

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148819.5 - phase: 0
(83 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
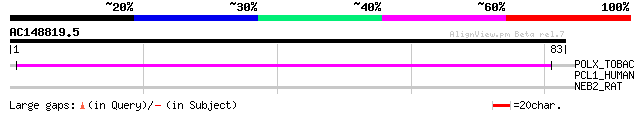
POLX_TOBAC (P10978) Retrovirus-related Pol polyprotein from tran... 46 2e-05
PCL1_HUMAN (Q9UHG3) Prenylcysteine oxidase precursor (EC 1.8.3.5) 29 1.9
NEB2_RAT (O35274) Neurabin-II (Neural tissue-specific F-actin bi... 28 3.2
>POLX_TOBAC (P10978) Retrovirus-related Pol polyprotein from
transposon TNT 1-94 [Contains: Protease (EC 3.4.23.-);
Reverse transcriptase (EC 2.7.7.49); Endonuclease]
Length = 1328
Score = 45.8 bits (107), Expect = 2e-05
Identities = 25/80 (31%), Positives = 40/80 (49%)
Query: 2 GTKWDIEKFIGDSDFGLWKVKIEVVLIQQKCAKARKGKILFSDTMSQEDKTKMADNARSF 61
G K+++ KF GD+ F W+ ++ +LIQQ K DTM ED + + A S
Sbjct: 3 GVKYEVAKFNGDNGFSTWQRRMRDLLIQQGLHKVLDVDSKKPDTMKAEDWADLDERAASA 62
Query: 62 FVLCLRDKVLREVVKDTTIK 81
L L D V+ ++ + T +
Sbjct: 63 IRLHLSDDVVNNIIDEDTAR 82
>PCL1_HUMAN (Q9UHG3) Prenylcysteine oxidase precursor (EC 1.8.3.5)
Length = 505
Score = 29.3 bits (64), Expect = 1.9
Identities = 19/57 (33%), Positives = 26/57 (45%), Gaps = 5/57 (8%)
Query: 2 GTKWDIEKFIGD-----SDFGLWKVKIEVVLIQQKCAKARKGKILFSDTMSQEDKTK 53
G DI F+G SD GLW V+ L+ +A K ++ M E+KTK
Sbjct: 226 GQSTDINAFVGAVSLSCSDSGLWAVEGGNKLVCSGLLQASKSNLISGSVMYIEEKTK 282
>NEB2_RAT (O35274) Neurabin-II (Neural tissue-specific F-actin
binding protein II) (Protein phosphatase 1 regulatory
subunit 9B) (Spinophilin) (p130) (PP1bp134)
Length = 817
Score = 28.5 bits (62), Expect = 3.2
Identities = 17/55 (30%), Positives = 28/55 (50%)
Query: 14 SDFGLWKVKIEVVLIQQKCAKARKGKILFSDTMSQEDKTKMADNARSFFVLCLRD 68
+D GL K+ I V + + A R G+I +D + + D T + +SF LR+
Sbjct: 517 ADMGLEKLGIFVKTVTEGGAAHRDGRIQVNDLLVEVDGTSLVGVTQSFAASVLRN 571
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.323 0.137 0.400
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 8,552,201
Number of Sequences: 164201
Number of extensions: 251873
Number of successful extensions: 747
Number of sequences better than 10.0: 3
Number of HSP's better than 10.0 without gapping: 2
Number of HSP's successfully gapped in prelim test: 1
Number of HSP's that attempted gapping in prelim test: 745
Number of HSP's gapped (non-prelim): 3
length of query: 83
length of database: 59,974,054
effective HSP length: 59
effective length of query: 24
effective length of database: 50,286,195
effective search space: 1206868680
effective search space used: 1206868680
T: 11
A: 40
X1: 16 ( 7.5 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.5 bits)
S2: 58 (26.9 bits)
Medicago: description of AC148819.5


