

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148969.3 + phase: 2 /pseudo
(595 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
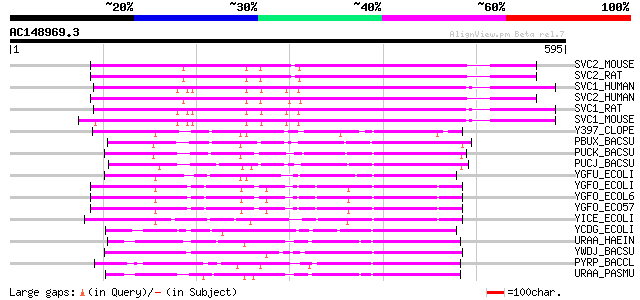
Score E
Sequences producing significant alignments: (bits) Value
SVC2_MOUSE (Q9EPR4) Solute carrier family 23, member 2 (Sodium-d... 270 6e-72
SVC2_RAT (Q9WTW8) Solute carrier family 23, member 2 (Sodium-dep... 270 8e-72
SVC1_HUMAN (Q9UHI7) Solute carrier family 23, member 1 (Sodium-d... 268 3e-71
SVC2_HUMAN (Q9UGH3) Solute carrier family 23, member 2 (Sodium-d... 267 5e-71
SVC1_RAT (Q9WTW7) Solute carrier family 23, member 1 (Sodium-dep... 264 6e-70
SVC1_MOUSE (Q9Z2J0) Solute carrier family 23, member 1 (Sodium-d... 263 7e-70
Y397_CLOPE (P50487) Putative purine permease CPE0397 115 2e-25
PBUX_BACSU (P42086) Xanthine permease 109 2e-23
PUCK_BACSU (O32140) Uric acid permease pucK 107 1e-22
PUCJ_BACSU (O32139) Uric acid permease pucJ 106 2e-22
YGFU_ECOLI (Q46821) Putative purine permease ygfU 105 3e-22
YGFO_ECOLI (P67444) Putative purine permease ygfO 100 1e-20
YGFO_ECOL6 (P67445) Putative purine permease ygfO 100 1e-20
YGFO_ECO57 (P67446) Putative purine permease ygfO 100 1e-20
YICE_ECOLI (P27432) Putative purine permease yicE 96 2e-19
YCDG_ECOLI (P75892) Putative purine permease ycdG 87 9e-17
URAA_HAEIN (P45117) Probable uracil permease (Uracil transporter) 80 1e-14
YWDJ_BACSU (P39618) Putative purine permease ywdJ 78 7e-14
PYRP_BACCL (P41006) Uracil permease (Uracil transporter) 77 1e-13
URAA_PASMU (Q9CPL9) Probable uracil permease (Uracil transporter) 76 2e-13
>SVC2_MOUSE (Q9EPR4) Solute carrier family 23, member 2
(Sodium-dependent vitamin C transporter 2) (mSVCT2)
(Na(+)/L-ascorbic acid transporter 2) (Yolk sac
permease-like molecule 2)
Length = 647
Score = 270 bits (691), Expect = 6e-72
Identities = 160/524 (30%), Positives = 268/524 (50%), Gaps = 72/524 (13%)
Query: 87 SPLNYELTDSPALVFLAVYGIQHYLSIIGSLILTPLVIAPAMGASHDE--TAAMVCTVLL 144
S + Y + D P G+QHYL+ I P ++A AM D+ T+ ++ T+
Sbjct: 86 SDMIYTIEDVPPWYLCIFLGLQHYLTCFSGTIAVPFLLADAMCVGDDQWATSQLIGTIFF 145
Query: 145 VSGVTTLLHTIFGSRLPLIQGPSFVYLAPVLAIINSPEFQ-------------ELNENKF 191
G+TTLL T FG RLPL Q +F +LAP AI++ +++ EL E+ +
Sbjct: 146 CVGITTLLQTTFGCRLPLFQASAFAFLAPARAILSLDKWKCNTTEITVANGTAELLEHIW 205
Query: 192 KHIMKELQGAIIIGSAFQTLLGYTGLMSLLVRFINPVVVSSTIAAVGLFL*LARVLRSVQ 251
++E+QGAII+ S + ++G GL L+R+I P+ ++ T+A +GL A R+ +
Sbjct: 206 HPRIQEIQGAIIMSSLIEVVIGLLGLPGALLRYIGPLTITPTVALIGLSGFQAAGERAGK 265
Query: 252 Y------RY*CLLFFVLYLRKI-------------SVFGHHIFQIYAVPLGLAVTWTFAF 292
+ +L F Y R + + + +F+++ + L + V+W F
Sbjct: 266 HWGIAMLTIFLVLLFSQYARNVKFPLPIYKSKKGWTAYKFQLFKMFPIILAILVSWLLCF 325
Query: 293 LLTENGRMKHCQVNTSD------------TMTSPPWFRFPYPLQWGTPVFNWKMAIVMCV 340
+ T N++D + PWF+ PYP QWG P + I M
Sbjct: 326 IFTVTDVFPS---NSTDYGYYARTDARKGVLLVAPWFKVPYPFQWGMPTVSAAGVIGMLS 382
Query: 341 VSLISSVDSVGTYHTSSLLAASGPPTPGVLSRGIGLEGFSSLLAGLWGTGMGSTTLTENV 400
+ S ++S+G Y+ + L+ + PP ++RGI +EG S +L G++GTG GST+ + N+
Sbjct: 383 AVVASIIESIGDYYACARLSCAPPPPIHAINRGIFVEGLSCVLDGIFGTGNGSTSSSPNI 442
Query: 401 HTIAGTKMGSRRPVQLGACLLIVLSLFGKVGGFIASIPEAMVAGLLCIMWAMLTALGLSN 460
+ TK+GSRR +Q GA L++ L + GK AS+P+ ++ L C ++ M+TA+GLSN
Sbjct: 443 GVLGITKVGSRRVIQYGAALMLGLGMVGKFSALFASLPDPVLGALFCTLFGMITAVGLSN 502
Query: 461 LRYTETGSSRNIIIVGLSLFFSLSIPAYFQQYESSPESNFSVPSYFQPYIVTSHGPFRSK 520
L++ + SSRN+ ++G S+FF L +P+Y +Q P +
Sbjct: 503 LQFIDLNSSRNLFVLGFSIFFGLVLPSYLRQ-----------------------NPLVTG 539
Query: 521 YEELNYVLNMIFSLHMVIAFLVALILDNTVPGSKQERELYGWSK 564
++ +LN++ + M + VA ILDNT+PG+ +ER + W K
Sbjct: 540 ITGIDQILNVLLTTAMFVGGCVAFILDNTIPGTPEERGIKKWKK 583
>SVC2_RAT (Q9WTW8) Solute carrier family 23, member 2
(Sodium-dependent vitamin C transporter 2)
(Na(+)/L-ascorbic acid transporter 2)
Length = 592
Score = 270 bits (690), Expect = 8e-72
Identities = 161/524 (30%), Positives = 268/524 (50%), Gaps = 72/524 (13%)
Query: 87 SPLNYELTDSPALVFLAVYGIQHYLSIIGSLILTPLVIAPAMGASHDE--TAAMVCTVLL 144
S + Y + D P G+QHYL+ I P ++A AM D+ T+ ++ T+
Sbjct: 31 SDMIYTIEDVPPWYLCIFLGLQHYLTCFSGTIAVPFLLADAMCVGDDQWATSQLIGTIFF 90
Query: 145 VSGVTTLLHTIFGSRLPLIQGPSFVYLAPVLAIINSPEFQ-------------ELNENKF 191
G+TTLL T FG RLPL Q +F +LAP AI++ +++ EL E+ +
Sbjct: 91 CVGITTLLQTTFGCRLPLFQASAFAFLAPARAILSLDKWKCNTTEITVANGTAELLEHIW 150
Query: 192 KHIMKELQGAIIIGSAFQTLLGYTGLMSLLVRFINPVVVSSTIAAVGLFL*LARVLRSVQ 251
++E+QGAII+ S + ++G GL L+R+I P+ ++ T+A +GL A R+ +
Sbjct: 151 HPRIQEIQGAIIMSSLIEVVIGLLGLPGALLRYIGPLTITPTVALIGLSGFQAAGERAGK 210
