

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148755.7 - phase: 0
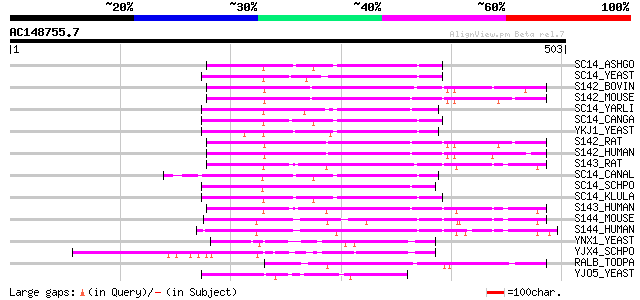
(503 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
SC14_ASHGO (Q75DK1) SEC14 cytosolic factor (Phosphatidylinositol... 94 6e-19
SC14_YEAST (P24280) SEC14 cytosolic factor (Phosphatidylinositol... 94 1e-18
S142_BOVIN (P58875) SEC14-like protein 2 (Alpha-tocopherol assoc... 93 2e-18
S142_MOUSE (Q99J08) SEC14-like protein 2 (Alpha-tocopherol assoc... 91 9e-18
SC14_YARLI (P45816) SEC14 cytosolic factor (Phosphatidylinositol... 90 2e-17
SC14_CANGA (P53989) SEC14 cytosolic factor (Phosphatidylinositol... 90 2e-17
YKJ1_YEAST (P33324) 36.1 kDa protein in BUD2-MIF2 intergenic region 89 2e-17
S142_RAT (Q99MS0) SEC14-like protein 2 (Alpha-tocopherol associa... 89 2e-17
S142_HUMAN (O76054) SEC14-like protein 2 (Alpha-tocopherol assoc... 89 2e-17
S143_RAT (Q9Z1J8) SEC14-like protein 3 (45 kDa secretory protein... 89 3e-17
SC14_CANAL (P46250) SEC14 cytosolic factor (Phosphatidylinositol... 89 3e-17
SC14_SCHPO (Q10137) Sec14 cytosolic factor (Phosphatidylinositol... 88 6e-17
SC14_KLULA (P24859) SEC14 cytosolic factor (Phosphatidylinositol... 87 1e-16
S143_HUMAN (Q9UDX4) SEC14-like protein 3 (Tocopherol-associated ... 87 1e-16
S144_MOUSE (Q8R0F9) SEC14-like protein 4 83 1e-15
S144_HUMAN (Q9UDX3) SEC14-like protein 4 (Tocopherol-associated ... 82 3e-15
YNX1_YEAST (P53860) Hypothetical 40.7 kDa protein in CSL4-URE2 i... 74 1e-12
YJX4_SCHPO (Q9UU99) Protein C23B6.04c in chromosome III 74 1e-12
RALB_TODPA (P49193) Retinal-binding protein (RALBP) 67 1e-10
YJO5_YEAST (P47008) Hypothetical 34.4 kDa protein in IDS2-MPI2 i... 67 1e-10
>SC14_ASHGO (Q75DK1) SEC14 cytosolic factor
(Phosphatidylinositol/phosphatidylcholine transfer
protein) (PI/PC TP)
Length = 311
Score = 94.4 bits (233), Expect = 6e-19
Identities = 76/224 (33%), Positives = 101/224 (44%), Gaps = 16/224 (7%)
Query: 179 DVILLKFLRARDFKVKEAFTMIKNTILWRKEFGIEELMDEKLGDELEKVV-----YMHGF 233
D LL+FLRAR F V A M +N WRKE G++ + ++ +E V Y H
Sbjct: 56 DSTLLRFLRARKFDVAAARAMFENCEKWRKENGVDTIFEDFHYEEKPLVAKFYPQYYHKT 115
Query: 234 DKEGHPVCYNIYGEFQNKELYNKTFSDEEKRHNFLKWRIQF-----LEKSIRNLDFNHGG 288
DK+G PV G E+Y T +E+ L W + L S R D
Sbjct: 116 DKDGRPVYIEELGAVNLTEMYKIT--TQERMLKNLIWEYESFSRYRLPASSRQADCLVET 173
Query: 289 VCTIVHVNDLKDSPGPGKWELRQATKQALQLFQDNYPEFVAKQVFINVPWWYLAVNRMIS 348
CTI+ DLK ++ ++A + Q+ YPE + K IN P+ + A R+
Sbjct: 174 SCTIL---DLKGISISAAAQVLSYVREASNIGQNYYPERMGKFYMINAPFGFSAAFRLFK 230
Query: 349 PFLTQRTKSKFVFAGPSKSTETLLSYIAPEQLPVKYGGLSKDGE 392
PFL T SK G S E LL I E LPVK+GG S E
Sbjct: 231 PFLDPVTVSKIFILGSSYQKE-LLKQIPAENLPVKFGGQSDVSE 273
>SC14_YEAST (P24280) SEC14 cytosolic factor
(Phosphatidylinositol/phosphatidylcholine transfer
protein) (PI/PC TP)
Length = 303
Score = 93.6 bits (231), Expect = 1e-18
Identities = 71/230 (30%), Positives = 106/230 (45%), Gaps = 20/230 (8%)
