

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148775.11 - phase: 0 /pseudo/partial
(467 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
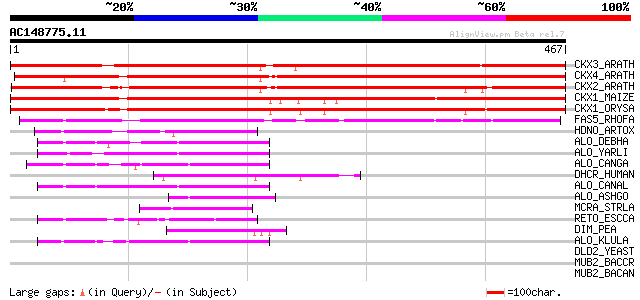
Score E
Sequences producing significant alignments: (bits) Value
CKX3_ARATH (Q9LTS3) Cytokinin dehydrogenase 3 precursor (EC 1.5.... 481 e-135
CKX4_ARATH (Q9FUJ2) Cytokinin dehydrogenase 4 precursor (EC 1.5.... 467 e-131
CKX2_ARATH (Q9FUJ3) Cytokinin dehydrogenase 2 precursor (EC 1.5.... 465 e-131
CKX1_MAIZE (Q9T0N8) Cytokinin dehydrogenase 1 precursor (EC 1.5.... 429 e-119
CKX1_ORYSA (Q9LDE6) Probable cytokinin dehydrogenase precursor (... 416 e-116
FAS5_RHOFA (P46377) Hypothetical 47.9 kDa oxidoreductase in fasc... 205 2e-52
HDNO_ARTOX (P08159) 6-hydroxy-D-nicotine oxidase (EC 1.5.3.6) (6... 65 4e-10
ALO_DEBHA (Q6BZA0) D-arabinono-1,4-lactone oxidase (EC 1.1.3.37)... 60 1e-08
ALO_YARLI (Q6CG88) D-arabinono-1,4-lactone oxidase (EC 1.1.3.37)... 59 3e-08
ALO_CANGA (Q6FS20) D-arabinono-1,4-lactone oxidase (EC 1.1.3.37)... 52 4e-06
DHCR_HUMAN (Q15392) 24-dehydrocholesterol reductase precursor (E... 51 7e-06
ALO_CANAL (O93852) D-arabinono-1,4-lactone oxidase (EC 1.1.3.37)... 48 6e-05
ALO_ASHGO (Q752Y3) D-arabinono-1,4-lactone oxidase (EC 1.1.3.37)... 47 8e-05
MCRA_STRLA (P43485) Mitomycin radical oxidase (EC 1.5.3.-) 45 4e-04
RETO_ESCCA (P30986) Reticuline oxidase precursor (EC 1.21.3.3) (... 44 7e-04
DIM_PEA (P93472) Cell elongation protein diminuto 44 7e-04
ALO_KLULA (Q6CSY3) D-arabinono-1,4-lactone oxidase (EC 1.1.3.37)... 44 9e-04
DLD2_YEAST (P46681) D-lactate dehydrogenase [cytochrome] 2, mito... 42 0.004
MUB2_BACCR (Q815R9) UDP-N-acetylenolpyruvoylglucosamine reductas... 41 0.006
MUB2_BACAN (Q81XC5) UDP-N-acetylenolpyruvoylglucosamine reductas... 41 0.006
>CKX3_ARATH (Q9LTS3) Cytokinin dehydrogenase 3 precursor (EC
1.5.99.12) (Cytokinin oxidase 3) (CKO 3)
Length = 523
Score = 481 bits (1238), Expect = e-135
Identities = 251/486 (51%), Positives = 331/486 (67%), Gaps = 34/486 (6%)
Query: 1 KLSRNPQILSQASTDYGHIIHENPFAVLEPTSISDIANLINYSNSLPHSFTISPRGQAHS 60
KL+ + + A+TD+GH+ P AVL P+S+ DI +LI S SF ++ RG HS
Sbjct: 47 KLTSSSSSVESAATDFGHVTKIFPSAVLIPSSVEDITDLIKLSFDSQLSFPLAARGHGHS 106
Query: 61 VLGQAMTQNGIVVNMTQLNWYRNGSGIVVSDCDVKNPLGC-YVDVGGEQLWIDVLNATLK 119
GQA ++G+VVNM + V D +K C YVDV LWI+VLN TL+
Sbjct: 107 HRGQASAKDGVVVNMRSM---------VNRDRGIKVSRTCLYVDVDAAWLWIEVLNKTLE 157
Query: 120 HGLTPLSWTDYLYLSVGGTLSNAGIGGQTFRFGPQISNVLELDVITGQGNIVTCSQEKNS 179
GLTP+SWTDYLYL+VGGTLSN GI GQTFR+GPQI+NVLE+DVITG+G I TCS++ NS
Sbjct: 158 LGLTPVSWTDYLYLTVGGTLSNGGISGQTFRYGPQITNVLEMDVITGKGEIATCSKDMNS 217
Query: 180 EVFYAVLGGLGQFGVITRARILLGPAPTRA---------------NQEHLISFDRRNDSN 224
++F+AVLGGLGQFG+ITRARI L AP RA +QE +IS ++
Sbjct: 218 DLFFAVLGGLGQFGIITRARIKLEVAPKRAKWLRFLYIDFSEFTRDQERVIS-----KTD 272
Query: 225 GADYVEGMLLLNKPP---LILSFYPPSDHPRITSLVTQYGITYVLELVKYYDTNSQANIT 281
G D++EG ++++ P ++YPPSDH RI S+V ++ + Y LE+VKYYD SQ +
Sbjct: 273 GVDFLEGSIMVDHGPPDNWRSTYYPPSDHLRIASMVKRHRVIYCLEVVKYYDETSQYTVN 332
Query: 282 EEVDNLVKGLKFVPTFMFQKDVTYEEFLDRVHSEELILRSKGLWDVPHPWLNMFIPKSRI 341
EE++ L L V FM++KDVTY +FL+RV + EL L+SKG WDVPHPWLN+F+PK++I