Query: 252 Y------RY*CLLFFVLYLRKI-------------SVFGHHIFQIYAVPLGLAVTWTFAF 292
+ +L F Y R + + + +F+++ + L + V+W F
Sbjct: 211 HWGIAMLTIFLVLLFSQYARNVKFPLPIYKSKKGWTAYKLQLFKMFPIILAILVSWLLCF 270
Query: 293 LLTENGRMKHCQVNTSD------------TMTSPPWFRFPYPLQWGTPVFNWKMAIVMCV 340
+ T N++D + PWF+ PYP QWG P + I M
Sbjct: 271 IFTVTDVFPS---NSTDYGYYARTDARKGVLLVAPWFKVPYPFQWGMPTVSAAGVIGMLS 327
Query: 341 VSLISSVDSVGTYHTSSLLAASGPPTPGVLSRGIGLEGFSSLLAGLWGTGMGSTTLTENV 400
+ S ++S+G Y+ + L+ + PP ++RGI +EG S +L G++GTG GST+ + N+
Sbjct: 328 AVVASIIESIGDYYACARLSCAPPPPIHAINRGIFVEGLSCVLDGVFGTGNGSTSSSPNI 387
Query: 401 HTIAGTKMGSRRPVQLGACLLIVLSLFGKVGGFIASIPEAMVAGLLCIMWAMLTALGLSN 460
+ TK+GSRR +Q GA L++ L + GK AS+P+ ++ L C ++ M+TA+GLSN
Sbjct: 388 GVLGITKVGSRRVIQYGAALMLGLGMIGKFSALFASLPDPVLGALFCTLFGMITAVGLSN 447
Query: 461 LRYTETGSSRNIIIVGLSLFFSLSIPAYFQQYESSPESNFSVPSYFQPYIVTSHGPFRSK 520
L++ + SSRN+ ++G S+FF L +P+Y +Q P +
Sbjct: 448 LQFIDLNSSRNLFVLGFSIFFGLVLPSYLRQ-----------------------NPLVTG 484
Query: 521 YEELNYVLNMIFSLHMVIAFLVALILDNTVPGSKQERELYGWSK 564
++ VLN++ + M + VA ILDNT+PG+ +ER + W K
Sbjct: 485 ITGIDQVLNVLLTTAMFVGGCVAFILDNTIPGTPEERGIKKWKK 528
>SVC1_HUMAN (Q9UHI7) Solute carrier family 23, member 1
(Sodium-dependent vitamin C transporter 1) (hSVCT1)
(Na(+)/L-ascorbic acid transporter 1) (Yolk sac
permease-like molecule 3)
Length = 598
Score = 268 bits (685), Expect = 3e-71
Identities = 166/540 (30%), Positives = 268/540 (48%), Gaps = 66/540 (12%)
Query: 91 YELTDSPALVFLAVYGIQHYLSIIGSLILTPLVIAPAMGASHDE--TAAMVCTVLLVSGV 148
Y++ D P + G QHYL+ I P ++A A+ HD+ + ++ T+ G+
Sbjct: 33 YKIEDVPPWYLCILLGFQHYLTCFSGTIAVPFLLAEALCVGHDQHMVSQLIGTIFTCVGI 92
Query: 149 TTLLHTIFGSRLPLIQGPSFVYLAPVLAII-----NSPEFQELNEN-----KFKHI---- 194
TTL+ T G RLPL Q +F +L P AI+ P +E+ N HI
Sbjct: 93 TTLIQTTVGIRLPLFQASAFAFLVPAKAILALERWKCPPEEEIYGNWSLPLNTSHIWHPR 152
Query: 195 MKELQGAIIIGSAFQTLLGYTGLMSLLVRFINPVVVSSTIAAVGLFL*LARVLRSVQY-- 252
++E+QGAI++ S + ++G GL L+ +I P+ V+ T++ +GL + A R+ +
Sbjct: 153 IREVQGAIMVSSVVEVVIGLLGLPGALLNYIGPLTVTPTVSLIGLSVFQAAGDRAGSHWG 212
Query: 253 ----RY*CLLFFVLYLRKIS-------------VFGHHIFQIYAVPLGLAVTWTFAFLLT 295
++ F YLR ++ + IF+++ + L + W ++LT
Sbjct: 213 ISACSILLIILFSQYLRNLTFLLPVYRWGKGLTLLRIQIFKMFPIMLAIMTVWLLCYVLT 272
Query: 296 ------ENGRMKHCQVNTS---DTMTSPPWFRFPYPLQWGTPVFNWKMAIVMCVVSLISS 346
+ + Q T D M PW R PYP QWG P + M +L
Sbjct: 273 LTDVLPTDPKAYGFQARTDARGDIMAIAPWIRIPYPCQWGLPTVTAAAVLGMFSATLAGI 332
Query: 347 VDSVGTYHTSSLLAASGPPTPGVLSRGIGLEGFSSLLAGLWGTGMGSTTLTENVHTIAGT 406
++S+G Y+ + LA + PP ++RGI EG ++AGL GTG GST+ + N+ + T
Sbjct: 333 IESIGDYYACARLAGAPPPPVHAINRGIFTEGICCIIAGLLGTGNGSTSSSPNIGVLGIT 392
Query: 407 KMGSRRPVQLGACLLIVLSLFGKVGGFIASIPEAMVAGLLCIMWAMLTALGLSNLRYTET 466
K+GSRR VQ GA +++VL GK AS+P+ ++ G+ C ++ M+TA+GLSNL++ +
Sbjct: 393 KVGSRRVVQYGAAIMLVLGTIGKFTALFASLPDPILGGMFCTLFGMITAVGLSNLQFVDM 452
Query: 467 GSSRNIIIVGLSLFFSLSIPAYFQQYESSPESNFSVPSYFQPYIVTSHGPFRSKYEELNY 526
SSRN+ ++G S+FF L++P Y ES+P G + E++
Sbjct: 453 NSSRNLFVLGFSMFFGLTLPNYL---ESNP------------------GAINTGILEVDQ 491
Query: 527 VLNMIFSLHMVIAFLVALILDNTVPGSKQERELYGWSKPNDARED-PFIVSEYGLPARVG 585
+L ++ + M + +A ILDNTVPGS +ER L W A D + Y P +G
Sbjct: 492 ILIVLLTTEMFVGGCLAFILDNTVPGSPEERGLIQWKAGAHANSDMSSSLKSYDFPIGMG 551
>SVC2_HUMAN (Q9UGH3) Solute carrier family 23, member 2
(Sodium-dependent vitamin C transporter 2) (hSVCT2)
(Na(+)/L-ascorbic acid transporter 2) (Yolk sac
permease-like molecule 2) (Nucleobase transporter-like 1
protein)
Length = 650
Score = 267 bits (683), Expect = 5e-71
Identities = 160/523 (30%), Positives = 269/523 (50%), Gaps = 68/523 (13%)
Query: 87 SPLNYELTDSPALVFLAVYGIQHYLSIIGSLILTPLVIAPAMGASHDE--TAAMVCTVLL 144
S + Y + D P G+QHYL+ I P ++A AM +D+ T+ ++ T+
Sbjct: 87 SDMIYTIEDVPPWYLCIFLGLQHYLTCFSGTIAVPFLLADAMCVGYDQWATSQLIGTIFF 146
Query: 145 VSGVTTLLHTIFGSRLPLIQGPSFVYLAPVLAIINSPEFQ-------------EL--NEN 189
G+TTLL T FG RLPL Q +F +LAP AI++ +++ EL E+
Sbjct: 147 CVGITTLLQTTFGCRLPLFQASAFAFLAPARAILSLDKWKCNTTDVSVANGTAELLHTEH 206
Query: 190 KFKHIMKELQGAIIIGSAFQTLLGYTGLMSLLVRFINPVVVSSTIAAVGLFL*LARVLRS 249
+ ++E+QGAII+ S + ++G GL L+++I P+ ++ T+A +GL A R+
Sbjct: 207 IWYPRIREIQGAIIMSSLIEVVIGLLGLPGALLKYIGPLTITPTVALIGLSGFQAAGERA 266
Query: 250 VQY------RY*CLLFFVLYLRKI-------------SVFGHHIFQIYAVPLGLAVTWTF 290
++ +L F Y R + + + +F+++ + L + V+W
Sbjct: 267 GKHWGIAMLTIFLVLLFSQYARNVKFPLPIYKSKKGWTAYKLQLFKMFPIILAILVSWLL 326
Query: 291 AFLLTENG-----RMKHCQVNTSDT----MTSPPWFRFPYPLQWGTPVFNWKMAIVMCVV 341
F+ T K+ +D + PWF+ PYP QWG P + I M
Sbjct: 327 CFIFTVTDVFPPDSTKYGFYARTDARQGVLLVAPWFKVPYPFQWGLPTVSAAGVIGMLSA 386