Query: 175 DETSDVILLKFLRARDFKVKEAFTMIKNTILWRKEFGIEELMDEKLGDELEKVV-----Y 229
+ D LL+FLRAR F V+ A M +N WRK++G + ++ + DE + Y
Sbjct: 50 ERLDDSTLLRFLRARKFDVQLAKEMFENCEKWRKDYGTDTILQDFHYDEKPLIAKFYPQY 109
Query: 230 MHGFDKEGHPVCYNIYGEFQNKELYNKTFSDEEKRHNF-------LKWRIQFLEKSIRNL 282
H DK+G PV + G E+ NK S+E N +++R+ ++ +L
Sbjct: 110 YHKTDKDGRPVYFEELGAVNLHEM-NKVTSEERMLKNLVWEYESVVQYRLPACSRAAGHL 168
Query: 283 DFNHGGVCTIVHVNDLKDSPGPGKWELRQATKQALQLFQDNYPEFVAKQVFINVPWWYLA 342
V T + DLK + + ++A + Q+ YPE + K IN P+ +
Sbjct: 169 ------VETSCTIMDLKGISISSAYSVMSYVREASYISQNYYPERMGKFYIINAPFGFST 222
Query: 343 VNRMISPFLTQRTKSKFVFAGPSKSTETLLSYIAPEQLPVKYGGLSKDGE 392
R+ PFL T SK G S E LL I E LPVK+GG S+ E
Sbjct: 223 AFRLFKPFLDPVTVSKIFILGSSYQKE-LLKQIPAENLPVKFGGKSEVDE 271
>S142_BOVIN (P58875) SEC14-like protein 2 (Alpha-tocopherol
associated protein) (TAP) (bTAP) (Fragment)
Length = 387
Score = 92.8 bits (229), Expect = 2e-18
Identities = 82/340 (24%), Positives = 151/340 (44%), Gaps = 36/340 (10%)
Query: 179 DVILLKFLRARDFKVKEAFTMIKNTILWRKEFGIEELMDEKLGDELEKVVY--MHGFDKE 236
D LL++LRAR+F ++++ M++ + +RK+ I+ +M + + +++ + M G+D E
Sbjct: 35 DYFLLRWLRARNFNLQKSEAMLRKHVEFRKQKDIDNIMSWQPPEVVQQYLSGGMCGYDLE 94
Query: 237 GHPVCYNIYGEFQNKELYNKTFSDEEKRHNFLKWRIQFLEKSIRNLDFNHGGVCTIVHVN 296
G P+ Y+I G K L + + + L++ +R + + +
Sbjct: 95 GSPIWYDIIGPLDAKGLLLSASKQDLFKTKMRDCEL-LLQECVRQTEKMGKKIEATTLIY 153
Query: 297 DLKDSPGPGKWELR-QATKQALQLFQDNYPEFVAKQVFINVPWWYLAVNRMISPFLTQRT 355
D + W+ +A + L +F++NYPE + + + P + ++ PFL++ T
Sbjct: 154 DCEGLGLKHLWKPAVEAYGEFLCMFEENYPETLKRLFIVKAPKLFPVAYNLVKPFLSEDT 213
Query: 356 KSKFVFAGPSKSTETLLSYIAPEQLPVKYGGLSKDGEFGN-------------------S 396
+ K G + E LL YI+P+QLPV+YGG D + GN
Sbjct: 214 RKKIQVLGANWK-EVLLKYISPDQLPVEYGGTMTDPD-GNPKCKSKINYGGDIPKKYYVR 271
Query: 397 DSVTE-----ITIRPASKHTVEFPVT-EKCLLSWEVRVIGWEVRYGAEFVPSNEGSYTVI 450
D V + + I S H VE+ + C+L W+ G ++ +G F+ + G
Sbjct: 272 DQVKQQYEHSVQISRGSSHQVEYEILFPGCVLRWQFMSDGSDIGFGI-FLKTKVGERQRA 330
Query: 451 VQKARKVASSEEAVLC----NSFKINEPGKVVLTIDNTSS 486
+ + S + S ++PG VL DNT S
Sbjct: 331 GEMREVLPSQRYSAHLVPEDGSLTCSDPGIYVLRFDNTYS 370
>S142_MOUSE (Q99J08) SEC14-like protein 2 (Alpha-tocopherol
associated protein) (TAP)
Length = 403
Score = 90.5 bits (223), Expect = 9e-18
Identities = 84/341 (24%), Positives = 151/341 (43%), Gaps = 38/341 (11%)
Query: 179 DVILLKFLRARDFKVKEAFTMIKNTILWRKEFGIEELMDEKLGDELEKVVY--MHGFDKE 236
D LL++LRAR F ++++ M++ + +RK+ I++++ + + +++ + G+D +
Sbjct: 35 DYFLLRWLRARSFDLQKSEAMLRKHVEFRKQKDIDKIISWQPPEVIQQYLSGGRCGYDLD 94
Query: 237 GHPVCYNIYGEFQNKELYNKTFSDEEKRHNFLKWRIQFLEKSIRNLDFNHGGVCTIVHVN 296
G PV Y+I G K L + R + L++ I+ + TI +
Sbjct: 95 GCPVWYDIIGPLDAKGLLFSASKQDLLRTKMRDCEL-LLQECIQQTTKLGKKIETITMIY 153
Query: 297 DLKDSPGPGKWELR-QATKQALQLFQDNYPEFVAKQVFINVPWWYLAVNRMISPFLTQRT 355
D + W+ +A + L +F++NYPE + + + P + +I PFL++ T
Sbjct: 154 DCEGLGLKHLWKPAVEAYGEFLTMFEENYPETLKRLFVVKAPKLFPVAYNLIKPFLSEDT 213