Sbjct: 333 EEMEELSDSLNHVRGFMYEKDVTYMDFLNRVRTGELNLKSKGQWDVPHPWLNLFVPKTQI 392
Query: 342 SDFNEGVFKGIILKQNISAGIVLVYPMNRNKWDDRMSAITPDEDVFYTLALLHAAKEMDE 401
S F++GVFKGIIL+ NI++G VLVYPMNRNKW+DRMSA P+EDVFY + L +A D
Sbjct: 393 SKFDDGVFKGIILRNNITSGPVLVYPMNRNKWNDRMSAAIPEEDVFYAVGFLRSA-GFDN 451
Query: 402 VKTFQAQNHQILQFCNDAGIKVKEYLSGNKTHQEWVEHFGAKWQQFEERKAKFDPKRILS 461
+ F +N +IL+FC DA + V +YL + + + WV HFG +W F ERK K+DPK ILS
Sbjct: 452 WEAFDQENMEILKFCEDANMGVIQYLPYHSSQEGWVRHFGPRWNIFVERKYKYDPKMILS 511
Query: 462 PGQGIF 467
PGQ IF
Sbjct: 512 PGQNIF 517
>CKX4_ARATH (Q9FUJ2) Cytokinin dehydrogenase 4 precursor (EC
1.5.99.12) (Cytokinin oxidase 4) (CKO 4)
Length = 524
Score = 467 bits (1202), Expect = e-131
Identities = 242/488 (49%), Positives = 321/488 (65%), Gaps = 34/488 (6%)
Query: 5 NPQILSQASTDYGHIIHENPFAVLEPTSISDIANLINYSN----------SLPHSFTISP 54
+P +S AS D+G+I ENP AVL P+S +++A L+ ++N S +F ++
Sbjct: 45 DPFSISAASHDFGNITDENPGAVLCPSSTTEVARLLRFANGGFSYNKGSTSPASTFKVAA 104
Query: 55 RGQAHSVLGQAMTQNGIVVNMTQLNWYRNGSGIVVSDCDVKNPLGCYVDVGGEQLWIDVL 114
RGQ HS+ GQA G+VVNMT L + +V+S G Y DV +W+DVL
Sbjct: 105 RGQGHSLRGQASAPGGVVVNMTCLAMAAKPAAVVISAD------GTYADVAAGTMWVDVL 158
Query: 115 NATLKHGLTPLSWTDYLYLSVGGTLSNAGIGGQTFRFGPQISNVLELDVITGQGNIVTCS 174
A + G++P++WTDYLYLSVGGTLSNAGIGGQTFR GPQISNV ELDVITG+G ++TCS
Sbjct: 159 KAAVDRGVSPVTWTDYLYLSVGGTLSNAGIGGQTFRHGPQISNVHELDVITGKGEMMTCS 218
Query: 175 QEKNSEVFYAVLGGLGQFGVITRARILLGPAPTRA---------------NQEHLISFDR 219
+ N E+FY VLGGLGQFG+ITRARI L APTR +QE LIS
Sbjct: 219 PKLNPELFYGVLGGLGQFGIITRARIALDHAPTRVKWSRILYSDFSAFKRDQERLISMT- 277
Query: 220 RNDSNGADYVEGMLLLNKPPLILSFYPPSDHPRITSLVTQYGITYVLELVKYYDTNSQAN 279
ND G D++EG L+++ + SF+P SD R+ SLV + I YVLE+ KYYD +
Sbjct: 278 -NDL-GVDFLEGQLMMSNGFVDTSFFPLSDQTRVASLVNDHRIIYVLEVAKYYDRTTLPI 335
Query: 280 ITEEVDNLVKGLKFVPTFMFQKDVTYEEFLDRVHSEELILRSKGLWDVPHPWLNMFIPKS 339
I + +D L + L F P FMF +DV Y +FL+RV +EE LRS GLW+VPHPWLN+F+P S
Sbjct: 336 IDQVIDTLSRTLGFAPGFMFVQDVPYFDFLNRVRNEEDKLRSLGLWEVPHPWLNIFVPGS 395
Query: 340 RISDFNEGVFKGIILKQNISAGIVLVYPMNRNKWDDRMSAITPDEDVFYTLALLHAAKEM 399
RI DF++GV G++L Q ++G+ L YP NRNKW++RMS +TPDEDVFY + LL +A
Sbjct: 396 RIQDFHDGVINGLLLNQTSTSGVTLFYPTNRNKWNNRMSTMTPDEDVFYVIGLLQSAGGS 455
Query: 400 DEVKTFQAQNHQILQFCNDAGIKVKEYLSGNKTHQEWVEHFGAKWQQFEERKAKFDPKRI 459
+ + N +++QFC ++GIK+KEYL ++WV+HFG KW F +K FDPKR+
Sbjct: 456 QNWQELENLNDKVIQFCENSGIKIKEYLMHYTRKEDWVKHFGPKWDDFLRKKIMFDPKRL 515
Query: 460 LSPGQGIF 467
LSPGQ IF
Sbjct: 516 LSPGQDIF 523
>CKX2_ARATH (Q9FUJ3) Cytokinin dehydrogenase 2 precursor (EC
1.5.99.12) (Cytokinin oxidase 2) (CKO 2)
Length = 501
Score = 465 bits (1197), Expect = e-131
Identities = 242/485 (49%), Positives = 331/485 (67%), Gaps = 37/485 (7%)
Query: 2 LSRNPQILSQASTDYGHIIHENPFAVLEPTSISDIANLINYSNSLPHSFTISPRGQAHSV 61
LS +P I+S AS D+G+I P V+ P+S +DI+ L+ Y+ + +F ++ RGQ HS+
Sbjct: 35 LSTDPSIISAASHDFGNITTVTPGGVICPSSTADISRLLQYAANGKSTFQVAARGQGHSL 94
Query: 62 LGQAMTQNGIVVNMTQLNWYRNGSGIVVSDCDVKNPLGCYVDVGGEQLWIDVLNATLKHG 121
GQA G++VNMT + + +VVS D K Y DV LW+DVL T + G
Sbjct: 95 NGQASVSGGVIVNMTCI------TDVVVSK-DKK-----YADVAAGTLWVDVLKKTAEKG 142
Query: 122 LTPLSWTDYLYLSVGGTLSNAGIGGQTFRFGPQISNVLELDVITGQGNIVTCSQEKNSEV 181