Query: 342 SLISSVDSVGTYHTSSLLAASGPPTPGVLSRGIGLEGFSSLLAGLWGTGMGSTTLTENVH 401
+ S ++S+G Y+ + L+ + PP ++RGI +EG S +L G++GTG GST+ + N+
Sbjct: 387 VVASIIESIGDYYACARLSCAPPPPIHAINRGIFVEGLSCVLDGIFGTGNGSTSSSPNIG 446
Query: 402 TIAGTKMGSRRPVQLGACLLIVLSLFGKVGGFIASIPEAMVAGLLCIMWAMLTALGLSNL 461
+ TK+GSRR +Q GA L++ L + GK AS+P+ ++ L C ++ M+TA+GLSNL
Sbjct: 447 VLGITKVGSRRVIQCGAALMLALGMIGKFSALFASLPDPVLGALFCTLFGMITAVGLSNL 506
Query: 462 RYTETGSSRNIIIVGLSLFFSLSIPAYFQQYESSPESNFSVPSYFQPYIVTSHGPFRSKY 521
++ + SSRN+ ++G S+FF L +P+Y +Q P +
Sbjct: 507 QFIDLNSSRNLFVLGFSIFFGLVLPSYLRQ-----------------------NPLVTGI 543
Query: 522 EELNYVLNMIFSLHMVIAFLVALILDNTVPGSKQERELYGWSK 564
++ VLN++ + M + VA ILDNT+PG+ +ER + W K
Sbjct: 544 TGIDQVLNVLLTTAMFVGGCVAFILDNTIPGTPEERGIRKWKK 586
>SVC1_RAT (Q9WTW7) Solute carrier family 23, member 1
(Sodium-dependent vitamin C transporter 1)
(Na(+)/L-ascorbic acid transporter 1)
Length = 604
Score = 264 bits (674), Expect = 6e-70
Identities = 163/540 (30%), Positives = 268/540 (49%), Gaps = 66/540 (12%)
Query: 91 YELTDSPALVFLAVYGIQHYLSIIGSLILTPLVIAPAMGASHDE--TAAMVCTVLLVSGV 148
Y++ D P + G QHYL+ I P ++A A+ D+ + ++ T+ G+
Sbjct: 40 YKIEDVPPWYLCILLGFQHYLTCFSGTIAVPFLLAEALCVGRDQHMISQLIGTIFTCVGI 99
Query: 149 TTLLHTIFGSRLPLIQGPSFVYLAPVLAII-----NSPEFQELNEN-----KFKHI---- 194
TTL+ T G RLPL Q +F +L P AI+ P +E+ N HI
Sbjct: 100 TTLIQTTVGIRLPLFQASAFAFLVPAKAILALERWKCPPEEEIYGNWSMPLNTSHIWHPR 159
Query: 195 MKELQGAIIIGSAFQTLLGYTGLMSLLVRFINPVVVSSTIAAVGLFL*LARVLRSVQY-- 252
++E+QGAI++ S + ++G GL L+ +I P+ V+ T++ +GL + A R+ +
Sbjct: 160 IREVQGAIMVSSVVEVVIGLLGLPGALLSYIGPLTVTPTVSLIGLSVFQAAGDRAGSHWG 219
Query: 253 ----RY*CLLFFVLYLRKIS-------------VFGHHIFQIYAVPLGLAVTWTFAFLLT 295
++ F YLR ++ +F IF+++ + L + W ++LT
Sbjct: 220 ISACSILLIVLFSQYLRNLTFLLPVYRWGKGLTLFRIQIFKMFPIVLAIMTVWLLCYVLT 279
Query: 296 ------ENGRMKHCQVNTS---DTMTSPPWFRFPYPLQWGTPVFNWKMAIVMCVVSLISS 346
+ + Q T D M PW R PYP QWG P + M +L
Sbjct: 280 LTDVLPADPTVYGFQARTDARGDIMAISPWIRIPYPCQWGLPTVTVAAVLGMFSATLAGI 339
Query: 347 VDSVGTYHTSSLLAASGPPTPGVLSRGIGLEGFSSLLAGLWGTGMGSTTLTENVHTIAGT 406
++S+G Y+ + LA + PP ++RGI EG ++AGL GTG GST+ + N+ + T
Sbjct: 340 IESIGDYYACARLAGAPPPPVHAINRGIFTEGVCCIIAGLLGTGNGSTSSSPNIGVLGIT 399
Query: 407 KMGSRRPVQLGACLLIVLSLFGKVGGFIASIPEAMVAGLLCIMWAMLTALGLSNLRYTET 466
K+GSRR VQ GA ++++L GK AS+P+ ++ G+ C ++ M+TA+GLSNL++ +
Sbjct: 400 KVGSRRVVQYGAGIMLILGAIGKFTALFASLPDPILGGMFCTLFGMITAVGLSNLQFVDM 459
Query: 467 GSSRNIIIVGLSLFFSLSIPAYFQQYESSPESNFSVPSYFQPYIVTSHGPFRSKYEELNY 526
SSRN+ ++G S+FF L++P Y +S+P G + E++
Sbjct: 460 NSSRNLFVLGFSMFFGLTLPNYL---DSNP------------------GAINTGVPEVDQ 498
Query: 527 VLNMIFSLHMVIAFLVALILDNTVPGSKQERELYGWSKPNDAREDPFI-VSEYGLPARVG 585
+L ++ + M + +A ILDNTVPGS +ER L W A + + Y P +G
Sbjct: 499 ILTVLLTTEMFVGGCLAFILDNTVPGSPEERGLIQWKAGAHANSETLASLKSYDFPFGMG 558
>SVC1_MOUSE (Q9Z2J0) Solute carrier family 23, member 1
(Sodium-dependent vitamin C transporter 1)
(Na(+)/L-ascorbic acid transporter 1) (Yolk sac
permease-like molecule 3)
Length = 605
Score = 263 bits (673), Expect = 7e-70
Identities = 169/565 (29%), Positives = 276/565 (47%), Gaps = 74/565 (13%)
Query: 74 MVDHDDLVLRRRPSPLN--------YELTDSPALVFLAVYGIQHYLSIIGSLILTPLVIA 125
+VD R R +PL Y++ D P + G QHYL+ I P ++A
Sbjct: 15 VVDSAGTSTRDRQAPLPTEPKFDMLYKIEDVPPWYLCILLGFQHYLTCFSGTIAVPFLLA 74
Query: 126 PAMGASHDE--TAAMVCTVLLVSGVTTLLHTIFGSRLPLIQGPSFVYLAPVLAII----- 178
A+ D+ + ++ T+ G+TTL+ T G RLPL Q +F +L P +I+
Sbjct: 75 EALCVGRDQHMVSQLIGTIFTCVGITTLIQTTVGIRLPLFQASAFAFLVPAKSILALERW 134
Query: 179 NSPEFQELNEN-----KFKHI----MKELQGAIIIGSAFQTLLGYTGLMSLLVRFINPVV 229
P +E+ N HI ++E+QGAI++ S + ++G GL L+ +I P+
Sbjct: 135 KCPSEEEIYGNWSMPLNTSHIWHPRIREVQGAIMVSSMVEVVIGLMGLPGALLSYIGPLT 194
Query: 230 VSSTIAAVGLFL*LARVLRSVQY------RY*CLLFFVLYLRKIS-------------VF 270
V+ T++ +GL + A R+ + ++ F YLR ++ +F
Sbjct: 195 VTPTVSLIGLSVFQAAGDRAGSHWGISACSILLIVLFSQYLRNLTFLLPVYRWGKGLTLF 254
Query: 271 GHHIFQIYAVPLGLAVTWTFAFLLT------ENGRMKHCQVNTS---DTMTSPPWFRFPY 321
IF+++ + L + W ++LT + + Q T D M PW R PY
Sbjct: 255 RVQIFKMFPIVLAIMTVWLLCYVLTLTDVLPADPTVYGFQARTDARGDIMAISPWIRIPY 314
Query: 322 PLQWGTPVFNWKMAIVMCVVSLISSVDSVGTYHTSSLLAASGPPTPGVLSRGIGLEGFSS 381
P QWG P + M +L ++S+G Y+ + LA + PP ++RGI EG
Sbjct: 315 PCQWGLPTVTVAAVLGMFSATLAGIIESIGDYYACARLAGAPPPPVHAINRGIFTEGICC 374
Query: 382 LLAGLWGTGMGSTTLTENVHTIAGTKMGSRRPVQLGACLLIVLSLFGKVGGFIASIPEAM 441
++AGL GTG GST+ + N+ + TK+GSRR VQ GA ++++L GK AS+P+ +
Sbjct: 375 IIAGLLGTGNGSTSSSPNIGVLGITKVGSRRVVQYGAGIMLILGAIGKFTALFASLPDPI 434
Query: 442 VAGLLCIMWAMLTALGLSNLRYTETGSSRNIIIVGLSLFFSLSIPAYFQQYESSPESNFS 501