Query: 356 KSKFVFAGPSKSTETLLSYIAPEQLPVKYGGLSKDGEFGN-------------------S 396
+ K + G + E LL +I+P+QLPV+YGG D + GN
Sbjct: 214 RRKIMVLG-ANWKEVLLKHISPDQLPVEYGGTMTDPD-GNPKCKSKINYGGDIPKQYYVR 271
Query: 397 DSVTE-----ITIRPASKHTVEFPVT-EKCLLSWEVRVIGWEVRYGAEFVP-----SNEG 445
D V + + + S H VE+ + C+L W+ G +V +G G
Sbjct: 272 DQVKQQYEHTVQVSRGSSHQVEYEILFPGCVLRWQFMSEGSDVGFGIFLKTKMGERQRAG 331
Query: 446 SYTVIVQKARKVASSEEAVLCNSFKINEPGKVVLTIDNTSS 486
T ++ R +S + +EPG VL DNT S
Sbjct: 332 EMTEVLPNQR--YNSHMVPEDGTLTCSEPGIYVLRFDNTYS 370
>SC14_YARLI (P45816) SEC14 cytosolic factor
(Phosphatidylinositol/phosphatidylcholine transfer
protein) (PI/PC TP)
Length = 492
Score = 89.7 bits (221), Expect = 2e-17
Identities = 72/225 (32%), Positives = 105/225 (46%), Gaps = 18/225 (8%)
Query: 175 DETSDVILLKFLRARDFKVKEAFTMIKNTILWRKEFGIEELMDEKLGDELEKVV-----Y 229
D T D LL+FLRAR F V A M +N WRKEFG ++++ E ++V Y
Sbjct: 50 DRTDDATLLRFLRARKFDVPLAQEMWENCEKWRKEFGTNTILEDFWYKEKKEVAKLYPQY 109
Query: 230 MHGFDKEGHPVCYNIYGEFQNKELYNKTFSDEEKRH------NFLKWRIQFLEKSIRNLD 283
H DK+G PV G+ E+Y T + R+ +F++ R+ + + +L
Sbjct: 110 YHKTDKDGRPVYVENVGKVNIHEMYKITTQERMLRNLVWEYESFVRHRLPACSRVVGHLI 169
Query: 284 FNHGGVCTIVHVNDLKDSPGPGKWELRQATKQALQLFQDNYPEFVAKQVFINVPWWYLAV 343
CTI+ DLK ++ K A + Q+ YPE + K IN P+ + V
Sbjct: 170 ETS---CTIL---DLKGVSLSSASQVYGFLKDASNIGQNYYPERMGKFYLINAPFGFSTV 223
Query: 344 NRMISPFLTQRTKSKFVFAGPSKSTETLLSYIAPEQLPVKYGGLS 388
+I FL T SK G S E LL+ + LP+K+GG S
Sbjct: 224 FSVIKRFLDPVTVSKIHVYG-SNYKEKLLAQVPAYNLPIKFGGQS 267
>SC14_CANGA (P53989) SEC14 cytosolic factor
(Phosphatidylinositol/phosphatidylcholine transfer
protein) (PI/PC TP)
Length = 302
Score = 89.7 bits (221), Expect = 2e-17
Identities = 71/228 (31%), Positives = 101/228 (44%), Gaps = 16/228 (7%)
Query: 175 DETSDVILLKFLRARDFKVKEAFTMIKNTILWRKEFGIEELMDEKLGDELEKVV-----Y 229
+ D LL+FLRAR F V A M +N WRKE+G +M + DE V Y
Sbjct: 49 ERLDDSTLLRFLRARKFDVALAKEMFENCEKWRKEYGTNTIMQDFHYDEKPLVAKYYPQY 108
Query: 230 MHGFDKEGHPVCYNIYGEFQNKELYNKTFSDEEKRHNFLKWRIQF-----LEKSIRNLDF 284
H DK+G PV + G E+ + + +E+ L W + L R +
Sbjct: 109 YHKTDKDGRPVYFEELGAVNLTEM--EKITTQERMLKNLVWEYESVVNYRLPACSRAAGY 166
Query: 285 NHGGVCTIVHVNDLKDSPGPGKWELRQATKQALQLFQDNYPEFVAKQVFINVPWWYLAVN 344
CT++ DLK + + ++A + Q+ YPE + K IN P+ +
Sbjct: 167 LVETSCTVM---DLKGISISSAYSVLSYVREASYISQNYYPERMGKFYLINAPFGFSTAF 223
Query: 345 RMISPFLTQRTKSKFVFAGPSKSTETLLSYIAPEQLPVKYGGLSKDGE 392
R+ PFL T SK G S +E LL I E LP K+GG S+ E
Sbjct: 224 RLFKPFLDPVTVSKIFILGSSYQSE-LLKQIPAENLPSKFGGKSEVDE 270
>YKJ1_YEAST (P33324) 36.1 kDa protein in BUD2-MIF2 intergenic region
Length = 310
Score = 89.4 bits (220), Expect = 2e-17
Identities = 68/229 (29%), Positives = 105/229 (45%), Gaps = 20/229 (8%)
Query: 175 DETSDVILLKFLRARDFKVKEAFTMIKNTILWRKEFG----IEELMDEKLGDELEKVV-- 228
+ D LL+FLRAR F + + M T WR+E+G IE+ + K ++ E++
Sbjct: 47 ERLDDSTLLRFLRARKFDINASVEMFVETERWREEYGANTIIEDYENNKEAEDKERIKLA 106