++P+SWTDYL+++VGGTLSN GIGGQ FR GP +SNVLELDVITG+G ++TCS++ N E+
Sbjct: 143 VSPVSWTDYLHITVGGTLSNGGIGGQVFRNGPLVSNVLELDVITGKGEMLTCSRQLNPEL 202
Query: 182 FYAVLGGLGQFGVITRARILLGPAPTRA---------------NQEHLISFDRRNDSNGA 226
FY VLGGLGQFG+ITRARI+L AP RA +QE LIS ND G
Sbjct: 203 FYGVLGGLGQFGIITRARIVLDHAPKRAKWFRMLYSDFTTFTKDQERLISM--ANDI-GV 259
Query: 227 DYVEGMLLLNKPPLILSFYPPSDHPRITSLVTQYGITYVLELVKYYDTNSQANITEEVDN 286
DY+EG + L+ + SF+PPSD ++ LV Q+GI YVLE+ KYYD + I++ +D
Sbjct: 260 DYLEGQIFLSNGVVDTSFFPPSDQSKVADLVKQHGIIYVLEVAKYYDDPNLPIISKVIDT 319
Query: 287 LVKGLKFVPTFMFQKDVTYEEFLDRVHSEELILRSKGLWDVPHPWLNMFIPKSRISDFNE 346
L K L ++P F+ DV Y +FL+RVH EE LRS GLW++PHPWLN+++PKSRI DF+
Sbjct: 320 LTKTLSYLPGFISMHDVAYFDFLNRVHVEENKLRSLGLWELPHPWLNLYVPKSRILDFHN 379
Query: 347 GVFKGIILKQNISAGIVLVYPMNRNKWDDRMSAITP--DEDVFYTLALLHAA--KEMDEV 402
GV K I+LKQ ++G+ L+YP NRNKWD+RMSA+ P DEDV Y + LL +A K++ EV
Sbjct: 380 GVVKDILLKQKSASGLALLYPTNRNKWDNRMSAMIPEIDEDVIYIIGLLQSATPKDLPEV 439
Query: 403 KTFQAQNHQILQFCNDAGIKVKEYLSGNKTHQEWVEHFGAKWQQFEERKAKFDPKRILSP 462
++ N +I++FC D+GIK+K+YL + ++W+EHFG+KW F +RK FDPK++LSP
Sbjct: 440 ---ESVNEKIIRFCKDSGIKIKQYLMHYTSKEDWIEHFGSKWDDFSKRKDLFDPKKLLSP 496
Query: 463 GQGIF 467
GQ IF
Sbjct: 497 GQDIF 501
>CKX1_MAIZE (Q9T0N8) Cytokinin dehydrogenase 1 precursor (EC
1.5.99.12) (Cytokinin oxidase 1) (CKO 1)
Length = 534
Score = 429 bits (1102), Expect = e-119
Identities = 240/495 (48%), Positives = 315/495 (63%), Gaps = 35/495 (7%)
Query: 1 KLSRNPQILSQASTDYGHIIHENPFAVLEPTSISDIANLINYSNSLPH-SFTISPRGQAH 59
KL + + ASTD+G+I P AVL P+S +D+ L++ +NS P +TI+ RG+ H
Sbjct: 46 KLRTDSNATAAASTDFGNITSALPAAVLYPSSTADLVALLSAANSTPGWPYTIAFRGRGH 105
Query: 60 SVLGQAMTQNGIVVNMTQLNWYRNGSGIVVSDCDVKNPLGCYVDVGGEQLWIDVLNATLK 119
S++GQA G+VVNM L I VS G YVD GGEQ+WIDVL A+L
Sbjct: 106 SLMGQAFAPGGVVVNMASLGDAAAPPRINVSAD------GRYVDAGGEQVWIDVLRASLA 159
Query: 120 HGLTPLSWTDYLYLSVGGTLSNAGIGGQTFRFGPQISNVLELDVITGQGNIVTCSQEKNS 179
G+ P SWTDYLYL+VGGTLSNAGI GQ FR GPQISNVLE+DVITG G +VTCS++ N+
Sbjct: 160 RGVAPRSWTDYLYLTVGGTLSNAGISGQAFRHGPQISNVLEMDVITGHGEMVTCSKQLNA 219
Query: 180 EVFYAVLGGLGQFGVITRARILLGPAPTRANQEHLISFD-------------RRNDSNGA 226
++F AVLGGLGQFGVITRARI + PAP RA L+ D R GA
Sbjct: 220 DLFDAVLGGLGQFGVITRARIAVEPAPARARWVRLVYTDFAAFSADQERLTAPRPGGGGA 279
Query: 227 -----DYVEGMLLLNKPPLI----LSFYPPSDHPRITSLVTQYGIT--YVLELVKYYD-- 273
YVEG + +N+ F+ +D RI +L + T Y +E YD
Sbjct: 280 SFGPMSYVEGSVFVNQSLATDLANTGFFTDADVARIVALAGERNATTVYSIEATLNYDNA 339
Query: 274 TNSQANITEEVDNLVKGLKFVPTFMFQKDVTYEEFLDRVHSEELILRSKGLWDVPHPWLN 333
T + A + +E+ +++ L +V F FQ+DV Y FLDRVH EE+ L GLW VPHPWLN
Sbjct: 340 TAAAAAVDQELASVLGTLSYVEGFAFQRDVAYAAFLDRVHGEEVALNKLGLWRVPHPWLN 399
Query: 334 MFIPKSRISDFNEGVFKGIILKQNISAGIVLVYPMNRNKWDDRMSAITPDEDVFYTLALL 393
MF+P+SRI+DF+ GVFKGI+ +I G ++VYP+N++ WDD MSA TP EDVFY ++LL
Sbjct: 400 MFVPRSRIADFDRGVFKGILQGTDI-VGPLIVYPLNKSMWDDGMSAATPSEDVFYAVSLL 458
Query: 394 HAAKEMDEVKTFQAQNHQILQFCNDAGIKVKEYLSGNKTHQEWVEHFG-AKWQQFEERKA 452
++ +++ Q QN +IL+FC+ AGI+ K YL+ + +WV HFG AKW +F E K
Sbjct: 459 FSSVAPNDLARLQEQNRRILRFCDLAGIQYKTYLARHTDRSDWVRHFGAAKWNRFVEMKN 518
Query: 453 KFDPKRILSPGQGIF 467
K+DPKR+LSPGQ IF
