

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148775.4 - phase: 0
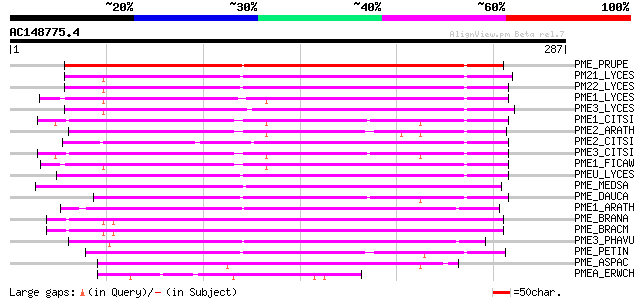
(287 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
PME_PRUPE (Q43062) Pectinesterase PPE8B precursor (EC 3.1.1.11) ... 212 7e-55
PM21_LYCES (P09607) Pectinesterase 2 precursor (EC 3.1.1.11) (Pe... 174 2e-43
PM22_LYCES (Q96575) Pectinesterase 2 precursor (EC 3.1.1.11) (Pe... 170 3e-42
PME1_LYCES (P14280) Pectinesterase 1 precursor (EC 3.1.1.11) (Pe... 166 5e-41
PME3_LYCES (Q96576) Pectinesterase 3 precursor (EC 3.1.1.11) (Pe... 166 8e-41
PME1_CITSI (O04886) Pectinesterase 1 precursor (EC 3.1.1.11) (Pe... 164 2e-40
PME2_ARATH (Q42534) Pectinesterase 2 precursor (EC 3.1.1.11) (Pe... 162 9e-40
PME2_CITSI (O04887) Pectinesterase 2 precursor (EC 3.1.1.11) (Pe... 160 3e-39
PME3_CITSI (P83948) Pectinesterase 3 precursor (EC 3.1.1.11) (Pe... 159 7e-39
PME1_FICAW (P83947) Pectinesterase precursor (EC 3.1.1.11) (Pect... 155 8e-38
PMEU_LYCES (Q43143) Pectinesterase U1 precursor (EC 3.1.1.11) (P... 151 2e-36
PME_MEDSA (Q42920) Pectinesterase precursor (EC 3.1.1.11) (Pecti... 146 5e-35
PME_DAUCA (P83218) Pectinesterase (EC 3.1.1.11) (Pectin methyles... 137 3e-32
PME1_ARATH (Q43867) Pectinesterase 1 precursor (EC 3.1.1.11) (Pe... 137 3e-32
PME_BRANA (P41510) Probable pectinesterase precursor (EC 3.1.1.1... 133 6e-31
PME_BRACM (Q42608) Pectinesterase (EC 3.1.1.11) (Pectin methyles... 128 1e-29
PME3_PHAVU (Q43111) Pectinesterase 3 precursor (EC 3.1.1.11) (Pe... 127 3e-29
PME_PETIN (Q43043) Pectinesterase precursor (EC 3.1.1.11) (Pecti... 124 2e-28
PME_ASPAC (Q12535) Pectinesterase precursor (EC 3.1.1.11) (Pecti... 63 1e-09
PMEA_ERWCH (P07863) Pectinesterase A precursor (EC 3.1.1.11) (Pe... 56 1e-07
>PME_PRUPE (Q43062) Pectinesterase PPE8B precursor (EC 3.1.1.11)
(Pectin methylesterase) (PE)
Length = 522
Score = 212 bits (540), Expect = 7e-55
Identities = 117/228 (51%), Positives = 147/228 (64%), Gaps = 3/228 (1%)
Query: 29 GKFPSWVKPGDRKLLQASAVPADAVVASDGSGNYMKIMDAVMAAPNGSKKRYVIHIKKGV 88
G+ PSWVK DRKLLQA V DA+VA DG+GN+ + DAV+AAP+ S +RYVI+IK+G
Sbjct: 186 GQIPSWVKTKDRKLLQADGVSVDAIVAQDGTGNFTNVTDAVLAAPDYSMRRYVIYIKRGT 245
Query: 89 YNEHVMINNSKSNLMMIGDGMGATVITGDLSWGRDKLDTSYTYTFGVEGLGFSAQDISFR 148
Y E+V I K NLMMIGDGM AT+I+G+ S+ D T + TF V G GF A+DI+F
Sbjct: 246 YKENVEIKKKKWNLMMIGDGMDATIISGNRSF-VDGWTTFRSATFAVSGRGFIARDITFE 304
Query: 149 NTAWPENHQAVALLSDSDTSVFYRCEISGFQDSLCANIKHHSIRIAKSEARLTSYLVRQL 208
NTA PE HQAVAL SDSD SVFYRC I G+QD+L + R K +
Sbjct: 305 NTAGPEKHQAVALRSDSDLSVFYRCNIRGYQDTLYTHTMRQFYRDCKISGTVDFIFGDAT 364
Query: 209 SSFKT-DILVRKGPTGQQNTITAQGGPEKPNLPFGFAFQFCNVCADPE 255
F+ IL +KG Q+N+ITAQG + PN P G + QFCN+ AD +
Sbjct: 365 VVFQNCQILAKKGLPNQKNSITAQGRKD-PNEPTGISIQFCNITADSD 411
>PM21_LYCES (P09607) Pectinesterase 2 precursor (EC 3.1.1.11)