Query: 229 -----YMHGFDKEGHPVCYNIYGEFQNKELYNKTFSDEEKRHNFLKWRIQFLEKSIRNLD 283
Y H DK+G P+ + G K++Y T ++++ N +K F +
Sbjct: 107 KMYPQYYHHVDKDGRPLYFEELGGINLKKMYKIT-TEKQMLRNLVKEYELFATYRVPACS 165
Query: 284 FNHGGV----CTIVHVNDLKDSPGPGKWELRQATKQALQLFQDNYPEFVAKQVFINVPWW 339
G + CT++ DLK + + K + Q+ YPE + K I+ P+
Sbjct: 166 RRAGYLIETSCTVL---DLKGISLSNAYHVLSYIKDVADISQNYYPERMGKFYIIHSPFG 222
Query: 340 YLAVNRMISPFLTQRTKSKFVFAGPSKSTETLLSYIAPEQLPVKYGGLS 388
+ + +M+ PFL T SK G S E LL I E LPVKYGG S
Sbjct: 223 FSTMFKMVKPFLDPVTVSKIFILGSSYKKE-LLKQIPIENLPVKYGGTS 270
>S142_RAT (Q99MS0) SEC14-like protein 2 (Alpha-tocopherol associated
protein) (TAP) (Supernatant protein factor) (SPF)
(Squalene transfer protein)
Length = 403
Score = 89.4 bits (220), Expect = 2e-17
Identities = 84/341 (24%), Positives = 149/341 (43%), Gaps = 38/341 (11%)
Query: 179 DVILLKFLRARDFKVKEAFTMIKNTILWRKEFGIEELMDEKLGDELEKVVY--MHGFDKE 236
D LL++LRAR F ++++ M++ + +RK+ I++++ + + +++ + G+D +
Sbjct: 35 DYFLLRWLRARSFDLQKSEAMLRKHVEFRKQKDIDKIISWQPPEVIQQYLSGGRCGYDLD 94
Query: 237 GHPVCYNIYGEFQNKELYNKTFSDEEKRHNFLKWRIQFLEKSIRNLDFNHGGVCTIVHVN 296
G PV Y+I G K L + R + E + + + TI +
Sbjct: 95 GCPVWYDIIGPLDAKGLLFSASKQDLLRTKMRDCELLLQECTQQTAKLGKK-IETITMIY 153
Query: 297 DLKDSPGPGKWELR-QATKQALQLFQDNYPEFVAKQVFINVPWWYLAVNRMISPFLTQRT 355
D + W+ +A + L +F++NYPE + + + P + +I PFL++ T
Sbjct: 154 DCEGLGLKHLWKPAVEAYGEFLTMFEENYPETLKRLFVVKAPKLFPVAYNLIKPFLSEDT 213
Query: 356 KSKFVFAGPSKSTETLLSYIAPEQLPVKYGGLSKDGEFGN-------------------S 396
+ K + G + E LL +I+P+QLPV+YGG D + GN
Sbjct: 214 RKKIMVLG-ANWKEVLLKHISPDQLPVEYGGTMTDPD-GNPKCKSKINYGGDIPKQYYVR 271
Query: 397 DSVTE-----ITIRPASKHTVEFPVT-EKCLLSWEVRVIGWEVRYGAEFVP-----SNEG 445
D V + + I S H VE+ + C+L W+ G +V +G G
Sbjct: 272 DQVKQQYEHSVQISRGSSHQVEYEILFPGCVLRWQFMSEGSDVGFGIFLKTKMGERQRAG 331
Query: 446 SYTVIVQKARKVASSEEAVLCNSFKINEPGKVVLTIDNTSS 486
T ++ R +S + +EPG VL DNT S
Sbjct: 332 EMTEVLPNQR--YNSHMVPEDGTLTCSEPGIYVLRFDNTYS 370
>S142_HUMAN (O76054) SEC14-like protein 2 (Alpha-tocopherol
associated protein) (TAP) (hTAP) (Supernatant protein
factor) (SPF) (Squalene transfer protein)
Length = 403
Score = 89.4 bits (220), Expect = 2e-17
Identities = 86/343 (25%), Positives = 152/343 (44%), Gaps = 42/343 (12%)
Query: 179 DVILLKFLRARDFKVKEAFTMIKNTILWRKEFGIEELMDEKLGDELEKVVY--MHGFDKE 236
D LL++LRAR F ++++ M++ + +RK+ I+ ++ + + +++ + M G+D +
Sbjct: 35 DYFLLRWLRARSFDLQKSEAMLRKHVEFRKQKDIDNIISWQPPEVIQQYLSGGMCGYDLD 94
Query: 237 GHPVCYNIYGEFQNKELYNKTFSDEEKRHNFLKWRIQFLEKSIRNLDFNHGGVCTIVHVN 296
G PV Y+I G K L + R + + E + + V TI +
Sbjct: 95 GCPVWYDIIGPLDAKGLLFSASKQDLLRTKMRECELLLQECAHQTTKLGRK-VETITIIY 153
Query: 297 DLKDSPGPGKWELR-QATKQALQLFQDNYPEFVAKQVFINVPWWYLAVNRMISPFLTQRT 355
D + W+ +A + L +F++NYPE + + + P + +I PFL++ T
Sbjct: 154 DCEGLGLKHLWKPAVEAYGEFLCMFEENYPETLKRLFVVKAPKLFPVAYNLIKPFLSEDT 213
Query: 356 KSKFVFAGPSKSTETLLSYIAPEQLPVKYGGLSKDGEFGN-------------------S 396
+ K + G + E LL +I+P+Q+PV+YGG D + GN