Sbjct: 519 KYDPKRLLSPGQDIF 533
>CKX1_ORYSA (Q9LDE6) Probable cytokinin dehydrogenase precursor (EC
1.5.99.12) (Cytokinin oxidase) (CKO)
Length = 532
Score = 416 bits (1068), Expect = e-116
Identities = 234/494 (47%), Positives = 308/494 (61%), Gaps = 35/494 (7%)
Query: 1 KLSRNPQILSQASTDYGHIIHENPFAVLEPTSISDIANLINYSNSLP-HSFTISPRGQAH 59
KL +P AS D+G+I P AVL P S D+A L+ + + P FT+S RG+ H
Sbjct: 46 KLRTDPNATVPASMDFGNITAALPAAVLFPGSPGDVAELLRAAYAAPGRPFTVSFRGRGH 105
Query: 60 SVLGQAMTQNGIVVNMTQLNWYRNGSGIVVSDCDVKNPLGCYVDVGGEQLWIDVLNATLK 119
S +GQA+ G+VV+M + G+ + D G YVD GGEQLW+DVL A L
Sbjct: 106 STMGQALAAGGVVVHMQSMGG--GGAPRINVSAD-----GAYVDAGGEQLWVDVLRAALA 158
Query: 120 HGLTPLSWTDYLYLSVGGTLSNAGIGGQTFRFGPQISNVLELDVITGQGNIVTCSQEKNS 179
G+ P SWTDYL+L+VGGTLSNAG+ GQT+R GPQISNVLELDVITG G VTCS+ NS
Sbjct: 159 RGVAPRSWTDYLHLTVGGTLSNAGVSGQTYRHGPQISNVLELDVITGHGETVTCSKAVNS 218
Query: 180 EVFYAVLGGLGQFGVITRARILLGPAPTRANQEHLISFD--------------RRNDSNG 225
++F AVLGGLGQFGVITRAR+ + PAP RA L+ D R + S+G
Sbjct: 219 DLFDAVLGGLGQFGVITRARVAVEPAPARARWVRLVYADFAAFSADQERLVAARPDGSHG 278
Query: 226 A-DYVEGMLLLNKPPLILS------FYPPSDHPRITSLVTQYGIT--YVLELVKYYDTN- 275
YVEG + L L ++ F+ +D R+ +L T Y +E Y N
Sbjct: 279 PWSYVEGAVYLAGRGLAVALKSSGGFFSDADAARVVALAAARNATAVYSIEATLNYAANA 338
Query: 276 SQANITEEVDNLVKGLKFVPTFMFQKDVTYEEFLDRVHSEELILRSKGLWDVPHPWLNMF 335
+ +++ V + L F F F +DVTYEEFLDRV+ EE L GLW VPHPWLN+F
Sbjct: 339 TPSSVDAAVAAALGDLHFEEGFSFSRDVTYEEFLDRVYGEEEALEKAGLWRVPHPWLNLF 398
Query: 336 IPKSRISDFNEGVFKGIILKQNISAGIVLVYPMNRNKWDDRMSAITP--DEDVFYTLALL 393
+P SRI+DF+ GVFKGI+ AG +++YP+N++KWD MSA+TP +E+VFY ++LL
Sbjct: 399 VPGSRIADFDRGVFKGILQTATDIAGPLIIYPVNKSKWDAAMSAVTPEGEEEVFYVVSLL 458
Query: 394 HAAKEMDEVKTFQAQNHQILQFCNDAGIKVKEYLSGNKTHQEWVEHFGAKWQQFEERKAK 453
+A D V +AQN +IL+FC+ AGI K YL+ + +WV HFGAKW +F +RK K
Sbjct: 459 FSAVAND-VAALEAQNRRILRFCDLAGIGYKAYLAHYDSRGDWVRHFGAKWDRFVQRKDK 517
Query: 454 FDPKRILSPGQGIF 467
+DPK++LSPGQ IF
Sbjct: 518 YDPKKLLSPGQDIF 531
>FAS5_RHOFA (P46377) Hypothetical 47.9 kDa oxidoreductase in
fasciation locus (ORF5)
Length = 438
Score = 205 bits (522), Expect = 2e-52
Identities = 147/457 (32%), Positives = 213/457 (46%), Gaps = 38/457 (8%)
Query: 9 LSQASTDYGHIIHENPFAVLEPTSISDIANLINYSNSLPHSFTISPRGQAHSVLGQAMTQ 68
L+ A D+G+ IH P V+ P +++D+ + Y+ + + +++ RG HS GQ
Sbjct: 12 LTSAGADFGNCIHAKPPVVVVPRTVADVQEALRYTAA--RNLSLAVRGSGHSTYGQCQAD 69
Query: 69 NGIVVNMTQLNWYRN-GSGIVVSDCDVKNPLGCYVDVGGEQLWIDVLNATLKHGLTPLSW 127
G+V++M + N + SG D V+ W DV+ ATL TP
Sbjct: 70 GGVVLDMKRFNTVHDVRSGQATIDAGVR--------------WSDVVAATLSRQQTPPVL 115
Query: 128 TDYLYLSVGGTLSNAGIGGQTFRFGPQISNVLELDVITGQGNIVTCSQEKNSEVFYAVLG 187
TDYL +VGGTLS G GG + FG Q NV L V+TG G+ CS NSE+F AV G
Sbjct: 116 TDYLGTTVGGTLSVGGFGGSSHGFGLQTDNVDSLAVVTGSGDFRECSAVSNSELFDAVRG 175
Query: 188 GLGQFGVITRARILLGPAPTRANQEHLISFDRRNDSNGADYVEGMLLLNKPPLILSFYPP 247
GLGQFGVI A I L A Q L SN ++ L L
Sbjct: 176 GLGQFGVIVNATIRLTAAHESVRQYKL------QYSNLGVFLGDQLRAMSNRLF------ 223
Query: 248 SDHPRITSLVTQYG-ITYVLELVKYYDTNSQANITEEVDNLVKGLKFVPTFMFQKDVTYE 306
DH + V G + Y L+L KY+ + + D L+ L++ + DV Y
Sbjct: 224 -DHVQGRIRVDADGHLRYRLDLAKYFTPPRR----PDDDALLSSLQYDSCAEYNSDVDYG 278