(Pectin methylesterase 2) (PE 2)
Length = 550
Score = 174 bits (441), Expect = 2e-43
Identities = 102/235 (43%), Positives = 139/235 (58%), Gaps = 5/235 (2%)
Query: 29 GKFPSWVKPGDRKLLQASA--VPADAVVASDGSGNYMKIMDAVMAAPNGSKKRYVIHIKK 86
GK PSWV DRKL+++S + A+AVVA DG+G Y + +AV AAP+ SK RYVI++K+
Sbjct: 213 GKMPSWVSSRDRKLMESSGKDIGANAVVAKDGTGKYRTLAEAVAAAPDKSKTRYVIYVKR 272
Query: 87 GVYNEHVMINNSKSNLMMIGDGMGATVITGDLSWGRDKLDTSYTYTFGVEGLGFSAQDIS 146
G Y E+V +++ K NLM+IGDGM AT+ITG L+ D T ++ T G GF QDI
Sbjct: 273 GTYKENVEVSSRKMNLMIIGDGMYATIITGSLN-VVDGSTTFHSATLAAVGKGFILQDIC 331
Query: 147 FRNTAWPENHQAVALLSDSDTSVFYRCEISGFQDSLCANIKHHSIRIAKSEARLTSYLVR 206
+NTA P HQAVAL +D SV RC I +QD+L A+ + R + +
Sbjct: 332 IQNTAGPAKHQAVALRVGADKSVINRCRIDAYQDTLYAHSQRQFYRDSYVTGTIDFIFGN 391
Query: 207 QLSSF-KTDILVRKGPTGQQNTITAQGGPEKPNLPFGFAFQFCNVCADPEFLPFV 260
F K ++ RK QQN +TAQG + PN G + QFC++ A P+ P V
Sbjct: 392 AAVVFQKCQLVARKPGKYQQNMVTAQGRTD-PNQATGTSIQFCDIIASPDLKPVV 445
>PM22_LYCES (Q96575) Pectinesterase 2 precursor (EC 3.1.1.11)
(Pectin methylesterase 2) (PE 2)
Length = 550
Score = 170 bits (431), Expect = 3e-42
Identities = 99/233 (42%), Positives = 138/233 (58%), Gaps = 5/233 (2%)
Query: 29 GKFPSWVKPGDRKLLQASA--VPADAVVASDGSGNYMKIMDAVMAAPNGSKKRYVIHIKK 86
GK PSWV DRKL+++S + A+AVVA DG+G Y + +AV AAP+ SK RYVI++K+
Sbjct: 213 GKMPSWVSSRDRKLMESSGKDIGANAVVAKDGTGKYRTLAEAVAAAPDKSKTRYVIYVKR 272
Query: 87 GVYNEHVMINNSKSNLMMIGDGMGATVITGDLSWGRDKLDTSYTYTFGVEGLGFSAQDIS 146
G+Y E+V +++ K LM++GDGM AT+ITG+L+ D T ++ T G GF QDI
Sbjct: 273 GIYKENVEVSSRKMKLMIVGDGMHATIITGNLN-VVDGSTTFHSATLAAVGKGFILQDIC 331
Query: 147 FRNTAWPENHQAVALLSDSDTSVFYRCEISGFQDSLCANIKHHSIRIAKSEARLTSYLVR 206
+NTA P HQAVAL +D SV RC I +QD+L A+ + R + +
Sbjct: 332 IQNTAGPAKHQAVALRVGADKSVINRCRIDAYQDTLYAHSQRQFYRDSYVTGTIDFIFGN 391
Query: 207 QLSSF-KTDILVRKGPTGQQNTITAQGGPEKPNLPFGFAFQFCNVCADPEFLP 258
F K ++ RK QQN +TAQG + PN G + QFCN+ A + P
Sbjct: 392 AAVVFQKCKLVARKPGKYQQNMVTAQGRTD-PNQATGTSIQFCNIIASSDLEP 443
>PME1_LYCES (P14280) Pectinesterase 1 precursor (EC 3.1.1.11)
(Pectin methylesterase 1) (PE 1)
Length = 546
Score = 166 bits (421), Expect = 5e-41
Identities = 103/249 (41%), Positives = 142/249 (56%), Gaps = 13/249 (5%)
Query: 16 TSSSDILDTITQKGKFPSWVKPGDRKLLQASA--VPADAVVASDGSGNYMKIMDAVMAAP 73
T D+ T+ GK PSWV DRKL+++S + A+AVVA DG+G+Y + +AV AAP
Sbjct: 198 TQDEDVFMTVL--GKMPSWVSSMDRKLMESSGKDIIANAVVAQDGTGDYQTLAEAVAAAP 255
Query: 74 NGSKKRYVIHIKKGVYNEHVMINNSKSNLMMIGDGMGATVITGDLSWGRDKLDTSYTY-- 131
+ SK RYVI++K+G Y E+V + ++K NLM++GDGM AT ITG L + +D S T+
Sbjct: 256 DKSKTRYVIYVKRGTYKENVEVASNKMNLMIVGDGMYATTITGSL----NVVDGSTTFRS 311
Query: 132 -TFGVEGLGFSAQDISFRNTAWPENHQAVALLSDSDTSVFYRCEISGFQDSLCANIKHHS 190
T G GF QDI +NTA P QAVAL +D SV RC I +QD+L A+ +
Sbjct: 312 ATLAAVGQGFILQDICIQNTAGPAKDQAVALRVGADMSVINRCRIDAYQDTLYAHSQRQF 371