Sbjct: 214 RKKIMVLG-ANWKEVLLKHISPDQVPVEYGGTMTDPD-GNPKCKSKINYGGDIPRKYYVR 271
Query: 397 DSVTE-----ITIRPASKHTVEFPVT-EKCLLSWEVRVIGWEVRYG-------AEFVPSN 443
D V + + I S H VE+ + C+L W+ G +V +G E +
Sbjct: 272 DQVKQQYEHSVQISRGSSHQVEYEILFPGCVLRWQFMSDGADVGFGIFLKTKMGERQRAG 331
Query: 444 EGSYTVIVQKARKVASSEEAVLCNSFKINEPGKVVLTIDNTSS 486
E + + Q+ E+ L S +PG VL DNT S
Sbjct: 332 EMTEVLPNQRYNSHLVPEDGTLTCS----DPGIYVLRFDNTYS 370
>S143_RAT (Q9Z1J8) SEC14-like protein 3 (45 kDa secretory protein)
(rsec45)
Length = 400
Score = 89.0 bits (219), Expect = 3e-17
Identities = 88/348 (25%), Positives = 158/348 (45%), Gaps = 52/348 (14%)
Query: 179 DVILLKFLRARDFKVKEAFTMIKNTILWRKEFGIEELMDEKLGDELEKVV--YMHGFDKE 236
D LL++LRAR+F ++++ M++ + +RK I+ ++D + + ++K + + G+D++
Sbjct: 35 DYFLLRWLRARNFDLQKSEAMLRKYMEFRKTMDIDHILDWQPPEVIQKYMPGGLCGYDRD 94
Query: 237 GHPVCYNIYGEFQNKELYNKTFSDEEKRHNFLKWRIQFLEKSIRNLDFN----HGGVCTI 292
G PV Y+I G K L FS + + LK +++ E+ + D + TI
Sbjct: 95 GCPVWYDIIGPLDPKGL---LFS--VTKQDLLKTKMRDCERILHECDLQTERLGRKIETI 149
Query: 293 VHVNDLKDSPGPGKWE-LRQATKQALQLFQDNYPEFVAKQVFINVPWWYLAVNRMISPFL 351
V + D + W+ L + ++ L ++NYPE + + + + ++ PFL
Sbjct: 150 VMIFDCEGLGLKHFWKPLVEVYQEFFGLLEENYPETLKFMLIVKATKLFPVGYNLMKPFL 209
Query: 352 TQRTKSKFVFAGPSKSTETLLSYIAPEQLPVKYGGLSKDGEFGNSDSVTEIT-------- 403
++ T+ K V G S E LL I+PE+LP +GG D + GN +T+I
Sbjct: 210 SEDTRRKIVVLGNSWK-EGLLKLISPEELPAHFGGTLTDPD-GNPKCLTKINYGGEIPKS 267
Query: 404 ----------------IRPASKHTVEFPVT-EKCLLSWEVRVIGWEVRYGAEFVPSNEGS 446
I S H VE+ + C+L W+ G ++ +G F+ + G
Sbjct: 268 MYVRDQVKTQYEHSVQISRGSSHQVEYEILFPGCVLRWQFSSDGADIGFGV-FLKTKMGE 326
Query: 447 YTVIVQKARKVASSEEAVLCNSFKINEPGKV--------VLTIDNTSS 486
QKA ++ + N+ + E G + VL DNT S
Sbjct: 327 R----QKAGEMTEVLTSQRYNAHMVPEDGSLTCTEAGVYVLRFDNTYS 370
>SC14_CANAL (P46250) SEC14 cytosolic factor
(Phosphatidylinositol/phosphatidylcholine transfer
protein) (PI/PC TP)
Length = 301
Score = 88.6 bits (218), Expect = 3e-17
Identities = 73/259 (28%), Positives = 118/259 (45%), Gaps = 26/259 (10%)
Query: 140 SASVPPEQKPVVEKVEASLPLPPEQVSIYGIPLLADETSDVILLKFLRARDFKVKEAFTM 199
++++ PEQK +L + +Q++ G D D LL+FLRAR F +++A M
Sbjct: 26 TSNLTPEQK-------TTLDIFRQQLTELGYK---DRLDDASLLRFLRARKFDIQKAIDM 75
Query: 200 IKNTILWRKEFGIEELMDEKLGDELEKV-----VYMHGFDKEGHPVCYNIYGEFQNKELY 254
WR++FG+ ++ + +E V Y H DK+G PV + G+ ++
Sbjct: 76 FVACEKWREDFGVNTILKDFHYEEKPIVAKMYPTYYHKTDKDGRPVYFEELGKVDLVKML 135
Query: 255 NKTFSDEEKRHNFLKWRIQF-----LEKSIRNLDFNHGGVCTIVHVNDLKDSPGPGKWEL 309
T +E+ L W + L R + CT++ DL + +
Sbjct: 136 KIT--TQERMLKNLVWEYEAMCQYRLPACSRKAGYLVETSCTVL---DLSGISVTSAYNV 190
Query: 310 RQATKQALQLFQDNYPEFVAKQVFINVPWWYLAVNRMISPFLTQRTKSKFVFAGPSKSTE 369
++A ++ QD YPE + K IN P+ + ++ PFL T SK G S E
Sbjct: 191 IGYVREASKIGQDYYPERMGKFYLINAPFGFSTAFKLFKPFLDPVTVSKIHILGYSYKKE 250
Query: 370 TLLSYIAPEQLPVKYGGLS 388
LL I P+ LPVK+GG+S
Sbjct: 251 -LLKQIPPQNLPVKFGGMS 268
>SC14_SCHPO (Q10137) Sec14 cytosolic factor