Query: 307 EFLDRVHSEELILRSKGLWDVPHPWLNMFIPKSRISDFNEGVFKGIILKQNISAGIVLVY 366
+F++R+ +EL LR G W PHPW ++ IP +I F E + ++G+++VY
Sbjct: 279 DFINRMADQELDLRHTGEWFYPHPWASLLIPADKIEQFIETTSSSLTDDLG-NSGLIMVY 337
Query: 367 PMNRNKWDDRMSAITPDEDVFYTLALLHAAKEMDEVKTFQAQNHQILQFCNDAGIKVKEY 426
P+ I P D F+ LA+L A E + A N + + D G
Sbjct: 338 PIPTTPITAPFIPI-PHCDTFFMLAVLRTASPGAEARMI-ASNRLLYEQARDVGGVAYAV 395
Query: 427 LSGNKTHQEWVEHFGAKWQQFEERKAKFDPKRILSPG 463
+ + +W HFG++WQ K +FDP RIL+PG
Sbjct: 396 NAVPMSPGDWCTHFGSRWQAIARAKRRFDPYRILAPG 432
>HDNO_ARTOX (P08159) 6-hydroxy-D-nicotine oxidase (EC 1.5.3.6)
(6-HDNO)
Length = 458
Score = 65.1 bits (157), Expect = 4e-10
Identities = 47/189 (24%), Positives = 83/189 (43%), Gaps = 19/189 (10%)
Query: 22 ENPFAVLEPTSISDIANLINYSNSLPHSFTISPRGQAHSVLGQAMTQNGIVVNMTQLNWY 81
+ P + S D+A + Y+ + IS R H+ G A GIV+++ +N
Sbjct: 36 QRPSLIARCLSAGDVAKSVRYA--CDNGLEISVRSGGHNPNGYATNDGGIVLDLRLMNSI 93
Query: 82 RNGSGIVVSDCDVKNPLGCYVDVGGEQLWIDVLNATLKHGLTPLSWTDYLYLSVG--GTL 139
+ G +GG + D++ K GL ++ ++ VG G
Sbjct: 94 HIDTA------------GSRARIGGGVISGDLVKEAAKFGLAAVTG---MHPKVGFCGLA 138
Query: 140 SNAGIGGQTFRFGPQISNVLELDVITGQGNIVTCSQEKNSEVFYAVLGGLGQFGVITRAR 199
N G+G T ++G N+L ++T G+++ CS ++ E+F+AV G FGV+T
Sbjct: 139 LNGGVGFLTPKYGLASDNILGATLVTATGDVIYCSDDERPELFWAVRGAGPNFGVVTEVE 198
Query: 200 ILLGPAPTR 208
+ L P +
Sbjct: 199 VQLYELPRK 207
>ALO_DEBHA (Q6BZA0) D-arabinono-1,4-lactone oxidase (EC 1.1.3.37)
(ALO) (L-galactono-gamma-lactone oxidase)
Length = 557
Score = 60.1 bits (144), Expect = 1e-08
Identities = 53/206 (25%), Positives = 89/206 (42%), Gaps = 24/206 (11%)
Query: 24 PFAVLEPTSISDIANLINYSNSLPHSFTISPRGQAHSVLGQAMTQNGIVVNMTQLNWY-- 81
P A+ +P ++ +I L++ + + TI G HS MT+ + N+ + N
Sbjct: 30 PQAIFQPRTVDEIRELVDQARI--NGKTIMTVGSGHSPSDMTMTKEWLC-NLDRFNQVLK 86
Query: 82 --------RNGSGIVVSDCDVKNPLGCYVDVGGEQLWIDVLNATLKHG-LTPLSWTDYLY 132
RNG G V D+ GC I LN LK L +
Sbjct: 87 KEEFSGPTRNGEGEEVKFVDLTVQAGCR---------IYELNRYLKENELAIQNLGSISD 137
Query: 133 LSVGGTLSNAGIGGQTFRFGPQISNVLELDVITGQGNIVTCSQEKNSEVFYAVLGGLGQF 192
S+ G +S G G T G V+ ++++ G ++TCS +N+++F A + LG+
Sbjct: 138 QSMAGVIST-GTHGSTQYHGLVSQQVVSIEIMNSAGKLITCSSMENTQLFKAAMLSLGKI 196
Query: 193 GVITRARILLGPAPTRANQEHLISFD 218
G+IT + P T +++ +I FD
Sbjct: 197 GIITHVTLRTIPKYTIKSKQEIIKFD 222
>ALO_YARLI (Q6CG88) D-arabinono-1,4-lactone oxidase (EC 1.1.3.37)
(ALO) (L-galactono-gamma-lactone oxidase)
Length = 526
Score = 58.5 bits (140), Expect = 3e-08
Identities = 54/197 (27%), Positives = 84/197 (42%), Gaps = 15/197 (7%)
Query: 24 PFAVLEPTSISDIANLINYSNSLPHSFTISPRGQAHSVLGQAMTQNGIVVNMTQLNWYRN 83
P +P SI ++ ++ + L TI G AHS MT W N
Sbjct: 26 PSLYFQPASIEELQAIVTRARDLGK--TIMVVGSAHSPSDLTMTSQ----------WLVN 73
Query: 84 GSGIVVSDCDVKNPLGCYVDVGGEQ-LWIDVLNATLKH-GLTPLSWTDYLYLSVGGTLSN 141
+ + + G Y DV E + I LN LK GL + SV G +S
Sbjct: 74 LDKLSKAVSFKPHTSGLYTDVTVEAGIRIHQLNEVLKRKGLAMQNLGSISDQSVAGIIST 133
Query: 142 AGIGGQTFRFGPQISNVLELDVITGQGNIVTCSQEKNSEVFYAVLGGLGQFGVITRARIL 201
G G + G ++ L ++ G ++TCS ++N +F A L LG+ G+I A +
Sbjct: 134 -GTHGSSAYHGLVSQQIVSLTIMIASGELLTCSPDENPTLFRAALLSLGKLGIIVYATLR 192
Query: 202 LGPAPTRANQEHLISFD 218
PA T + +H+I+F+
Sbjct: 193 TVPAYTIHSTQHVITFE 209
