVCDG+H +NE Q D+W
Sbjct: 182 PVALQLHHAVCDGYHASLFMNEFQDIIHKVDDW 214
>CAT5_STAAU (P36883) Chloramphenicol acetyltransferase (EC 2.3.1.28)
(CAT)
Length = 209
Score = 172 bits (437), Expect = 4e-43
Identities = 80/204 (39%), Positives = 124/204 (60%), Gaps = 3/204 (1%)
Query: 8 YTTVDISQWHRKEHFEAFQSVAQCTYNQTVQLDITAFLKTVKKNKHKFYPAFIHILARLM 67
+ +++ W RKE+F + + Q TY+ T +LDIT +K ++ YPA IH + ++
Sbjct: 3 FNIINLETWDRKEYFNHYFN-QQTTYSVTKELDITLLKSMIKNKGYELYPALIHAIVSVI 61
Query: 68 NAHPEFRMAMK-DGELVIWDSVHPCYTVFHEQTETFSSLWSEYHDDFRQFLHIYSQDVAC 126
N + FR + +G L WD + P YTVF+++TE FS++W+E + F F + Y D+
Sbjct: 62 NRNKVFRTGINSEGNLGYWDKLEPLYTVFNKETENFSNIWTESNASFTLFYNSYKNDLIK 121
Query: 127 YGENLAYFPKGFI-ENMFFVSANPWVSFTSFDLNVANMDNFFAPVFTMGKYYTQGDKVLM 185
Y + FPK I EN +S PW+ F+SF+LN+ N F P+ T+GK+Y++ DK+ +
Sbjct: 122 YKDKNEMFPKKPIPENTVPISMIPWIDFSSFNLNIGNNSRFLLPIITIGKFYSKDDKIYL 181
Query: 186 PLAIQVHHAVCDGFHVGRMLNELQ 209
P +QVHHAVCDG+HV +NE Q
Sbjct: 182 PFPLQVHHAVCDGYHVSLFMNEFQ 205
>CAT_STRAC (P20074) Chloramphenicol acetyltransferase (EC 2.3.1.28)
(CAT)
Length = 220
Score = 172 bits (436), Expect = 5e-43
Identities = 82/211 (38%), Positives = 129/211 (60%), Gaps = 2/211 (0%)
Query: 1 MEKKITGYTTVDISQWHRKEHFEAFQSVAQCTYNQTVQLDITAFLKTVKKNKHKFYPAFI 60
M+ I +D+ W R++HF+ ++ CTY TV++D+TAF ++++ K Y A +
Sbjct: 1 MDAPIPTPAPIDLDTWPRRQHFDHYRRRVPCTYAMTVEVDVTAFAAALRRSPRKSYLAQV 60
Query: 61 HILARLMNAHPEFRMAMKD-GELVIWDSVHPCYTVFHEQTETFSSLWSEYHDDFRQFLHI 119
LA ++N H EFRM + G+ +W VHP +TVF+ + ETF+ LW+ Y DF F
Sbjct: 61 WALATVVNRHEEFRMCLNSSGDPAVWPVVHPAFTVFNPERETFACLWAPYDPDFGTFHDT 120
Query: 120 YSQDVACYGENLAYFPKGFIE-NMFFVSANPWVSFTSFDLNVANMDNFFAPVFTMGKYYT 178
+ +A + +FP+G N F VS+ PWVSFT F L++ + + AP+FT+G+Y
Sbjct: 121 AAPLLAEHSRATDFFPQGNPPPNAFDVSSLPWVSFTGFTLDIRDGWDHLAPIFTLGRYTE 180
Query: 179 QGDKVLMPLAIQVHHAVCDGFHVGRMLNELQ 209
+ ++L+PL++Q+HHA DGFH R+ NELQ
Sbjct: 181 RDTRLLLPLSVQIHHAAADGFHTARLTNELQ 211
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.326 0.137 0.446
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 26,711,086
Number of Sequences: 164201
Number of extensions: 1052042
Number of successful extensions: 2722
Number of sequences better than 10.0: 28
Number of HSP's better than 10.0 without gapping: 26
Number of HSP's successfully gapped in prelim test: 2
Number of HSP's that attempted gapping in prelim test: 2650
Number of HSP's gapped (non-prelim): 30
length of query: 219
length of database: 59,974,054
effective HSP length: 106
effective length of query: 113
effective length of database: 42,568,748
effective search space: 4810268524
effective search space used: 4810268524
T: 11
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.7 bits)
S2: 63 (28.9 bits)
Medicago: description of AC148969.8