(Phosphatidylinositol/phosphatidyl-choline transfer
protein) (PI/PC TP) (Sporulation-specific protein 20)
Length = 286
Score = 87.8 bits (216), Expect = 6e-17
Identities = 64/217 (29%), Positives = 96/217 (43%), Gaps = 6/217 (2%)
Query: 175 DETSDVILLKFLRARDFKVKEAFTMIKNTILWRKEFGIEELMDEKLGDELEKV-----VY 229
+ D LL+FLRAR F ++++ M WRKEFG+++L+ DE E V +
Sbjct: 46 ERLDDATLLRFLRARKFNLQQSLEMFIKCEKWRKEFGVDDLIKNFHYDEKEAVSKYYPQF 105
Query: 230 MHGFDKEGHPVCYNIYGEFQNKELYNKTFSDEEKRHNFLKWRIQFLEKSIRNLDFNHGGV 289
H D +G PV G K+LY T + ++ ++ + L++ G +
Sbjct: 106 YHKTDIDGRPVYVEQLGNIDLKKLYQITTPERMMQNLVYEYEMLALKRFPACSRKAGGLI 165
Query: 290 CTIVHVNDLKDSPGPGKWELRQATKQALQLFQDNYPEFVAKQVFINVPWWYLAVNRMISP 349
T + DLK + +QA + QD YPE + K IN PW + + +I
Sbjct: 166 ETSCTIMDLKGVGITSIHSVYSYIRQASSISQDYYPERMGKFYVINAPWGFSSAFNLIKG 225
Query: 350 FLTQRTKSKFVFAGPSKSTETLLSYIAPEQLPVKYGG 386
FL + T K G S LL I + LP K GG
Sbjct: 226 FLDEATVKKIHILG-SNYKSALLEQIPADNLPAKLGG 261
>SC14_KLULA (P24859) SEC14 cytosolic factor
(Phosphatidylinositol/phosphatidylcholine transfer
protein) (PI/PC TP)
Length = 301
Score = 86.7 bits (213), Expect = 1e-16
Identities = 68/228 (29%), Positives = 101/228 (43%), Gaps = 16/228 (7%)
Query: 175 DETSDVILLKFLRARDFKVKEAFTMIKNTILWRKEFGIEELMDEKLGDELEKVV-----Y 229
+ D LL+FLRAR F ++ + M +N WRKEFG++ + ++ +E V Y
Sbjct: 49 ERLDDSTLLRFLRARKFDLEASKIMYENCEKWRKEFGVDTIFEDFHYEEKPLVAKYYPQY 108
Query: 230 MHGFDKEGHPVCYNIYGEFQNKELYNKTFSDEEKRHNFLKWRIQF-----LEKSIRNLDF 284
H D +G PV G ++Y T +E+ L W + L R +
Sbjct: 109 YHKTDNDGRPVYIEELGSVNLTQMYKIT--TQERMLKNLVWEYEAFVRYRLPACSRKAGY 166
Query: 285 NHGGVCTIVHVNDLKDSPGPGKWELRQATKQALQLFQDNYPEFVAKQVFINVPWWYLAVN 344
CTI+ DLK ++ ++A + Q+ YPE + K IN P+ +
Sbjct: 167 LVETSCTIL---DLKGISISSAAQVLSYVREASNIGQNYYPERMGKFYLINAPFGFSTAF 223
Query: 345 RMISPFLTQRTKSKFVFAGPSKSTETLLSYIAPEQLPVKYGGLSKDGE 392
R+ PFL T SK G S + LL I E LP K+GG S+ E
Sbjct: 224 RLFKPFLDPVTVSKIFILGSSYQKD-LLKQIPAENLPKKFGGQSEVSE 270
>S143_HUMAN (Q9UDX4) SEC14-like protein 3 (Tocopherol-associated
protein 2)
Length = 400
Score = 86.7 bits (213), Expect = 1e-16
Identities = 84/348 (24%), Positives = 159/348 (45%), Gaps = 52/348 (14%)
Query: 179 DVILLKFLRARDFKVKEAFTMIKNTILWRKEFGIEELMDEKLGDELEKVV--YMHGFDKE 236
D LL++LRAR+F ++++ +++ + +RK I+ ++D + + ++K + + G+D++
Sbjct: 35 DYFLLRWLRARNFDLQKSEALLRKYMEFRKTMDIDHILDWQPPEVIQKYMPGGLCGYDRD 94
Query: 237 GHPVCYNIYGEFQNKELYNKTFSDEEKRHNFLKWRIQFLEKSIRNLDFN----HGGVCTI 292
G PV Y+I G K L FS + + LK +++ E+ + D + TI
Sbjct: 95 GCPVWYDIIGPLDPKGL---LFS--VTKQDLLKTKMRDCERILHECDLQTERLGKKIETI 149
Query: 293 VHVNDLKDSPGPGKWE-LRQATKQALQLFQDNYPEFVAKQVFINVPWWYLAVNRMISPFL 351
V + D + W+ L + ++ L ++NYPE + + + + ++ PFL
Sbjct: 150 VMIFDCEGLGLKHFWKPLVEVYQEFFGLLEENYPETLKFMLIVKATKLFPVGYNLMKPFL 209
Query: 352 TQRTKSKFVFAGPSKSTETLLSYIAPEQLPVKYGGLSKDGEFGNSDSVTEIT-------- 403
++ T+ K + G + E LL I+PE+LP ++GG D + GN +T+I