Query: 191 IRIAKSEARLTSYLVRQLSSF-KTDILVRKGPTGQQNTITAQGGPEKPNLPFGFAFQFCN 249
R + + F K ++ RK QQN +TAQG + PN G + QFCN
Sbjct: 372 YRDSYVTGTVDFIFGNAAVVFQKCQLVARKPGKYQQNMVTAQGRTD-PNQATGTSIQFCN 430
Query: 250 VCADPEFLP 258
+ A + P
Sbjct: 431 IIASSDLEP 439
>PME3_LYCES (Q96576) Pectinesterase 3 precursor (EC 3.1.1.11)
(Pectin methylesterase 3) (PE 3)
Length = 544
Score = 166 bits (419), Expect = 8e-41
Identities = 97/236 (41%), Positives = 136/236 (57%), Gaps = 6/236 (2%)
Query: 29 GKFPSWVKPGDRKLLQASA--VPADAVVASDGSGNYMKIMDAVMAAPNGSKKRYVIHIKK 86
GK P WV DRKL+++S + A+ VVA DG+G+Y + +AV AAP+ +K RYVI++K
Sbjct: 208 GKMPYWVSSRDRKLMESSGKDIIANRVVAQDGTGDYQTLAEAVAAAPDKNKTRYVIYVKM 267
Query: 87 GVYNEHVMINNSKSNLMMIGDGMGATVITGDLSWGRDKLDTSYTYTFGVEGLGFSAQDIS 146
G+Y E+V++ K NLM++GDGM AT+ITG L+ T + T G GF QDI
Sbjct: 268 GIYKENVVVTKKKMNLMIVGDGMNATIITGSLNVVDG--STFPSNTLAAVGQGFILQDIC 325
Query: 147 FRNTAWPENHQAVALLSDSDTSVFYRCEISGFQDSLCANIKHHSIRIAKSEARLTSYLVR 206
+NTA PE QAVAL +D SV RC I +QD+L A+ + R + +
Sbjct: 326 IQNTAGPEKDQAVALRVGADMSVINRCRIDAYQDTLYAHSQRQFYRDSYVTGTVDFIFGN 385
Query: 207 QLSSF-KTDILVRKGPTGQQNTITAQGGPEKPNLPFGFAFQFCNVCADPEFLPFVN 261
F K I+ RK Q+N +TAQG + PN G + QFC++ A P+ P +N
Sbjct: 386 AAVVFQKCQIVARKPNKRQKNMVTAQGRTD-PNQATGTSIQFCDIIASPDLEPVMN 440
>PME1_CITSI (O04886) Pectinesterase 1 precursor (EC 3.1.1.11)
(Pectin methylesterase) (PE)
Length = 584
Score = 164 bits (416), Expect = 2e-40
Identities = 103/251 (41%), Positives = 145/251 (57%), Gaps = 14/251 (5%)
Query: 15 RTSSSDIL--DTITQKGKFPSWVKPGDRKLLQASAVPADAVVASDGSGNYMKIMDAVMAA 72
RTS++ L +T T G +P+W+ PGDR+LLQ+S+V +AVVA+DGSGN+ + AV AA
Sbjct: 236 RTSNNRKLTEETSTVDG-WPAWLSPGDRRLLQSSSVTPNAVVAADGSGNFKTVAAAVAAA 294
Query: 73 PNGSKKRYVIHIKKGVYNEHVMINNSKSNLMMIGDGMGATVITGDLSWGRDKLDTSYTY- 131
P G KRY+I IK GVY E+V + N+M IGDG T+ITG R+ +D S T+
Sbjct: 295 PQGGTKRYIIRIKAGVYRENVEVTKKHKNIMFIGDGRTRTIITG----SRNVVDGSTTFK 350
Query: 132 --TFGVEGLGFSAQDISFRNTAWPENHQAVALLSDSDTSVFYRCEISGFQDSLCANIKHH 189
T V G GF A+DI+F+NTA P HQAVAL +D S FY C++ +QD+L + +
Sbjct: 351 SATAAVVGEGFLARDITFQNTAGPSKHQAVALRVGADLSAFYNCDMLAYQDTLYVH-SNR 409
Query: 190 SIRIAKSEARLTSYLVRQLSSF--KTDILVRKGPTGQQNTITAQGGPEKPNLPFGFAFQF 247
+ A ++ ++ DI RK +GQ+N +TAQG + PN G Q
Sbjct: 410 QFFVNCLIAGTVDFIFGNAAAVLQNCDIHARKPNSGQKNMVTAQGRTD-PNQNTGIVIQK 468
Query: 248 CNVCADPEFLP 258
+ A + P
Sbjct: 469 SRIGATSDLKP 479
>PME2_ARATH (Q42534) Pectinesterase 2 precursor (EC 3.1.1.11)
(Pectin methylesterase 2) (PE 2)
Length = 587
Score = 162 bits (410), Expect = 9e-40
Identities = 97/235 (41%), Positives = 135/235 (57%), Gaps = 17/235 (7%)
Query: 31 FPSWVKPGDRKLLQASAVPADAVVASDGSGNYMKIMDAVMAAPNGSKKRYVIHIKKGVYN 90
+P W+ GDR+LLQ S + ADA VA DGSG++ + AV AAP S KR+VIHIK GVY
Sbjct: 256 WPKWLSVGDRRLLQGSTIKADATVADDGSGDFTTVAAAVAAAPEKSNKRFVIHIKAGVYR 315
Query: 91 EHVMINNSKSNLMMIGDGMGATVITGDLSWGRDKLDTSYTY---TFGVEGLGFSAQDISF 147