+ G+ C ++ M+TA+GLSNL++ + SSRN+ ++G S+FF L++P Y +S+P
Sbjct: 435 LGGMFCTLFGMITAVGLSNLQFVDMNSSRNLFVLGFSMFFGLTLPNYL---DSNP----- 486
Query: 502 VPSYFQPYIVTSHGPFRSKYEELNYVLNMIFSLHMVIAFLVALILDNTVPGSKQERELYG 561
G + E++ +L ++ + M + +A ILDNTVPGS +ER L
Sbjct: 487 -------------GAINTGIPEVDQILTVLLTTEMFVGGCLAFILDNTVPGSPEERGLIQ 533
Query: 562 WSKPNDAR-EDPFIVSEYGLPARVG 585
W A E + Y P +G
Sbjct: 534 WKAGAHANSETSASLKSYDFPFGMG 558
>Y397_CLOPE (P50487) Putative purine permease CPE0397
Length = 452
Score = 115 bits (289), Expect = 2e-25
Identities = 102/418 (24%), Positives = 191/418 (45%), Gaps = 55/418 (13%)
Query: 89 LNYELTDSPALVFLAVYGIQHYLSIIGSLILTPLVIAPAMGASHDETAAMVCTVLLVSGV 148
L Y + D L ++G+QH + G +I+ PLVIA ++G T A++ +L SG+
Sbjct: 13 LIYGVDDDLDLPKKVLFGLQHIFAAFGGIIVVPLVIATSLGFDSKVTTALISASILGSGL 72
Query: 149 TTLLHT----IFGSRLPLIQGPSFVYLAPVLAIINSPEFQELNENKFKHIMKELQGAIII 204
T++ G+R+ I G F +++P +++ + + + GA I+
Sbjct: 73 ATIIQAKGVGKVGARVACIMGTDFTFVSPAISVGSVLG------------LPGIIGATIL 120
Query: 205 GSAFQTLLGYTGLMSLLVRFINPVVVSSTIAAVGLFL*LARV------LRSVQY----RY 254
GS F+ +L + + L++F P+V + +A +GL L + S Y
Sbjct: 121 GSLFEVILSF--FIKPLMKFFPPLVTGTVVALIGLTLLPVSIDWAAGGAGSANYASLENL 178
Query: 255 *CLLFFVLYLRKISVFGHHIFQIYAVPLGLAVTWTFAFLLTENGRMKHCQVNTSDTMTSP 314
+F ++ ++ +G + ++ +G+ V + L G + V +
Sbjct: 179 AVAMFVLVITLLLNNYGKGMISSASILIGIVVGYIVCIPL---GLVDFTPVKEAS----- 230
Query: 315 PWFRFPYPLQWGTPVFNWKMAIVMCVVSLISSVDSVGTY----HTSSLLAASGPPTPGVL 370
W FP L++G F+ K + ++++ +VG TS++ GVL
Sbjct: 231 -WLSFPKILEFGV-TFDAKAVMAFIPAYFVATIGTVGCLKAIGETSNIDIGDKRVAAGVL 288
Query: 371 SRGIGLEGFSSLLAGLWGTGMGSTTLTENVHTIAGTKMGSRRPVQLGACLLIVLSLFGKV 430
S G+G S L GL G+ +T+ ++N+ I+ TK+ SR + LL++L KV
Sbjct: 289 SDGVG-----SALGGLVGS-CPNTSFSQNIGIISLTKVASRHVAVMAGILLVILGFLPKV 342
Query: 431 GGFIASIPEAMVAGLLCIMWAMLTALG---LSNLRYTETGSSRNIIIVGLSLFFSLSI 485
I IP ++ G+ +M+ + A G LSN++ TE RN++I+ +S+ L +
Sbjct: 343 AAIITGIPNPVLGGVGIMMFGTVAAAGIRTLSNIKLTE----RNLLIIAISMGLGLGV 396
>PBUX_BACSU (P42086) Xanthine permease
Length = 438
Score = 109 bits (273), Expect = 2e-23
Identities = 98/411 (23%), Positives = 180/411 (42%), Gaps = 50/411 (12%)
Query: 106 GIQHYLSIIGSLILTPLVIAPAMGASHDETAAMVCTVLLVSGVTTLL----HTIFGSRLP 161
GIQH L++ I+ PL++ AMG + ++ +V + + GV TLL + FG LP
Sbjct: 12 GIQHVLAMYAGAIVVPLIVGKAMGLTVEQLTYLVSIDIFMCGVATLLQVWSNRFFGIGLP 71
Query: 162 LIQGPSFVYLAPVLAIINSPEFQELNENKFKHIMKELQGAIIIGSAFQTLLGYTGLMSLL 221
++ G +F ++P++AI + ++ + + G+II L+ + L
Sbjct: 72 VVLGCTFTAVSPMIAIGS------------EYGVSTVYGSIIASGILVILISF--FFGKL 117
Query: 222 VRFINPVVVSSTIAAVGLFL*LARV--------------LRSVQYRY*CLLFFVLYLRKI 267
V F PVV S + +G+ L + L ++ + L VL R
Sbjct: 118 VSFFPPVVTGSVVTIIGITLMPVAMNNMAGGEGSADFGDLSNLALAFTVLSIIVLLYR-- 175
Query: 268 SVFGHHIFQIYAVPLGLAVTWTFAFLLTENGRMKHCQVNTSDTMTSPPWFRFPYPLQWGT 327
F + ++ +G+ + A+ + G+++ D ++ + P +G
Sbjct: 176 --FTKGFIKSVSILIGILIGTFIAYFM---GKVQF------DNVSDAAVVQMIQPFYFGA 224
Query: 328 PVFNWKMAIVMCVVSLISSVDSVGTYHTSSLLAASGPPTPGVLSRGIGLEGFSSLLAGLW 387
P F+ I M +V+++S V+S G Y L + T LS+G EG + LL G++
Sbjct: 225 PSFHAAPIITMSIVAIVSLVESTGVYFALGDL-TNRRLTEIDLSKGYRAEGLAVLLGGIF 283
Query: 388 GTGMGSTTLTENVHTIAGTKMGSRRPVQLGACLLIVLSLFGKVGGFIASIPEAMVAGLLC 447
T ++NV + T + + + +L+ LF K+ F IP A++ G +
Sbjct: 284 -NAFPYTAFSQNVGLVQLTGIKKNAVIVVTGVILMAFGLFPKIAAFTTIIPSAVLGGAMV 342
Query: 448 IMWAMLTALGLSNLRYTETGSSRNIIIVGLSLFFSLS---IPAYFQQYESS 495
M+ M+ A G+ L + N++IV S+ L +P F+Q S+
Sbjct: 343 AMFGMVIAYGIKMLSRIDFAKQENLLIVACSVGLGLGVTVVPDIFKQLPSA 393
>PUCK_BACSU (O32140) Uric acid permease pucK
Length = 430
Score = 107 bits (266), Expect = 1e-22
Identities = 96/409 (23%), Positives = 176/409 (42%), Gaps = 50/409 (12%)
Query: 102 LAVYGIQHYLSIIGSLILTPLVIAPAMGASHDETAAMVCTVLLVSGVTTLL----HTIFG 157
L + G+QH L++ IL PL++ A+G + + ++ L + G TLL + FG
Sbjct: 10 LMMLGLQHMLAMYAGAILVPLIVGAAIGLNAGQLTYLIAIDLFMCGAATLLQLWRNRYFG 69
Query: 158 SRLPLIQGPSFVYLAPVLAIINSPEFQELNENKFKHIMKELQGAIIIGSAFQTLLGYTGL 217
LP++ G +F + P+++I ++ + + + GAII L G
Sbjct: 70 IGLPVVLGCTFTAVGPMISIGST------------YGVPAIYGAIIAAGLIVVLAA--GF 115
Query: 218 MSLLVRFINPVVVSSTIAAVGLFL*LARV--------------LRSVQYRY*CLLFFVLY 263
LVRF PVV S + +G+ L + L +V + F +L
Sbjct: 116 FGKLVRFFPPVVTGSVVMIIGISLIPTAMNNLAGGEGSKEFGSLDNVLLGFGVTAFILL- 174
Query: 264 LRKISVFGHHIFQIYAVPLGLAVTWTFAFLLTENGRMKHCQVNTSDTMTSPPWFRFPYPL 323
+ F + A+ LGL A+ + + V+ S+ + + W P
Sbjct: 175 ---LFYFFKGFIRSIAILLGLIAGTAAAYFMGK--------VDFSEVLEAS-WLHVPSLF 222
Query: 324 QWGTPVFNWKMAIVMCVVSLISSVDSVGTYHTSSLLAASGPPTPGVLSRGIGLEGFSSLL 383