Sbjct: 210 SEDTRRKIIVLG-NNWKEGLLKLISPEELPAQFGGTLTDPD-GNPKCLTKINYGGEIPKS 267
Query: 404 ----------------IRPASKHTVEFPVT-EKCLLSWEVRVIGWEVRYGAEFVPSNEGS 446
I S H VE+ + C+L W+ G ++ +G F+ + G
Sbjct: 268 MYVRDQVKTQYEHSVQINRGSSHQVEYEILFPGCVLRWQFSSDGADIGFGV-FLKTKMGE 326
Query: 447 YTVIVQKARKVASSEEAVLCNSFKINEPGKV--------VLTIDNTSS 486
Q+A ++ + N+ + E G + VL DNT S
Sbjct: 327 R----QRAGEMTDVLPSQRYNAHMVPEDGNLTCSEAGVYVLRFDNTYS 370
>S144_MOUSE (Q8R0F9) SEC14-like protein 4
Length = 403
Score = 83.2 bits (204), Expect = 1e-15
Identities = 84/357 (23%), Positives = 156/357 (43%), Gaps = 64/357 (17%)
Query: 176 ETSDVILLKFLRARDFKVKEAFTMIKNTILWRKEFGIEELMDEKLGD--ELEKVVYMHGF 233
+ D LL++LRAR+F +K++ M++ + +R + +++++ + + +L + G+
Sbjct: 32 KADDYFLLRWLRARNFDLKKSEDMLRKHVEFRNQQNLDQILTWQAPEVIQLYDSGGLSGY 91
Query: 234 DKEGHPVCYNIYGEFQNKELYNKTFSDEEKRHNFLKWRIQFLEKSIRNLDFNH----GGV 289
D EG PV ++I G K L+ + + ++ RI+ E + + +
Sbjct: 92 DYEGCPVWFDIIGTMDPKGLFMSA-----SKQDMIRKRIKVCEMLLHECELQSQKLGRKI 146
Query: 290 CTIVHVNDLKDSPGPGKWELRQATKQALQLFQD-------NYPEFVAKQVFINVPWWYLA 342
+V V D++ LR K A++++Q NYPE V + I P +
Sbjct: 147 ERMVMVFDMEG------LSLRHLWKPAVEVYQQFFAILEANYPETVKNLIIIRAPKLFPV 200
Query: 343 VNRMISPFLTQRTKSKFVFAGPSKSTETLLSYIAPEQLPVKYGGLSKDGEFGNSDSVTEI 402
++ F+ + T+ K V G + E L+ +++P+QLPV++GG D + GN +T+I
Sbjct: 201 AFNLVKSFMGEETQKKIVILGGNWKQE-LVKFVSPDQLPVEFGGTMTDPD-GNPKCLTKI 258
Query: 403 TI--------------RP----------ASKHTVEFPVT-EKCLLSWEVRVIGWEVRYGA 437
RP S H VE + C+L W+ G ++ +G
Sbjct: 259 NYGGEVPKRYYLSNQERPQYEHSVVVGRGSSHQVENEILFPGCVLRWQFASDGGDIGFGV 318
Query: 438 EFVPSNEGSYTVIVQKARKVASSEEAVLCNSFKINEPGKV--------VLTIDNTSS 486
F+ + G QKA ++ + N+ + E G + VL DNT S
Sbjct: 319 -FLKTRMGER----QKAGEMVEVLPSQRYNAHMVPEDGSLNCLKAGVYVLRFDNTYS 370
>S144_HUMAN (Q9UDX3) SEC14-like protein 4 (Tocopherol-associated
protein 3)
Length = 406
Score = 82.0 bits (201), Expect = 3e-15
Identities = 87/372 (23%), Positives = 164/372 (43%), Gaps = 61/372 (16%)
Query: 170 IPLLADETSDVILLKFLRARDFKVKEAFTMIKNTILWRKEFGIEELMDEKLGD--ELEKV 227
+P+L + D LL++LRAR+F ++++ M++ + +RK+ ++ ++ + + +L
Sbjct: 27 LPILPN-ADDYFLLRWLRARNFDLQKSEDMLRRHMEFRKQQDLDNIVTWQPPEVIQLYDS 85
Query: 228 VYMHGFDKEGHPVCYNIYGEFQNKELYNKTFSDEEKRHNFLKWRIQFLEKSIRNLDFNHG 287
+ G+D EG PV +NI G K L + + ++ RI+ E + +
Sbjct: 86 GGLCGYDYEGCPVYFNIIGSLDPKGLLLSA-----SKQDMIRKRIKVCELLLHECELQTQ 140
Query: 288 GVCTIVH----VNDLKDSPGPGKWE-LRQATKQALQLFQDNYPEFVAKQVFINVPWWYLA 342
+ + V D++ W+ + +Q + + NYPE + + I P +
Sbjct: 141 KLGRKIEMALMVFDMEGLSLKHLWKPAVEVYQQFFSILEANYPETLKNLIVIRAPKLFPV 200
Query: 343 VNRMISPFLTQRTKSKFVFAGPSKSTETLLSYIAPEQLPVKYGGLSKDGEFGNSDSVTEI 402
++ F+++ T+ K V G + E L +I+P+QLPV++GG D + GN +T+I
Sbjct: 201 AFNLVKSFMSEETRRKIVILGDNWKQE-LTKFISPDQLPVEFGGTMTDPD-GNPKCLTKI 258
Query: 403 T--------------IRPASKHT--------------VEFPVTEKCLLSWEVRVIGWEVR 434
+R +HT + FP C+L W+ G ++
Sbjct: 259 NYGGEVPKSYYLCEQVRLQYEHTRSVGRGSSLQVENEILFP---GCVLRWQFASDGGDIG 315