E+V + K+N+M +GDG G T+ITG R+ +D S T+ T G F A+DI+F
Sbjct: 316 ENVEVTKKKTNIMFLGDGRGKTIITG----SRNVVDGSTTFHSATVAAVGERFLARDITF 371
Query: 148 RNTAWPENHQAVALLSDSDTSVFYRCEISGFQDSLCANIKHHSIRIAKSEARLT---SYL 204
+NTA P HQAVAL SD S FY+C++ +QD+L HS R + +T ++
Sbjct: 372 QNTAGPSKHQAVALRVGSDFSAFYQCDMFAYQDTLYV----HSNRQFFVKCHITGTVDFI 427
Query: 205 VRQLSSF--KTDILVRKGPTGQQNTITAQGGPEKPNLPFGFAFQFCNVCADPEFL 257
++ DI R+ +GQ+N +TAQG + PN G Q C + + L
Sbjct: 428 FGNAAAVLQDCDINARRPNSGQKNMVTAQGRSD-PNQNTGIVIQNCRIGGTSDLL 481
>PME2_CITSI (O04887) Pectinesterase 2 precursor (EC 3.1.1.11)
(Pectin methylesterase) (PE)
Length = 510
Score = 160 bits (406), Expect = 3e-39
Identities = 98/231 (42%), Positives = 125/231 (53%), Gaps = 5/231 (2%)
Query: 28 KGKFPSWVKPGDRKLLQASAVPADAVVASDGSGNYMKIMDAVMAAPNGSKKRYVIHIKKG 87
K FP+WVKPGDRKLLQ + A+ VVA DGSGN I +AV AA RYVI+IK G
Sbjct: 180 KDGFPTWVKPGDRKLLQTTP-RANIVVAQDGSGNVKTIQEAVAAASRAGGSRYVIYIKAG 238
Query: 88 VYNEHVMINNSKSNLMMIGDGMGATVITGDLSWGRDKLDTSYTYTFGVEGLGFSAQDISF 147
YNE++ + N+M +GDG+G T+ITG S G T + T V G F A+DI+
Sbjct: 239 TYNENIEVK--LKNIMFVGDGIGKTIITGSKSVGGGAT-TFKSATVAVVGDNFIARDITI 295
Query: 148 RNTAWPENHQAVALLSDSDTSVFYRCEISGFQDSLCANIKHHSIRIAKSEARLTSYLVRQ 207
RNTA P NHQAVAL S SD SVFYRC G+QD+L + + R +
Sbjct: 296 RNTAGPNNHQAVALRSGSDLSVFYRCSFEGYQDTLYVHSQRQFYRECDIYGTVDFIFGNA 355
Query: 208 LSSFKTDILVRKGPTGQQNTITAQGGPEKPNLPFGFAFQFCNVCADPEFLP 258
+ + + P + NT+TAQG + PN G C V A + P
Sbjct: 356 AVVLQNCNIFARKPPNRTNTLTAQGRTD-PNQSTGIIIHNCRVTAASDLKP 405
>PME3_CITSI (P83948) Pectinesterase 3 precursor (EC 3.1.1.11)
(Pectin methylesterase 3) (PE 3)
Length = 584
Score = 159 bits (402), Expect = 7e-39
Identities = 100/251 (39%), Positives = 143/251 (56%), Gaps = 14/251 (5%)
Query: 15 RTSSSDIL--DTITQKGKFPSWVKPGDRKLLQASAVPADAVVASDGSGNYMKIMDAVMAA 72
RTS++ L +T T G +P+W+ GDR+LLQ+S+V + VVA+DGSGN+ + +V AA
Sbjct: 236 RTSNNRKLIEETSTVDG-WPAWLSTGDRRLLQSSSVTPNVVVAADGSGNFKTVAASVAAA 294
Query: 73 PNGSKKRYVIHIKKGVYNEHVMINNSKSNLMMIGDGMGATVITGDLSWGRDKLDTSYTY- 131
P G KRY+I IK GVY E+V + N+M IGDG T+ITG R+ +D S T+
Sbjct: 295 PQGGTKRYIIRIKAGVYRENVEVTKKHKNIMFIGDGRTRTIITG----SRNVVDGSTTFK 350
Query: 132 --TFGVEGLGFSAQDISFRNTAWPENHQAVALLSDSDTSVFYRCEISGFQDSLCANIKHH 189
T V G GF A+DI+F+NTA P HQAVAL +D S FY C++ +QD+L + +
Sbjct: 351 SATVAVVGEGFLARDITFQNTAGPSKHQAVALRVGADLSAFYNCDMLAYQDTLYVH-SNR 409
Query: 190 SIRIAKSEARLTSYLVRQLSSF--KTDILVRKGPTGQQNTITAQGGPEKPNLPFGFAFQF 247
+ A ++ ++ DI RK +GQ+N +TAQG + PN G Q
Sbjct: 410 QFFVNCLIAGTVDFIFGNAAAVLQNCDIHARKPNSGQKNMVTAQGRAD-PNQNTGIVIQK 468
Query: 248 CNVCADPEFLP 258
+ A + P
Sbjct: 469 SRIGATSDLKP 479
>PME1_FICAW (P83947) Pectinesterase precursor (EC 3.1.1.11) (Pectin
methylesterase) (PE)
Length = 545
Score = 155 bits (393), Expect = 8e-38
Identities = 95/248 (38%), Positives = 139/248 (55%), Gaps = 13/248 (5%)