+G P F + M +V+++S V+S G Y + + + + L +G EG + LL
Sbjct: 223 YFGPPTFELPAVVTMLLVAIVSLVESTGVYFALADIT-NRRLSEKDLEKGYRAEGLAILL 281
Query: 384 AGLWGTGMGSTTLTENVHTIAGTKMGSRRPVQLGACLLIVLSLFGKVGGFIASIPEAMVA 443
GL+ T ++NV + +KM S + + +L+ + L K IP ++
Sbjct: 282 GGLFNA-FPYTAFSQNVGIVQLSKMKSVNVIAITGIILVAIGLVPKAAALTTVIPTPVLG 340
Query: 444 GLLCIMWAMLTALGLSNLRYTETGSSRNIIIVGLSLFFSL---SIPAYF 489
G + +M+ M+ + G+ L + S N++I+ S+ L ++PA F
Sbjct: 341 GAMIVMFGMVISYGIKMLSSVDLDSQGNLLIIASSVSLGLGATTVPALF 389
>PUCJ_BACSU (O32139) Uric acid permease pucJ
Length = 449
Score = 106 bits (265), Expect = 2e-22
Identities = 97/412 (23%), Positives = 181/412 (43%), Gaps = 55/412 (13%)
Query: 107 IQHYLSIIGSLILTPLVIAPAMGASHDETAAMVCTVLLVSGVTTLLHTIFGSR----LPL 162
+QH L++ IL PL++ A+ + ++ + ++ LL GV TLL T+ G+ LP+
Sbjct: 13 LQHVLAMYAGAILVPLLVGRALNVTTEQLSYLLAIDLLTCGVATLLQTLRGTYIGIGLPV 72
Query: 163 IQGPSFVYLAPVLAIINSPEFQELNENKFKHIMKELQGAIIIGSAFQTLLG-YTGLMSLL 221
+ G SFV + P++AI ++ + + + G+II F L + G +++L
Sbjct: 73 MLGSSFVAVTPMIAIGSN------------YGIHAIYGSIIAAGVFIFLFARFFGKLTVL 120
Query: 222 VRFINPVVVSSTIAAVGLFL*LARVLR---------SVQYRY*CL---------LFFVLY 263
PVV + + +GL L V S Y L L +L
Sbjct: 121 ---FPPVVTGTVVTLIGLSLVPTGVKNMAGGEKINGSANPEYGSLENLLLSVGVLVLILV 177
Query: 264 LRKISVFGHHIFQIYAVPLGLAVTWTFAFLLTENGRMKHCQVNTSDTMTSPPWFRFPYPL 323
L + F + +V +G+A A ++ G++ V T P+F+ P P
Sbjct: 178 LNR---FLKGFARTLSVLIGIAAGTAAAAIM---GKVSFSSV------TEAPFFQIPKPF 225
Query: 324 QWGTPVFNWKMAIVMCVVSLISSVDSVGTYHTSSLLAASGPPTPGVLSRGIGLEGFSSLL 383
+G P F + M +V ++ V+S G ++ + P T L +G EG + L+
Sbjct: 226 YFGAPAFEIGPILTMLIVGIVIIVESTGVFYAIGKICGR-PLTDKDLVKGYRAEGIAILI 284
Query: 384 AGLWGTGMGSTTLTENVHTIAGTKMGSRRPVQLGACLLIVLSLFGKVGGFIASIPEAMVA 443
GL+ T +N + TK+ +R V C+L+ L L K+ +++P A++
Sbjct: 285 GGLFNA-FPYNTFAQNAGLLQLTKVKTRNIVVTAGCILVCLGLIPKIAALASAVPAAVLG 343
Query: 444 GLLCIMWAMLTALGLSNLRYTETGSSRNIIIVGLSLFFSL---SIPAYFQQY 492
G +M+ M+ A G+ L + + +++ + S+ + + P F ++
Sbjct: 344 GATVVMFGMVIASGVKMLSTADLKNQYHLLTIACSIALGIGASTAPGIFAEF 395
>YGFU_ECOLI (Q46821) Putative purine permease ygfU
Length = 482
Score = 105 bits (263), Expect = 3e-22
Identities = 94/398 (23%), Positives = 176/398 (43%), Gaps = 47/398 (11%)
Query: 102 LAVYGIQHYLSIIGSLILTPLVIAPAMGASHDETAAMVCTVLLVSGVTTLLHTI-----F 156
L + G+QH L + + PL+I +G S + A ++ + L G+ TLL I
Sbjct: 30 LIILGLQHVLVMYAGAVAVPLMIGDRLGLSKEAIAMLISSDLFCCGIVTLLQCIGIGRFM 89
Query: 157 GSRLPLIQGPSFVYLAPVLAIINSPEFQELNENKFKHIMKELQGAIIIGSAFQTLLGYTG 216
G RLP+I +F + P++AI +P+ L + GA I TLL
Sbjct: 90 GIRLPVIMSVTFAAVTPMIAIGMNPDIGLLG----------IFGATIAAGFITTLL--AP 137
Query: 217 LMSLLVRFINPVVVSSTIAAVGLFL*LARV------LRSVQY--------RY*CLLFFVL 262
L+ L+ P+V I ++GL + + + QY + L+F +L
Sbjct: 138 LIGRLMPLFPPLVTGVVITSIGLSIIQVGIDWAAGGKGNPQYGNPVYLGISFAVLIFILL 197
Query: 263 YLRKISVFGHHIFQIYAVPLGLAVTWTFAFLLTENGRMKHCQVNTSDTMTSPPWFRFPYP 322
R F ++ + + G ++W +VN S + WF P
Sbjct: 198 ITRYAKGFMSNVAVLLGIVFGFLLSWMMN------------EVNLSG-LHDASWFAIVTP 244
Query: 323 LQWGTPVFNWKMAIVMCVVSLISSVDSVGTYHTSSLLAASGPPTPGVLSRGIGLEGFSSL 382
+ +G P+F+ + M V +I ++S+G + + + ++ RG+ ++G ++
Sbjct: 245 MSFGMPIFDPVSILTMTAVLIIVFIESMGMFLALGEIVGRKLSSHDII-RGLRVDGVGTM 303
Query: 383 LAGLWGTGMGSTTLTENVHTIAGTKMGSRRPVQLGACLLIVLSLFGKVGGFIASIPEAMV 442
+ G + + T+ ++NV ++ T++ SR +LI+ + K+ +ASIP+ ++
Sbjct: 304 IGGTFNS-FPHTSFSQNVGLVSVTRVHSRWVCISSGIILILFGMVPKMAVLVASIPQFVL 362
Query: 443 AGLLCIMWAMLTALGLSNL-RYTETGSSRNIIIVGLSL 479
G +M+ M+ A G+ L R T + N+ IV +SL
Sbjct: 363 GGAGLVMFGMVLATGIRILSRCNYTTNRYNLYIVAISL 400
>YGFO_ECOLI (P67444) Putative purine permease ygfO
Length = 485
Score = 100 bits (249), Expect = 1e-20
Identities = 104/423 (24%), Positives = 191/423 (44%), Gaps = 45/423 (10%)
Query: 87 SPLNYELTDSPALVFLAVYGIQHYLSIIGSLILTPLVIAPAMGASHDETAAMVCTVLLVS 146
S L +EL D P V I H L+I ++ L++ A+ S + TA +V ++ S
Sbjct: 28 SDLIFELEDRPPFHQALVGAITHLLAIFVPMVTPALIVGAALQLSAETTAYLVSMAMIAS 87
Query: 147 GVTTLLHT----IFGSRLPLIQGPSFVYLAPVLAIINSPEFQELNENKFKHIMKELQGAI 202
G+ T L I GS L IQ +F ++ ++A+ +S + +E IM L G
Sbjct: 88 GIGTWLQVNRYGIVGSGLLSIQSVNFSFVTVMIALGSSMKSDGFHEEL---IMSSLLGVS 144
Query: 203 IIGSAFQTLLGYTGLMSLLVRFINPVVVSSTIAAVGLFL*LARVL------------RSV 250
+G+ ++G + ++ L R I P V + +GL L ++
Sbjct: 145 FVGAFL--VVGSSFILPYLRRVITPTVSGIVVLMIGLSLIKVGIIDFGGGFAAKSSGTFG 202
Query: 251 QYRY*CLLFFVLYLRKISVFGHH-----IFQIYAVPLGLAVTWTFAFLLTENGRMKHCQV 305
Y + + VL I V G + + ++ + +GL V + + L V
Sbjct: 203 NYEHLGVGLLVL----IVVIGFNCCRSPLLRMGGIAIGLCVGYIASLCLG--------MV 250
Query: 306 NTSDTMTSPPWFRFPYPLQWGTPVFNWKMAIVMCVVSLISSVDSVGTYHTSSLLAA---S 362
+ S +M + P P+P ++G F++ +V+ + L+S +++VG +++++