>ALO_CANGA (Q6FS20) D-arabinono-1,4-lactone oxidase (EC 1.1.3.37)
(ALO) (L-galactono-gamma-lactone oxidase)
Length = 525
Score = 51.6 bits (122), Expect = 4e-06
Identities = 48/207 (23%), Positives = 89/207 (42%), Gaps = 17/207 (8%)
Query: 15 DYGHIIHENPFAVLEPTSISDIANLINYSNSLPHSFTISPRGQAHSVLGQAMTQNGIVVN 74
++ I P +P+S+ ++ ++ + + TI G HS +T ++ N
Sbjct: 16 NWAGIYSSRPEWYFQPSSVDEVVEIVKAAKL--KNKTIVTVGSGHSPSNMCVTDEWMM-N 72
Query: 75 MTQLNWYRNGSGIVVSDCDVKNPLGCYVDV---GGEQLWIDVLNATLKHGLTPLSWTDYL 131
+ ++N + V+N Y DV GG +L+ + + G S
Sbjct: 73 LDKMNKLLDF---------VENEDKTYADVTIQGGTRLY-KIHKILREKGYAMQSLGSIS 122
Query: 132 YLSVGGTLSNAGIGGQTFRFGPQISNVLELDVITGQGNIVTCSQEKNSEVFYAVLGGLGQ 191
S+GG +S G F G S ++ L V+ G+G ++ ++ N EVF A LG+
Sbjct: 123 EQSIGGIISTGTHGSSPFH-GLVSSTIVNLTVVNGKGEVLFLDEKSNPEVFRAATLSLGK 181
Query: 192 FGVITRARILLGPAPTRANQEHLISFD 218
G+I A + + PA + + +I F+
Sbjct: 182 IGIIVGATVRVVPAFNIKSTQEVIKFE 208
>DHCR_HUMAN (Q15392) 24-dehydrocholesterol reductase precursor (EC
1.3.1.-) (3-beta-hydroxysterol delta-24-reductase)
(Seladin-1) (Diminuto/dwarf1 homolog)
Length = 516
Score = 50.8 bits (120), Expect = 7e-06
Identities = 42/190 (22%), Positives = 77/190 (40%), Gaps = 28/190 (14%)
Query: 122 LTPLSWT-----DYLYLSVGGTLSNAGIGGQTFRFGPQISNVLELDVITGQGNIVTCSQE 176
LT + WT + L+VGG + GI + ++G +++ G+ V C+
Sbjct: 146 LTSIGWTLPVLPELDDLTVGGLIMGTGIESSSHKYGLFQHICTAYELVLADGSFVRCTPS 205
Query: 177 KNSEVFYAVLGGLGQFGVITRARILLGPA---------PTRANQEHLISFDRRNDSNGAD 227
+NS++FYAV G G + A I + PA P R + F +
Sbjct: 206 ENSDLFYAVPWSCGTLGFLVAAEIRIIPAKKYVKLRFEPVRGLEAICAKFTHESQRQENH 265
Query: 228 YVEGMLLLNKPPLILS--FYPPSDHPRITSLVTQYGITYVLELVKYYDTNSQANITEEVD 285
+VEG+L +I++ ++ ++ S+ Y + + Y TN +
Sbjct: 266 FVEGLLYSLDEAVIMTGVMTDEAEPSKLNSIGNYYKPWFFKHVENYLKTNRE-------- 317
Query: 286 NLVKGLKFVP 295
GL+++P
Sbjct: 318 ----GLEYIP 323
>ALO_CANAL (O93852) D-arabinono-1,4-lactone oxidase (EC 1.1.3.37)
(ALO) (L-galactono-gamma-lactone oxidase)
Length = 557
Score = 47.8 bits (112), Expect = 6e-05
Identities = 51/198 (25%), Positives = 82/198 (40%), Gaps = 7/198 (3%)
Query: 24 PFAVLEPTSISDIANLINYSNSLPHSFTISPRGQAHSVLGQAMTQNGIVVNMTQLNWYRN 83
P A+ +P ++ +I LI + H TI G HS MT + N+ + N
Sbjct: 29 PQAIFQPRNVEEIQELIKQARL--HGKTIMTVGSGHSPSDLTMTTEWLC-NLDKFNHVLL 85
Query: 84 GSGIVVSDCDVKN-PLGCYVDVGGEQ-LWIDVLNATLK-HGLTPLSWTDYLYLSVGGTLS 140
+ P +VD+ E I LN LK + L + S+ G +S
Sbjct: 86 EEPYYAPKSPTDDTPEIKFVDLTVEAGTRIFELNEYLKRNNLAIQNLGSISDQSIAGLIS 145
Query: 141 NAGIGGQTFRFGPQISNVLELDVITGQGNIVTCSQEKNSEVFYAVLGGLGQFGVITRARI 200
G G T G V+ + + G ++TCS E F A+L LG+ G+IT +
Sbjct: 146 T-GTHGSTQYHGLVSQQVVSVKFLNSAGELITCSSVDKPEYFRAILLSLGKIGIITHVTL 204
Query: 201 LLGPAPTRANQEHLISFD 218
P T +++ +I+F+
Sbjct: 205 RTCPKYTIKSKQEIINFE 222
>ALO_ASHGO (Q752Y3) D-arabinono-1,4-lactone oxidase (EC 1.1.3.37)
(ALO) (L-galactono-gamma-lactone oxidase)
Length = 532
Score = 47.4 bits (111), Expect = 8e-05
Identities = 29/90 (32%), Positives = 46/90 (50%), Gaps = 1/90 (1%)
Query: 134 SVGGTLSNAGIGGQTFRFGPQISNVLELDVITGQGNIVTCSQEKNSEVFYAVLGGLGQFG 193
SVGG +S G + G S + L ++ G+G +V E EVF A + LG+ G
Sbjct: 129 SVGGIISTGTHGSSPYH-GLVSSQYVNLTLVNGRGELVFLDSEHEPEVFRAAMLSLGKLG 187