Query: 17 SSSDILDTITQKGKFPSWVKPGDRKLLQASA--VPADAVVASDGSGNYMKIMDAVMAAPN 74
+ S++++ +T G FP+WV GDR+LLQ + D VVA DGSG+Y + +AV A P+
Sbjct: 198 AKSNVIEPVT--GNFPTWVTAGDRRLLQTLGKDIEPDIVVAKDGSGDYETLNEAVAAIPD 255
Query: 75 GSKKRYVIHIKKGVYNEHVMINNSKSNLMMIGDGMGATVITGDLSWGRDKLDTSYTY--- 131
SKKR ++ ++ G+Y E+V K N+M++G+GM T+ITG R+ +D S T+
Sbjct: 256 NSKKRVIVLVRTGIYEENVDFGYQKKNVMLVGEGMDYTIITG----SRNVVDGSTTFDSA 311
Query: 132 TFGVEGLGFSAQDISFRNTAWPENHQAVALLSDSDTSVFYRCEISGFQDSLCANIKHHSI 191
T G GF AQDI F+NTA PE +QAVAL +D +V RC I +QD+L +
Sbjct: 312 TVAAVGDGFIAQDICFQNTAGPEKYQAVALRIGADETVINRCRIDAYQDTLYPHNYRQFY 371
Query: 192 RIAKSEARLTSYLVRQLSSFKT-DILVRKGPTGQQNTITAQGGPEKPNLPFGFAFQFCNV 250
R + F+ +++ RK GQ+NTITAQG + PN G + Q C +
Sbjct: 372 RDRNITGTVDFIFGNAAVVFQNCNLIPRKQMKGQENTITAQGRTD-PNQNTGTSIQNCEI 430
Query: 251 CADPEFLP 258
A + P
Sbjct: 431 FASADLEP 438
>PMEU_LYCES (Q43143) Pectinesterase U1 precursor (EC 3.1.1.11)
(Pectin methylesterase) (PE)
Length = 583
Score = 151 bits (382), Expect = 2e-36
Identities = 90/235 (38%), Positives = 128/235 (54%), Gaps = 3/235 (1%)
Query: 25 ITQKGKFPSWVKPGDRKLLQASAVPADAVVASDGSGNYMKIMDAVMAAPNGSKKRYVIHI 84
+ G++P W+ GDR+LLQ+S V D VVA+DGSG+Y + +AV AP S KRYVI I
Sbjct: 246 VEDNGEWPEWLSAGDRRLLQSSTVTPDVVVAADGSGDYKTVSEAVRKAPEKSSKRYVIRI 305
Query: 85 KKGVYNEHVMINNSKSNLMMIGDGMGATVITGDLSWGRDKLDTSYTYTFGVEGLGFSAQD 144
K GVY E+V + K+N+M +GDG T+IT + +D T ++ T A+D
Sbjct: 306 KAGVYRENVDVPKKKTNIMFMGDGKSNTIITASRN-VQDGSTTFHSATVVRVAGKVLARD 364
Query: 145 ISFRNTAWPENHQAVALLSDSDTSVFYRCEISGFQDSLCANIKHHSIRIAKSEARLTSYL 204
I+F+NTA HQAVAL SD S FYRC++ +QD+L + +
Sbjct: 365 ITFQNTAGASKHQAVALCVGSDLSAFYRCDMLAYQDTLYVHSNRQFFVQCLVAGTVDFIF 424
Query: 205 VRQLSSFK-TDILVRKGPTGQQNTITAQGGPEKPNLPFGFAFQFCNVCADPEFLP 258
+ F+ DI R+ +GQ+N +TAQG + PN G Q C + A + P
Sbjct: 425 GNGAAVFQDCDIHARRPGSGQKNMVTAQGRTD-PNQNTGIVIQKCRIGATSDLRP 478
>PME_MEDSA (Q42920) Pectinesterase precursor (EC 3.1.1.11) (Pectin
methylesterase) (PE) (P65)
Length = 447
Score = 146 bits (369), Expect = 5e-35
Identities = 85/241 (35%), Positives = 118/241 (48%), Gaps = 1/241 (0%)
Query: 14 FRTSSSDILDTITQKGKFPSWVKPGDRKLLQASAVPADAVVASDGSGNYMKIMDAVMAAP 73
F TS + + PSWV G R+LL V +AVVA DGSG + + DA+ P
Sbjct: 94 FDTSQYSVSRRLLSDDGIPSWVNDGHRRLLAGGNVQPNAVVAQDGSGQFKTLTDALKTVP 153
Query: 74 NGSKKRYVIHIKKGVYNEHVMINNSKSNLMMIGDGMGATVITGDLSWGRDKLDTSYTYTF 133
+ +VIH+K GVY E V + + + +IGDG T TG L++ D ++T T TF
Sbjct: 154 PKNAVPFVIHVKAGVYKETVNVAKEMNYVTVIGDGPTKTKFTGSLNYA-DGINTYNTATF 212
Query: 134 GVEGLGFSAQDISFRNTAWPENHQAVALLSDSDTSVFYRCEISGFQDSLCANIKHHSIRI 193
GV G F A+DI F NTA HQAVAL +D ++FY C++ GFQD+L + R
Sbjct: 213 GVNGANFMAKDIGFENTAGTGKHQAVALRVTADQAIFYNCQMDGFQDTLYVQSQRQFYRD 272
Query: 194 AKSEARLTSYLVRQLSSFKTDILVRKGPTGQQNTITAQGGPEKPNLPFGFAFQFCNVCAD 253
+ + F+ LV + P Q + GG EK N FQ + +
Sbjct: 273 CSISGTIDFVFGERFGVFQNCKLVCRLPAKGQQCLVTAGGREKQNSASALVFQSSHFTGE 332