Sbjct: 251 DFS-SMRNLPLITIPHPFKYGFS-FSFHQFLVVGTIYLLSVLEAVGDITATAMVSRRPIQ 308
Query: 363 GPPTPGVLSRGIGLEGFSSLLAGLWGTGMGSTTLTENVHTIAGTKMGSRRPVQLGACLLI 422
G L G+ +G S++A G+ + TT +N I T + SR + A +L+
Sbjct: 309 GEEYQSRLKGGVLADGLVSVIASAVGS-LPLTTFAQNNGVIQMTGVASRYVGRTIAVMLV 367
Query: 423 VLSLFGKVGGFIASIPEAMVAGLLCIMWAMLTALGLSNLRYTETGSSRNIIIVGLSLFFS 482
+L LF +GGF +IP A++ G + +M++M+ G+ + T R +IV SL
Sbjct: 368 ILGLFPMIGGFFTTIPSAVLGGAMTLMFSMIAIAGI-RIIITNGLKRRETLIVATSLGLG 426
Query: 483 LSI 485
L +
Sbjct: 427 LGV 429
>YGFO_ECOL6 (P67445) Putative purine permease ygfO
Length = 485
Score = 100 bits (249), Expect = 1e-20
Identities = 104/423 (24%), Positives = 191/423 (44%), Gaps = 45/423 (10%)
Query: 87 SPLNYELTDSPALVFLAVYGIQHYLSIIGSLILTPLVIAPAMGASHDETAAMVCTVLLVS 146
S L +EL D P V I H L+I ++ L++ A+ S + TA +V ++ S
Sbjct: 28 SDLIFELEDRPPFHQALVGAITHLLAIFVPMVTPALIVGAALQLSAETTAYLVSMAMIAS 87
Query: 147 GVTTLLHT----IFGSRLPLIQGPSFVYLAPVLAIINSPEFQELNENKFKHIMKELQGAI 202
G+ T L I GS L IQ +F ++ ++A+ +S + +E IM L G
Sbjct: 88 GIGTWLQVNRYGIVGSGLLSIQSVNFSFVTVMIALGSSMKSDGFHEEL---IMSSLLGVS 144
Query: 203 IIGSAFQTLLGYTGLMSLLVRFINPVVVSSTIAAVGLFL*LARVL------------RSV 250
+G+ ++G + ++ L R I P V + +GL L ++
Sbjct: 145 FVGAFL--VVGSSFILPYLRRVITPTVSGIVVLMIGLSLIKVGIIDFGGGFAAKSSGTFG 202
Query: 251 QYRY*CLLFFVLYLRKISVFGHH-----IFQIYAVPLGLAVTWTFAFLLTENGRMKHCQV 305
Y + + VL I V G + + ++ + +GL V + + L V
Sbjct: 203 NYEHLGVGLLVL----IVVIGFNCCRSPLLRMGGIAIGLCVGYIASLCLG--------MV 250
Query: 306 NTSDTMTSPPWFRFPYPLQWGTPVFNWKMAIVMCVVSLISSVDSVGTYHTSSLLAA---S 362
+ S +M + P P+P ++G F++ +V+ + L+S +++VG +++++
Sbjct: 251 DFS-SMRNLPLITIPHPFKYGFS-FSFHQFLVVGTIYLLSVLEAVGDITATAMVSRRPIQ 308
Query: 363 GPPTPGVLSRGIGLEGFSSLLAGLWGTGMGSTTLTENVHTIAGTKMGSRRPVQLGACLLI 422
G L G+ +G S++A G+ + TT +N I T + SR + A +L+
Sbjct: 309 GEEYQSRLKGGVLADGLVSVIASAVGS-LPLTTFAQNNGVIQMTGVASRYVGRTIAVMLV 367
Query: 423 VLSLFGKVGGFIASIPEAMVAGLLCIMWAMLTALGLSNLRYTETGSSRNIIIVGLSLFFS 482
+L LF +GGF +IP A++ G + +M++M+ G+ + T R +IV SL
Sbjct: 368 ILGLFPMIGGFFTTIPSAVLGGAMTLMFSMIAIAGI-RIIITNGLKRRETLIVATSLGLG 426
Query: 483 LSI 485
L +
Sbjct: 427 LGV 429
>YGFO_ECO57 (P67446) Putative purine permease ygfO
Length = 485
Score = 100 bits (249), Expect = 1e-20
Identities = 104/423 (24%), Positives = 191/423 (44%), Gaps = 45/423 (10%)
Query: 87 SPLNYELTDSPALVFLAVYGIQHYLSIIGSLILTPLVIAPAMGASHDETAAMVCTVLLVS 146
S L +EL D P V I H L+I ++ L++ A+ S + TA +V ++ S
Sbjct: 28 SDLIFELEDRPPFHQALVGAITHLLAIFVPMVTPALIVGAALQLSAETTAYLVSMAMIAS 87
Query: 147 GVTTLLHT----IFGSRLPLIQGPSFVYLAPVLAIINSPEFQELNENKFKHIMKELQGAI 202
G+ T L I GS L IQ +F ++ ++A+ +S + +E IM L G
Sbjct: 88 GIGTWLQVNRYGIVGSGLLSIQSVNFSFVTVMIALGSSMKSDGFHEEL---IMSSLLGVS 144
Query: 203 IIGSAFQTLLGYTGLMSLLVRFINPVVVSSTIAAVGLFL*LARVL------------RSV 250
+G+ ++G + ++ L R I P V + +GL L ++
Sbjct: 145 FVGAFL--VVGSSFILPYLRRVITPTVSGIVVLMIGLSLIKVGIIDFGGGFAAKSSGTFG 202
Query: 251 QYRY*CLLFFVLYLRKISVFGHH-----IFQIYAVPLGLAVTWTFAFLLTENGRMKHCQV 305
Y + + VL I V G + + ++ + +GL V + + L V
Sbjct: 203 NYEHLGVGLLVL----IVVIGFNCCRSPLLRMGGIAIGLCVGYIASLCLG--------MV 250
Query: 306 NTSDTMTSPPWFRFPYPLQWGTPVFNWKMAIVMCVVSLISSVDSVGTYHTSSLLAA---S 362
+ S +M + P P+P ++G F++ +V+ + L+S +++VG +++++
Sbjct: 251 DFS-SMRNLPLITIPHPFKYGFS-FSFHQFLVVGTIYLLSVLEAVGDITATAMVSRRPIQ 308
Query: 363 GPPTPGVLSRGIGLEGFSSLLAGLWGTGMGSTTLTENVHTIAGTKMGSRRPVQLGACLLI 422
G L G+ +G S++A G+ + TT +N I T + SR + A +L+
Sbjct: 309 GEEYQSRLKGGVLADGLVSVIASAVGS-LPLTTFAQNNGVIQMTGVASRYVGRTIAVMLV 367
Query: 423 VLSLFGKVGGFIASIPEAMVAGLLCIMWAMLTALGLSNLRYTETGSSRNIIIVGLSLFFS 482
+L LF +GGF +IP A++ G + +M++M+ G+ + T R +IV SL
Sbjct: 368 ILGLFPMIGGFFTTIPSAVLGGAMTLMFSMIAIAGI-RIIITNGLKRRETLIVATSLGLG 426
Query: 483 LSI 485
L +
Sbjct: 427 LGV 429
>YICE_ECOLI (P27432) Putative purine permease yicE
Length = 463
Score = 96.3 bits (238), Expect = 2e-19
Identities = 105/430 (24%), Positives = 185/430 (42%), Gaps = 46/430 (10%)
Query: 81 VLRRRPSPLNYELTDSPALVFLAVYGIQHYLSIIGSLILTPLVIAPAMGASHDETAAMVC 140
V + + S L Y L D P L QH L++ ++I L+I A+G +T ++
Sbjct: 14 VAQTQNSELIYRLEDRPPLPQTLFAACQHLLAMFVAVITPALLICQALGLPAQDTQHIIS 73
Query: 141 TVLLVSGVTTLLHTI----FGSRLPLIQGPSFVYLAPVLAIINSPEFQELNENKFKHIMK 196
L SGV +++ GS L IQG SF ++AP +I + +M
Sbjct: 74 MSLFASGVASIIQIKAWGPVGSGLLSIQGTSFNFVAP---LIMGGTALKTGGADVPTMMA 130
Query: 197 ELQGAIIIGSAFQTLLGYTGLMSLLVRFINPVVVSSTIAAVGLFL*LARVLRSVQYRY*C 256
L G +++ S + ++ + ++ L R I P+V + +GL L + L S+ Y
Sbjct: 131 ALFGTLMLASCTEMVI--SRVLHLARRIITPLVSGVVVMIIGLSL-IQVGLTSIGGGYAA 187