Query: 194 VITRARILLGPAPTRANQEHLISFDRRNDS 223
+I RA I + PA + + +I+F+ D+
Sbjct: 188 IIVRATIRVVPAFNIHSTQEVINFETLLDN 217
>MCRA_STRLA (P43485) Mitomycin radical oxidase (EC 1.5.3.-)
Length = 447
Score = 45.1 bits (105), Expect = 4e-04
Identities = 30/95 (31%), Positives = 45/95 (46%), Gaps = 1/95 (1%)
Query: 110 WIDVLNATLKHGLTPLSWTDYLYLSVGGTLSNAGIGGQTFRFGPQISNVLELDVITGQGN 169
W VL T HGL PL+ + +VG L G G RFG +V L ++T G
Sbjct: 100 WRKVLEHTAPHGLAPLNGSSPNVGAVG-YLVGGGAGLLGRRFGYAADHVRRLRLVTADGR 158
Query: 170 IVTCSQEKNSEVFYAVLGGLGQFGVITRARILLGP 204
+ + + ++F+AV GG FG++ + L P
Sbjct: 159 LRDVTAGTDPDLFWAVRGGKDNFGLVVGMEVDLFP 193
>RETO_ESCCA (P30986) Reticuline oxidase precursor (EC 1.21.3.3)
(Berberine-bridge-forming enzyme) (BBE)
(Tetrahydroprotoberberine synthase)
Length = 538
Score = 44.3 bits (103), Expect = 7e-04
Identities = 49/188 (26%), Positives = 84/188 (44%), Gaps = 17/188 (9%)
Query: 24 PFAVLEPTSISDIANLINYSNSLPHSFTISPRGQAHSVLGQAMTQNGIVVNMTQLNWYRN 83
P A++ P S +++N I S+TI R HS G + T + + + +N R
Sbjct: 71 PSAIILPGSKEELSNTIRCIRK--GSWTIRLRSGGHSYEGLSYTSDTPFILIDLMNLNR- 127
Query: 84 GSGIVVSDCDVKNPLGCYVDVGGE--QLWIDVLNATLKHGLTPLSWTDYLYLSVGGTLSN 141
V D + + +V+ G +L+ + ++ K G T W + GG +S
Sbjct: 128 ----VSIDLESET---AWVESGSTLGELYYAITESSSKLGFTA-GWCPTV--GTGGHISG 177
Query: 142 AGIGGQTFRFGPQISNVLELDVITGQGNIVTCSQEKNSEVFYAVL-GGLGQFGVITRARI 200
G G + ++G NV++ +I G I+ Q +VF+A+ GG G +G I +I
Sbjct: 178 GGFGMMSRKYGLAADNVVDAILIDANGAILD-RQAMGEDVFWAIRGGGGGVWGAIYAWKI 236
Query: 201 LLGPAPTR 208
L P P +
Sbjct: 237 KLLPVPEK 244
>DIM_PEA (P93472) Cell elongation protein diminuto
Length = 567
Score = 44.3 bits (103), Expect = 7e-04
Identities = 33/121 (27%), Positives = 57/121 (46%), Gaps = 20/121 (16%)
Query: 133 LSVGGTLSNAGIGGQTFRFGPQISNVLELDVITGQGNIVTCSQEKN-SEVFYAVLGGLGQ 191
L+VGG ++ GI G + ++G V+ ++I G++V +++ S++FYA+ G
Sbjct: 158 LTVGGLINGYGIEGSSHKYGLFSDTVVAFEIILADGSLVKATKDNEYSDLFYAIPWSQGT 217
Query: 192 FGVITRARILLGP---------APTRAN-----QEHLISF-----DRRNDSNGADYVEGM 232
G++ A + L P P N Q + SF D+ ND D+VE M
Sbjct: 218 LGLLVAAEVKLIPIKEYMKLTYKPVVGNLKDIAQAYSDSFAPRDGDQDNDEKVPDFVETM 277
Query: 233 L 233
+
Sbjct: 278 I 278
>ALO_KLULA (Q6CSY3) D-arabinono-1,4-lactone oxidase (EC 1.1.3.37)
(ALO) (L-galactono-gamma-lactone oxidase)
Length = 525
Score = 43.9 bits (102), Expect = 9e-04
Identities = 51/197 (25%), Positives = 82/197 (40%), Gaps = 15/197 (7%)
Query: 24 PFAVLEPTSISDIANLINYSNSLPHSFTISPRGQAHSVLGQAMTQNGIVVNMTQLNWYRN 83
P +P SI ++ ++ + + TI G HS +T ++ N+ LN
Sbjct: 27 PQLYFQPNSIDEVVQIVKAA--IEQGKTIVTVGSGHSPSDMCVTDQWLM-NLDNLN---- 79
Query: 84 GSGIVVSDCDVKNPLGCYVDVGGEQ-LWIDVLNATL-KHGLTPLSWTDYLYLSVGGTLSN 141
VV + K L Y DV E L I L+ L + G + SV G +S
Sbjct: 80 ---SVVEFKENKEEL--YADVTVEAGLRIYQLSEILAEKGYAIQNLGSISEQSVAGIIST 134
Query: 142 AGIGGQTFRFGPQISNVLELDVITGQGNIVTCSQEKNSEVFYAVLGGLGQFGVITRARIL 201
G + G S + L ++ G+G +V E + EVF A LG+ G+I +A I
Sbjct: 135 GTHGSSPYH-GLVSSQYVNLTIVNGKGEVVFLDSENSPEVFRAATLSLGKIGIIVKATIR 193
Query: 202 LGPAPTRANQEHLISFD 218
+ P + + +I F+
Sbjct: 194 VIPEFNIKSTQEVIHFE 210
>DLD2_YEAST (P46681) D-lactate dehydrogenase [cytochrome] 2,
mitochondrial precursor (EC 1.1.2.4) (D-lactate