Query: 254 P 254
P
Sbjct: 333 P 333
>PME_DAUCA (P83218) Pectinesterase (EC 3.1.1.11) (Pectin
methylesterase) (PE)
Length = 319
Score = 137 bits (345), Expect = 3e-32
Identities = 87/217 (40%), Positives = 119/217 (54%), Gaps = 5/217 (2%)
Query: 44 QASAVPADAVVASDGSGNYMKIMDAVMAAPNGSKKRYVIHIKKGVYNEHVMINNSKSNLM 103
Q+S V + VVA+DGSG+Y + +AV AAP SK RYVI IK GVY E+V + K N+M
Sbjct: 1 QSSTVTPNVVVAADGSGDYKTVSEAVAAAPEDSKTRYVIRIKAGVYRENVDVPKKKKNIM 60
Query: 104 MIGDGMGATVITGDLSWGRDKLDTSYTYTFGVEGLGFSAQDISFRNTAWPENHQAVALLS 163
+GDG +T+IT + +D T + T G GF A+DI+F+NTA HQAVAL
Sbjct: 61 FLGDGRTSTIITASKN-VQDGSTTFNSATVAAVGAGFLARDITFQNTAGAAKHQAVALRV 119
Query: 164 DSDTSVFYRCEISGFQDSLCANIKHHSIRIAKSEARLTSYLVRQLSSF--KTDILVRKGP 221
SD S FYRC+I +QDSL + + I A ++ + DI R+
Sbjct: 120 GSDLSAFYRCDILAYQDSLYVH-SNRQFFINCFIAGTVDFIFGNAAVVLQDCDIHARRPG 178
Query: 222 TGQQNTITAQGGPEKPNLPFGFAFQFCNVCADPEFLP 258
+GQ+N +TAQG + PN G Q + A + P
Sbjct: 179 SGQKNMVTAQGRTD-PNQNTGIVIQKSRIGATSDLQP 214
>PME1_ARATH (Q43867) Pectinesterase 1 precursor (EC 3.1.1.11)
(Pectin methylesterase 1) (PE 1)
Length = 586
Score = 137 bits (345), Expect = 3e-32
Identities = 84/228 (36%), Positives = 122/228 (52%), Gaps = 6/228 (2%)
Query: 27 QKGKFPSWVKPGDRKLLQASAVPADAVVASDGSGNYMKIMDAVMAAPNGSKKRYVIHIKK 86
Q F W + R+LLQ + + D VA DG+G+ + + +AV P S K +VI++K
Sbjct: 259 QSVDFEKWAR---RRLLQTAGLKPDVTVAGDGTGDVLTVNEAVAKVPKKSLKMFVIYVKS 315
Query: 87 GVYNEHVMINNSKSNLMMIGDGMGATVITGDLSWGRDKLDTSYTYTFGVEGLGFSAQDIS 146
G Y E+V+++ SK N+M+ GDG G T+I+G ++ D T T TF ++G GF +DI
Sbjct: 316 GTYVENVVMDKSKWNVMIYGDGKGKTIISGSKNF-VDGTPTYETATFAIQGKGFIMKDIG 374
Query: 147 FRNTAWPENHQAVALLSDSDTSVFYRCEISGFQDSLCANIKHHSIRIAKSEARLTSYLVR 206
NTA HQAVA S SD SV+Y+C GFQD+L + R +
Sbjct: 375 IINTAGAAKHQAVAFRSGSDFSVYYQCSFDGFQDTLYPHSNRQFYRDCDVTGTIDFIFGS 434
Query: 207 QLSSFK-TDILVRKGPTGQQNTITAQGGPEKPNLPFGFAFQFCNVCAD 253
F+ I+ R+ + Q NTITAQ G + PN G + Q C + A+
Sbjct: 435 AAVVFQGCKIMPRQPLSNQFNTITAQ-GKKDPNQSSGMSIQRCTISAN 481
>PME_BRANA (P41510) Probable pectinesterase precursor (EC 3.1.1.11)
(Pectin methylesterase) (PE)
Length = 584
Score = 133 bits (334), Expect = 6e-31
Identities = 91/257 (35%), Positives = 120/257 (46%), Gaps = 22/257 (8%)
Query: 20 DILDTITQKGKFPSWVKPGDRKLLQASA---VPADA----------------VVASDGSG 60
D+L+ + QKG P W DRKL+ + PAD VVA DGSG
Sbjct: 223 DLLEDLDQKG-LPKWHSDKDRKLMAQAGRPGAPADEGIGEGGGGGGKIKPTHVVAKDGSG 281
Query: 61 NYMKIMDAVMAAPNGSKKRYVIHIKKGVYNEHVMINNSKSNLMMIGDGMGATVITGDLSW 120
+ I +AV A P + R +I+IK GVY E V I +N+ M GDG T+IT D S
Sbjct: 282 QFKTISEAVKACPEKNPGRCIIYIKAGVYKEQVTIPKKVNNVFMFGDGATQTIITFDRSV 341
Query: 121 GRDK-LDTSYTYTFGVEGLGFSAQDISFRNTAWPENHQAVALLSDSDTSVFYRCEISGFQ 179
G TS + T VE GF A+ I F+NTA P HQAVA + D +V + C G+Q
Sbjct: 342 GLSPGTTTSLSGTVQVESEGFMAKWIGFQNTAGPLGHQAVAFRVNGDRAVIFNCRFDGYQ 401
Query: 180 DSLCANIKHHSIRIAKSEARLTSYLVRQLSSFKTD-ILVRKGPTGQQNTITAQGGPEKPN 238