Query: 257 ------------------LLFFVLYLRKISVFGHHIFQIYAVPLGLAVTWTFAFLLTENG 298
L +L R+ + + + A+ G A+ W L N
Sbjct: 188 MSDNTFGAPKNLLLAGVVLALIILLNRQRNPYLRVASLVIAMAAGYALAWFMGMLPESNE 247
Query: 299 RMKHCQVNTSDTMTSPPWFRFPYPLQWGTPVFNWKMAIVMCVVSLISSVDSVGTYHTSSL 358
M + P PL +G + W + + + +V +I+S++++G +S
Sbjct: 248 PM------------TQELIMVPTPLYYGLGI-EWSLLLPLMLVFMITSLETIGDITATSD 294
Query: 359 LA---ASGPPTPGVLSRGIGLEGFSSLLAGLWGTGMGSTTLTENVHTIAGTKMGSRRPVQ 415
++ SGP L G+ G +S ++ ++ T ++ +N I T + SR
Sbjct: 295 VSEQPVSGPLYMKRLKGGVLANGLNSFVSAVFNT-FPNSCFGQNNGVIQLTGVASRYVGF 353
Query: 416 LGACLLIVLSLFGKVGGFIASIPEAMVAGLLCIMWAMLTALGLSNLRYTETGSSRNIIIV 475
+ A +LIVL LF V GF+ IPE ++ G +M+ + A G+ + E + R I+I+
Sbjct: 354 VVALMLIVLGLFPAVSGFVQHIPEPVLGGATLVMFGTIAASGV-RIVSREPLNRRAILII 412
Query: 476 GLSLFFSLSI 485
LSL L +
Sbjct: 413 ALSLAVGLGV 422
>YCDG_ECOLI (P75892) Putative purine permease ycdG
Length = 442
Score = 87.4 bits (215), Expect = 9e-17
Identities = 95/386 (24%), Positives = 177/386 (45%), Gaps = 35/386 (9%)
Query: 103 AVYGIQHYLSIIGSLILTPLVIAPAMGASHDETAAMVCTVLLVSGVTTLLHT-IFGSRLP 161
AV G+QH +++ G+ +L P+++ S +L+SG+ TLL I G R+P
Sbjct: 33 AVMGVQHAVAMFGATVLMPILMGLDPNLS-----------ILMSGIGTLLFFFITGGRVP 81
Query: 162 LIQGPSFVYLAPVLAIINSPEFQELNENKFKHIMKELQGAIIIGSAFQTLLGYTGLMSLL 221
G S ++ V+A Q +N N I L G I G + T++G +M +
Sbjct: 82 SYLGSSAAFVGVVIAATGF-NGQGINPN----ISIALGGIIACGLVY-TVIGLV-VMKIG 134
Query: 222 VRFIN----PVVVSSTIAAVGLFL*LARV--LRSVQYRY*CLLFFVLYLRKISVFGHHIF 275
R+I PVV + + A+GL L V + + + + VL + ++VF +
Sbjct: 135 TRWIERLMPPVVTGAVVMAIGLNLAPIAVKSVSASAFDSWMAVMTVLCIGLVAVFTRGMI 194
Query: 276 QIYAVPLGLAVTWTFAFLLTENGRMKHCQVNTSDTMTSPPWFRFPYPLQWGTPVFNWKMA 335
Q + +GL V ++T + T ++ WF P+ + TP FN +
Sbjct: 195 QRLLILVGLIVACLLYGVMTNVLGLGKAVDFT--LVSHAAWFGLPH---FSTPAFNGQAM 249
Query: 336 IVMCVVSLISSVDSVGTYHTSSLLAASGPPTPGVLSRGIGLEGFSSLLAGLWGTGMGSTT 395
+++ V++I +++G H ++ +G + R +G +++L+G G G G TT
Sbjct: 250 MLIAPVAVILVAENLG--HLKAVAGMTGRNMDPYMGRAFVGDGLATMLSGSVG-GSGVTT 306
Query: 396 LTENVHTIAGTKMGSRRPVQLGACLLIVLSLFGKVGGFIASIPEAMVAGLLCIMWAMLTA 455
EN+ +A TK+ S A + ++L K G I +IP A++ G +++ ++
Sbjct: 307 YAENIGVMAVTKVYSTLVFVAAAVIAMLLGFSPKFGALIHTIPAAVIGGASIVVFGLIAV 366
Query: 456 LG--LSNLRYTETGSSRNIIIVGLSL 479
G + + + N+I+V ++L
Sbjct: 367 AGARIWVQNRVDLSQNGNLIMVAVTL 392
>URAA_HAEIN (P45117) Probable uracil permease (Uracil transporter)
Length = 414
Score = 80.5 bits (197), Expect = 1e-14
Identities = 93/390 (23%), Positives = 163/390 (40%), Gaps = 46/390 (11%)
Query: 106 GIQHYLSIIGSLILTPLVIAPAMGASHDETAAMVCTVLLVSGVTTLLHTIF-GSRLPLIQ 164
G+Q G+L+L PL+ T T LL +GV TLL G ++P+
Sbjct: 22 GLQMLFVAFGALVLVPLI-----------TGLDSNTALLTAGVGTLLFQFCTGKQVPIFL 70
Query: 165 GPSFVYLAPVLAIINSPEFQELNENKFKHIMKELQGAIIIGSAFQTLLGYTGLMSLLVRF 224
SF ++AP+ + + M L ++ A TL+ G +L RF
Sbjct: 71 ASSFAFIAPIQYGVQTWGIATT--------MGGLAFTGLVYFALSTLVKLRGAEALQ-RF 121
Query: 225 INPVVVSSTIAAVGLFL*LARVLRSV------QYRY*CLLFFVLYLRKISV--FGHHIFQ 276
PVVV I +G+ L V S+ Y L+ V L +SV F + +
Sbjct: 122 FPPVVVGPVIIIIGMGLAPIAVDMSLGKNSAYAYNDAVLVSMVTLLTTLSVAVFAKGLMK 181
Query: 277 IYAVPLGLAVTWTFAFLLTENGRMKHCQVNTSDTMTSPPWFRFPYPLQWGTPVFNWKMAI 336
+ + G+ + L + + PWF P + TP FN + +
Sbjct: 182 LIPIMFGITAGYILCLFLG---------LINFQPVIDAPWFSLP---KLTTPEFNLEAIL 229
Query: 337 VMCVVSLISSVDSVGTYHT-SSLLAASGPPTPGVLSRGIGLEGFSSLLAGLWGTGMGSTT 395
M +++ +V+ VG SS+ PG+ +G +G ++ A L G G +TT
Sbjct: 230 YMLPIAIAPAVEHVGGIMAISSVTGKDFLKKPGLHRTLLG-DGIATAAASLVG-GPPNTT 287
Query: 396 LTENVHTIAGTKMGSRRPVQLGACLLIVLSLFGKVGGFIASIPEAMVAGLLCIMWAMLTA 455
E + T+ + + A I +S GKVG F+++IP ++ G++ +++ +
Sbjct: 288 YAEVTGAVMLTRNFNPNIMTWAAVWAIAISFCGKVGAFLSTIPTIVMGGIMMLVFGSIAV 347
Query: 456 LGLSNL--RYTETGSSRNIIIVGLSLFFSL 483
+G+S L + +RN+ I+ + + F +
Sbjct: 348 VGMSTLIRGKVDVTEARNLCIISVVMTFGI 377
>YWDJ_BACSU (P39618) Putative purine permease ywdJ
Length = 460
Score = 77.8 bits (190), Expect = 7e-14
Identities = 88/400 (22%), Positives = 171/400 (42%), Gaps = 32/400 (8%)
Query: 102 LAVYGIQHYLSIIGSLILTPLVIAPAMGASHDETAAMVCTVLLVSGVTTLLHTIFGSRLP 161
L + +Q II + I+ P+ +A + H ++A ++ + V G+ ++ + G RLP
Sbjct: 3 LVLGALQWTAFIIAAAIVVPVAVAQSFHLDHSDSARLIQSTFFVLGIAAVIQCLKGHRLP 62
Query: 162 LIQGPSFVY--LAPVLAIINSPEFQELNENKFKHIMKELQGAIIIGSAFQTLLGYTGLMS 219
+ + P+ ++ + + A + F + ++ LQGA+++ + LL ++
Sbjct: 63 INESPAGLWWGVYTIYAGLTGTVFATYGDT-----LRGLQGALLVSAVCFFLLSVFKVID 117
Query: 220 LLVRFINPVV--VSSTIAAVGLFL*LARVLRSVQYRY*CLLFFVLYLRKISVFGHHI--- 274
L + PVV V + + L + + + + YR + V L + + I
Sbjct: 118 RLAKLFTPVVTGVYLLLLVMQLSQPIIKGILGIGYRQDGVDGLVFGLALVVIAAAFIMTN 177