ferricytochrome C oxidoreductase) (D-LCR) (Actin
interacting protein 2)
Length = 530
Score = 41.6 bits (96), Expect = 0.004
Identities = 46/195 (23%), Positives = 84/195 (42%), Gaps = 20/195 (10%)
Query: 27 VLEPTSISDIANLINYSNSLPHSFTISPRGQAHSVLGQAMTQ-NGIVVNMTQLNWYRNGS 85
VL P S+ ++ ++NY N + P+G ++G ++ + +++++ LN R+
Sbjct: 105 VLRPKSVEKVSLILNYCND--EKIAVVPQGGNTGLVGGSVPIFDELILSLANLNKIRDFD 162
Query: 86 GIV-VSDCDVKNPLGCYVDVGGEQLWIDVLNATLKHGLTPLSWTDYLYLSVGGTLSNAGI 144
+ + CD L + EQ ++ L+ K VGG ++
Sbjct: 163 PVSGILKCDAGVILENANNYVMEQNYMFPLDLGAKGSC-----------HVGGVVATNAG 211
Query: 145 GGQTFRFGPQISNVLELDVITGQGNIVTC--SQEKNS---EVFYAVLGGLGQFGVITRAR 199
G + R+G +VL L+V+ G IV S K++ ++ +G G G+IT
Sbjct: 212 GLRLLRYGSLHGSVLGLEVVMPNGQIVNSMHSMRKDNTGYDLKQLFIGSEGTIGIITGVS 271
Query: 200 ILLGPAPTRANQEHL 214
IL P P N +L
Sbjct: 272 ILTVPKPKAFNVSYL 286
>MUB2_BACCR (Q815R9) UDP-N-acetylenolpyruvoylglucosamine reductase 2
(EC 1.1.1.158) (UDP-N-acetylmuramate dehydrogenase 2)
Length = 305
Score = 41.2 bits (95), Expect = 0.006
Identities = 42/149 (28%), Positives = 69/149 (46%), Gaps = 24/149 (16%)
Query: 30 PTSISDIANLINYSNSLPHSFTISPRGQAHSVLGQAMTQNGIVVNMTQLNWYRNGSGIVV 89
PT+ +I +I Y+N ++ ++ G +V+ + GI V++ + +G+ V
Sbjct: 43 PTNYDEIQEVIKYANE--YNIPVTFLGNGSNVIIKDGGIRGITVSLIHI------TGVTV 94
Query: 90 SDCDVKNPLGCYVDVGGEQLWIDVLNATLKHGLTPLSWTDYLYLSVGGTL-SNAGIGGQT 148
+ + G + IDV L H LT L + + SVGG L NAG
Sbjct: 95 TGTTIVAQCGAAI--------IDVSRIALDHNLTGLEFACGIPGSVGGALYMNAG----- 141
Query: 149 FRFGPQISNVL-ELDVITGQGNIVTCSQE 176
+G +IS VL E V+TG G + T ++E
Sbjct: 142 -AYGGEISFVLTEAVVMTGDGELRTLTKE 169
>MUB2_BACAN (Q81XC5) UDP-N-acetylenolpyruvoylglucosamine reductase 2
(EC 1.1.1.158) (UDP-N-acetylmuramate dehydrogenase 2)
Length = 305
Score = 41.2 bits (95), Expect = 0.006
Identities = 42/149 (28%), Positives = 69/149 (46%), Gaps = 24/149 (16%)
Query: 30 PTSISDIANLINYSNSLPHSFTISPRGQAHSVLGQAMTQNGIVVNMTQLNWYRNGSGIVV 89
PT+ +I +I Y+N ++ ++ G +V+ + GI V++ + +G+ V
Sbjct: 43 PTNYDEIQEVIKYANK--YNIPVTFLGNGSNVIIKDGGIRGITVSLIHI------TGVTV 94
Query: 90 SDCDVKNPLGCYVDVGGEQLWIDVLNATLKHGLTPLSWTDYLYLSVGGTL-SNAGIGGQT 148
+ + G + IDV L H LT L + + SVGG L NAG
Sbjct: 95 TGTTIVAQCGAAI--------IDVSRIALDHNLTGLEFACGIPGSVGGALYMNAG----- 141
Query: 149 FRFGPQISNVL-ELDVITGQGNIVTCSQE 176
+G +IS VL E V+TG G + T ++E
Sbjct: 142 -AYGGEISFVLTEAVVMTGDGELRTLTKE 169
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.319 0.138 0.414
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 58,677,295
Number of Sequences: 164201
Number of extensions: 2602736
Number of successful extensions: 5753
Number of sequences better than 10.0: 45
Number of HSP's better than 10.0 without gapping: 25
Number of HSP's successfully gapped in prelim test: 20
Number of HSP's that attempted gapping in prelim test: 5694
Number of HSP's gapped (non-prelim): 52
length of query: 467
length of database: 59,974,054
effective HSP length: 114
effective length of query: 353
effective length of database: 41,255,140
effective search space: 14563064420
effective search space used: 14563064420
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 68 (30.8 bits)
Medicago: description of AC148775.11