D+L N R + + + + IL RKG GQ N +TA G +
Sbjct: 402 DTLYVNNGRQFYRNIVVSGTVDFIFGKSATVIQNSLILCRKGSPGQTNHVTADGNEKGKA 461
Query: 239 LPFGFAFQFCNVCADPE 255
+ G C + AD E
Sbjct: 462 VKIGIVLHNCRIMADKE 478
>PME_BRACM (Q42608) Pectinesterase (EC 3.1.1.11) (Pectin
methylesterase) (PE) (Fragment)
Length = 571
Score = 128 bits (322), Expect = 1e-29
Identities = 90/257 (35%), Positives = 120/257 (46%), Gaps = 22/257 (8%)
Query: 20 DILDTITQKGKFPSWVKPGDRKLLQASA---VPADA----------------VVASDGSG 60
D+L+ + QKG P W DRKL+ + PAD VVA DGSG
Sbjct: 210 DLLEDLDQKG-LPKWHSDKDRKLMAQAGRPGAPADEGIGEGGGGGGKIKPTHVVAKDGSG 268
Query: 61 NYMKIMDAVMAAPNGSKKRYVIHIKKGVYNEHVMINNSKSNLMMIGDGMGATVITGDLSW 120
+ I +AV A P + R +I+IK GVY E V I +N+ M GDG T+IT D S
Sbjct: 269 QFKTISEAVKACPEKNPGRCIIYIKAGVYKEQVTIPKKVNNVFMFGDGATQTIITFDRSV 328
Query: 121 GRDK-LDTSYTYTFGVEGLGFSAQDISFRNTAWPENHQAVALLSDSDTSVFYRCEISGFQ 179
G TS + T VE GF A+ I F+NTA P +QAVA + D +V + C G+Q
Sbjct: 329 GLSPGTTTSLSGTVQVESEGFMAKWIGFQNTAGPLGNQAVAFRVNGDRAVIFNCRFDGYQ 388
Query: 180 DSLCANIKHHSIRIAKSEARLTSYLVRQLSSFKTD-ILVRKGPTGQQNTITAQGGPEKPN 238
D+L N R + + + + IL RKG GQ N +TA G +
Sbjct: 389 DTLYVNNGRQFYRNIVVSGTVDFINGKSATVIQNSLILCRKGSPGQTNHVTADGKQKGKA 448
Query: 239 LPFGFAFQFCNVCADPE 255
+ G C + AD E
Sbjct: 449 VKIGIVLHNCRIMADKE 465
>PME3_PHAVU (Q43111) Pectinesterase 3 precursor (EC 3.1.1.11)
(Pectin methylesterase 3) (PE 3)
Length = 581
Score = 127 bits (319), Expect = 3e-29
Identities = 81/219 (36%), Positives = 117/219 (52%), Gaps = 5/219 (2%)
Query: 31 FPSWVKPGDRKLLQASAVPA--DAVVASDGSGNYMKIMDAVMAAPNGSKKRYVIHIKKGV 88
FP W+ +R+LL+ + DAVVA DGSG + I +A+ S++R+ +++K+G
Sbjct: 251 FPEWLGAAERRLLEEKNNDSTPDAVVAKDGSGQFKTIGEALKLVKKKSEERFSVYVKEGR 310
Query: 89 YNEHVMINNSKSNLMMIGDGMGATVITGDLSWGRDKLDTSYTYTFGVEGLGFSAQDISFR 148
Y E++ ++ + N+M+ GDG T + G ++ D T T TF V+G GF A+DI F
Sbjct: 311 YVENIDLDKNTWNVMIYGDGKDKTFVVGSRNF-MDGTPTFETATFAVKGKGFIAKDIGFV 369
Query: 149 NTAWPENHQAVALLSDSDTSVFYRCEISGFQDSLCANIKHHSIRIAKSEARLTSYLVRQL 208
N A HQAVAL S SD SVF+RC GFQD+L A+ R +
Sbjct: 370 NNAGASKHQAVALRSGSDRSVFFRCSFDGFQDTLYAHSNRQFYRDCDITGTIDFIFGNAA 429
Query: 209 SSFKT-DILVRKGPTGQQNTITAQGGPEKPNLPFGFAFQ 246
F++ I+ R+ Q NTITAQ G + PN G Q
Sbjct: 430 VVFQSCKIMPRQPLPNQFNTITAQ-GKKDPNQNTGIIIQ 467
>PME_PETIN (Q43043) Pectinesterase precursor (EC 3.1.1.11) (Pectin
methylesterase) (PE)
Length = 374
Score = 124 bits (312), Expect = 2e-28
Identities = 80/222 (36%), Positives = 118/222 (53%), Gaps = 11/222 (4%)
Query: 40 RKLLQASAVPADAVVASDGSGNYMKIMDAVMAAPNGSKKRYVIHIKKGVYNEHVMINNSK 99
R+LLQ S +A VA DGSG Y I +A+ A P + + ++I IK GVY E++ I S
Sbjct: 47 RRLLQISNAKPNATVALDGSGQYKTIKEALDAVPKKNTEPFIIFIKAGVYKEYIDIPKSM 106
Query: 100 SNLMMIGDGMGATVITGDLSWGRDKLDTSYTYTFGVEGLGFSAQDISFRNTAWPENHQAV 159
+N+++IG+G T ITG+ S +D T +T T GV G F A++I F NTA PE QAV