Query: 275 -----FQIYAVPLGLAVTWTFAFLLTENGRMKHCQVNTSDTMTSPPWFRFPYPLQWGTPV 329
F+ Y++ L L W L G K + D + F+ P +GTP+
Sbjct: 178 SNIMFFKQYSILLALFGGWV---LFAAAGAAK--PIEMPDRL-----FQLPSLFPFGTPL 227
Query: 330 FNWKMAIVMCVVSLISSVDSVGTYHTSSLLAA--SGPPTPGVLSRGIGLEG-FSSLLAGL 386
FN + I ++++ V+ + + + S P R G FS LL+GL
Sbjct: 228 FNSGLIITSIFITILLIVNMLASMKVVDIAMKKFSKQPDGKHHERHAGFAASFSHLLSGL 287
Query: 387 WGTGMGSTTLTENVHTIAGTKMGSRRPVQLGACLLIVLSLFGKVGGFIASIPEAMVAGLL 446
G + ++ I TKM S++P LG+ L+IV+S+ AS+P + +
Sbjct: 288 TGA-IAPVPISGAAGFIETTKMPSKKPFMLGSILVIVISVIPFFMNTFASLPSPVGFAVN 346
Query: 447 CIMWAMLTALGLSNL-RYTETGSSRNIIIVGLSLFFSLSI 485
++++ + L + Y + S R I+G+SL + I
Sbjct: 347 FVVFSAMGGLAFAEFDSYEKEESKRVRSIIGISLLTGVGI 386
>PYRP_BACCL (P41006) Uracil permease (Uracil transporter)
Length = 432
Score = 77.0 bits (188), Expect = 1e-13
Identities = 94/411 (22%), Positives = 172/411 (40%), Gaps = 54/411 (13%)
Query: 92 ELTDSPALVFLAVYGIQHYLSIIGSLILTPLVIAPAMGASHDETAAMVCTVLLVSGVTTL 151
++ D P + +QH ++ G+ IL P ++ D + A LL SG+ TL
Sbjct: 7 DIQDRPTVGQWITLSLQHLFAMFGATILVPYLVGL------DPSIA-----LLTSGLGTL 55
Query: 152 LHTIFGS-RLPLIQGPSFVYLAPVLAIINSPEFQELNENKFKHIMKELQGAIIIGSAFQT 210
+ ++P G SF Y+AP++A + F + A+II A
Sbjct: 56 AFLLITKWQVPAYLGSSFAYIAPIIAAKTAGGPGAAMIGSFLAGLVYGVVALIIKKA--- 112
Query: 211 LLGYTGLMSLLVRFINPVVVSSTIAAVGLFL*---LARVLRSVQYRY*CLLFFVLYLRKI 267
GY +M LL PVVV I +GL L + + +Y L F V +
Sbjct: 113 --GYRWVMKLLP----PVVVGPVIIVIGLGLAGTAVGMAMNGPDGKYSLLHFSVALVTLA 166
Query: 268 -----SVFGHHIFQIYAVPLGLAVTWTFAFLLTENGRMKHCQVNTSDTMTSPPWFRFP-- 320
SV + + V +G+ V + +A + K + + WF +P
Sbjct: 167 ATIVCSVLARGMLSLIPVLVGIVVGYLYALAVGLVDLSK---------VAAAKWFEWPDF 217
Query: 321 ------YPLQWGTPVFNWKMAIVMCVVSLISSVDSVGTYHTSSLLAASGPPTPGVLSRGI 374
YP++ W++ ++M V++++ + +G S + L R I
Sbjct: 218 LIPFADYPVR-----VTWEIVMLMVPVAIVTLSEHIGHQLVLSKVVGRDLIQKPGLHRSI 272
Query: 375 GLEGFSSLLAGLWGTGMGSTTLTENVHTIAGTKMGSRRPVQLGACLLIVLSLFGKVGGFI 434
+G +++++ L G G TT EN+ +A T++ S + A + I GK+ I
Sbjct: 273 LGDGTATMISALLG-GPPKTTYGENIGVLAITRVYSVYVLAGAAVIAIAFGFVGKITALI 331
Query: 435 ASIPEAMVAGLLCIMWAMLTALGLSNL--RYTETGSSRNIIIVGLSLFFSL 483
+SIP ++ G+ +++ ++ + GL L + G +RN++I + L +
Sbjct: 332 SSIPTPVMGGVSILLFGIIASSGLRMLIDSRVDFGQTRNLVIASVILVIGI 382
>URAA_PASMU (Q9CPL9) Probable uracil permease (Uracil transporter)
Length = 417
Score = 76.3 bits (186), Expect = 2e-13
Identities = 88/396 (22%), Positives = 166/396 (41%), Gaps = 52/396 (13%)
Query: 103 AVYGIQHYLSIIGSLILTPLVIAPAMGASHDETAAMVCTVLLVSGVTTLLHTIF-GSRLP 161
A G+Q G+L+L PL+ T T LL +G+ TLL + G ++P
Sbjct: 18 AFVGLQMLFVAFGALVLVPLI-----------TGLNANTALLTAGIGTLLFQLCTGRQVP 66
Query: 162 LIQGPSFVYLAPVLAIINSPEFQELNENKFKHIMKELQGAIIIGS---AFQTLLGYTGLM 218
+ SF ++AP+ + + I + G + G A TL+ G
Sbjct: 67 IFLASSFAFIAPIQYGVTT-----------WGIATTMGGLVFTGLVYFALSTLVKIKGA- 114
Query: 219 SLLVRFINPVVVSSTIAAVGLFL*LARVLRSV----QYRY*CLLFF----VLYLRKISVF 270
L + PVVV I +G+ L V ++ Y+Y +F +L ++VF
Sbjct: 115 GALQKVFPPVVVGPVIIIIGMGLAPVAVDMALGKNSTYQYNDAVFVSMATLLTTLGVAVF 174
Query: 271 GHHIFQIYAVPLGLAVTWTFAFLLTENGRMKHCQVNTSDTMTSPPWFRFPYPLQWGTPVF 330
+ ++ + G+ V + L + + PWF P + TP F
Sbjct: 175 AKGMMKLIPIMFGIVVGYILCLFLG---------LINFQPVIDAPWFSVP---EITTPEF 222
Query: 331 NWKMAIVMCVVSLISSVDSVGTYHT-SSLLAASGPPTPGVLSRGIGLEGFSSLLAGLWGT 389
+ + + +++ +V+ VG SS+ PG+ +G +G ++ A G
Sbjct: 223 KLEAILYLLPIAIAPAVEHVGGIMAISSVTGKDFLQKPGLHRTLLG-DGIATSAASFLG- 280
Query: 390 GMGSTTLTENVHTIAGTKMGSRRPVQLGACLLIVLSLFGKVGGFIASIPEAMVAGLLCIM 449
G +TT E + T+ + + + A I +S GKVG F+++IP ++ G++ ++
Sbjct: 281 GPPNTTYAEVTGAVMLTRNFNPKIMTWAAVWAIAISFCGKVGAFLSTIPTIVMGGIMMLV 340
Query: 450 WAMLTALGLSNL--RYTETGSSRNIIIVGLSLFFSL 483
+ + +G+S L + +RN+ I+ + + F +
Sbjct: 341 FGSIAVVGMSTLIRGKVDVTEARNLCIISVVMTFGI 376
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.325 0.140 0.428
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 70,001,381
Number of Sequences: 164201
Number of extensions: 3046277
Number of successful extensions: 10118
Number of sequences better than 10.0: 44
Number of HSP's better than 10.0 without gapping: 25
Number of HSP's successfully gapped in prelim test: 19
Number of HSP's that attempted gapping in prelim test: 10020
Number of HSP's gapped (non-prelim): 61
length of query: 595
length of database: 59,974,054
effective HSP length: 116
effective length of query: 479
effective length of database: 40,926,738
effective search space: 19603907502
effective search space used: 19603907502
T: 11
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.6 bits)
S2: 69 (31.2 bits)
Medicago: description of AC148969.3