Sbjct: 107 TNVVLIGEGPTKTKITGNKS-VKDGPSTFHTTTVGVNGANFVAKNIGFENTAGPEKEQAV 165
Query: 160 ALLSDSDTSVFYRCEISGFQDSLCANIKHHSIRIAKSEARLTSYLVRQLSSFKT-----D 214
AL +D ++ Y C+I G+QD+L H+ R + +T + + +
Sbjct: 166 ALRVSADKAIIYNCQIDGYQDTLYV----HTYRQFYRDCTITGTVDFIFGNGEAVLQNCK 221
Query: 215 ILVRKGPTGQQNTITAQGGPEKPNLPFGFAFQFCNVCADPEF 256
++VRK Q +TAQG E P Q C + D ++
Sbjct: 222 VIVRKPAQNQSCMVTAQGRTE-PIQKGAIVLQNCEIKPDTDY 262
>PME_ASPAC (Q12535) Pectinesterase precursor (EC 3.1.1.11) (Pectin
methylesterase) (PE)
Length = 331
Score = 62.8 bits (151), Expect = 1e-09
Identities = 49/196 (25%), Positives = 85/196 (43%), Gaps = 11/196 (5%)
Query: 46 SAVPADAVVASDGSGNYMKIMDAVMAAPNGSKKRYVIHIKKGVYNEHVMINNSKSNLMMI 105
+ P+ A+V + G+Y I DA+ A + I I++G Y+E V + +++
Sbjct: 21 TTAPSGAIVVAKSGGDYTTIGDAIDALSTSTTDTQTIFIEEGTYDEQVYLPAMTGKVIIY 80
Query: 106 GDGMGA-------TVITGDLSWGRDKLDTSYTYTFGVEGLGFSAQDISFRNTAWPENHQA 158
G IT +S+ T TF + +G +++ NT HQA
Sbjct: 81 GQTENTDSYADNLVTITHAISYEDAGESDDLTATFRNKAVGSQVYNLNIANTCGQACHQA 140
Query: 159 VALLSDSDTSVFYRCEISGFQDSLCANIKHHSIRIAKSEARLTSYLVRQLSSF--KTDIL 216
+AL + +D +Y C +G+QD+L A + + E + + ++ DI
Sbjct: 141 LALSAWADQQGYYGCNFTGYQDTLLAQTGNQLYINSYIEGAVDFIFGQHARAWFQNVDIR 200
Query: 217 VRKGPTGQQNTITAQG 232
V +GPT +ITA G
Sbjct: 201 VVEGPTSA--SITANG 214
>PMEA_ERWCH (P07863) Pectinesterase A precursor (EC 3.1.1.11)
(Pectin methylesterase A) (PE A)
Length = 366
Score = 55.8 bits (133), Expect = 1e-07
Identities = 51/160 (31%), Positives = 75/160 (46%), Gaps = 26/160 (16%)
Query: 46 SAVPADAVVASDGSGN--YMKIMDAVMAAPNGSKKRYVIHIKKGVYNEHVMINNSKSNLM 103
+A +AVV+ S + I DA+ +AP GS +VI IK GVYNE + I +++NL+
Sbjct: 24 AATTYNAVVSKSSSDGKTFKTIADAIASAPAGSTP-FVILIKNGVYNERLTI--TRNNLL 80
Query: 104 MIGDGMGATVI-----TGDLSWGRDKLDTSYTYTFGVEGLGFSAQDISFRNT-AWPENH- 156
+ G+ VI G L K T+ + T + FSAQ ++ RN +P N
Sbjct: 81 LKGESRNGAVIAAATAAGTLKSDGSKWGTAGSSTITISAKDFSAQSLTIRNDFDFPANQA 140
Query: 157 ------------QAVAL--LSDSDTSVFYRCEISGFQDSL 182
QAVAL D + F + G+QD+L
Sbjct: 141 KSDSDSSKIKDTQAVALYVTKSGDRAYFKDVSLVGYQDTL 180
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.320 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 33,214,110
Number of Sequences: 164201
Number of extensions: 1371776
Number of successful extensions: 3403
Number of sequences better than 10.0: 23
Number of HSP's better than 10.0 without gapping: 20
Number of HSP's successfully gapped in prelim test: 3
Number of HSP's that attempted gapping in prelim test: 3330
Number of HSP's gapped (non-prelim): 23
length of query: 287
length of database: 59,974,054
effective HSP length: 109
effective length of query: 178
effective length of database: 42,076,145
effective search space: 7489553810
effective search space used: 7489553810
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 65 (29.6 bits)
Medicago: description of AC148775.4