Query: 435 YGAEFVPSNEGSYTVIVQKARKVASSEEAVLCNSFKINEPGKV--------VLTIDNTSS 486
+G F+ + G Q AR++ + N+ + E G + VL DNT S
Sbjct: 316 FGV-FLKTKMGEQ----QSAREMTEVLPSQRYNAHMVPEDGSLTCLQAGVYVLRFDNTYS 370
Query: 487 R--KKKLLYRLK 496
R KKL Y ++
Sbjct: 371 RMHAKKLSYTVE 382
>YNX1_YEAST (P53860) Hypothetical 40.7 kDa protein in CSL4-URE2
intergenic region
Length = 351
Score = 73.6 bits (179), Expect = 1e-12
Identities = 60/221 (27%), Positives = 97/221 (43%), Gaps = 39/221 (17%)
Query: 183 LKFLRARDFKVKEAFTMIKNTILWRKEFGIEELMDEKLGDELE-----------KVVYMH 231
L++LRA + +K+ I T+ WR+EFGI L +E GD++ K V +
Sbjct: 91 LRYLRATKWVLKDCIDRITMTLAWRREFGISHLGEEH-GDKITADLVAVENESGKQVIL- 148
Query: 232 GFDKEGHPVCYNIYGEFQNKELYNKTFSDEEKRHNFLKWRIQFLEKSIRNLDFNHGGVCT 291
G++ + P+ Y G K H ++ + LE+ I DF G +
Sbjct: 149 GYENDARPILYLKPGRQNTKT-----------SHRQVQHLVFMLERVI---DFMPAGQDS 194
Query: 292 IVHVNDLKDSPG----PGKWELRQ--ATKQALQLFQDNYPEFVAKQVFINVPWWYLAVNR 345
+ + D KD P PG ++ K+ L + Q +YPE + K + N+PW +
Sbjct: 195 LALLIDFKDYPDVPKVPGNSKIPPIGVGKEVLHILQTHYPERLGKALLTNIPWLAWTFLK 254
Query: 346 MISPFLTQRTKSKFVFAGPSKSTETLLSYIAPEQLPVKYGG 386
+I PF+ T+ K VF E + Y+ +L YGG
Sbjct: 255 LIHPFIDPLTREKLVF------DEPFVKYVPKNELDSLYGG 289
>YJX4_SCHPO (Q9UU99) Protein C23B6.04c in chromosome III
Length = 1008
Score = 73.6 bits (179), Expect = 1e-12
Identities = 88/371 (23%), Positives = 152/371 (40%), Gaps = 64/371 (17%)
Query: 58 PNNENNSLQELQNLIQQAFNNHAFSAPPLIKEQKQSTTTTVAAEPAQENKYQLEDKKENV 117
P+ + L + + F S P + + AA QE + ++
Sbjct: 474 PSTPSKVLPAMSPKVAPKFQGGRSSTAPSTPNKVLPAASAKAAPKLQEKAASFDIPNKST 533
Query: 118 VSSVEDDGAKTVEAIEESIVAVSAS---VPPEQKP------VVEKVEASLPLPP------ 162
+ S + A ++A E S S + P E P + +A+ P+ P
Sbjct: 534 IISTKSTVASPIKANENSPALKSKASFEFPKELDPDGTWNAPIPYPDANCPMAPTYRNLT 593
Query: 163 -EQVSIYG-----------IPLLADET--SDVI-----------LLKFLRARDFKVKEAF 197
EQ +Y IP+ ++ + +D+I +L++LRA + V A
Sbjct: 594 SEQEEMYEEVLKYCLELKEIPVASNSSKKTDLIELERLWLTRECILRYLRATKWHVSNAK 653
Query: 198 TMIKNTILWRKEFGIEELMDEKLGDE--LEKVVYMHGFDKEGHPVCYNIYGEFQNKELYN 255
I +T++WR+ FG+ + +++ +E K V + G+DK+G P C +Y QN +
Sbjct: 654 KRIVDTLVWRRHFGVNNMDPDEIQEENATGKQVLL-GYDKDGRP-CLYLYPARQNTKT-- 709
Query: 256 KTFSDEEKRHNFLKWRIQFLEKSIRNLDFNHGGVCTIVHVNDLKDSPGPGKWELRQATKQ 315
S + RH LE +I D GV T+ + + K S + Q K+
Sbjct: 710 ---SPLQIRHLVFS-----LECAI---DLMPPGVETLALLINFKSSSNRSNPSVGQG-KE 757
Query: 316 ALQLFQDNYPEFVAKQVFINVPWWYLAVNRMISPFLTQRTKSKFVFAGPSKSTETLLSYI 375
L + Q +Y E + + + IN+PW ++ISPF+ T+ K F E L Y+
Sbjct: 758 VLNILQTHYCERLGRALVINIPWAVWGFFKLISPFIDPITREKLKF------NEPLDRYV 811
Query: 376 APEQLPVKYGG 386
+QL +GG
Sbjct: 812 PKDQLDSNFGG 822
>RALB_TODPA (P49193) Retinal-binding protein (RALBP)
Length = 342
Score = 67.0 bits (162), Expect = 1e-10
Identities = 70/284 (24%), Positives = 120/284 (41%), Gaps = 41/284 (14%)
Query: 232 GFDKEGHPVCYNIYGEFQNKEL-YNKTFSDEEKRHNFLKWRIQFLEKSIRNLDFNH---G 287
G DK+G + +G K + Y+ SD EK ++ EK +++L+ G
Sbjct: 31 GHDKDGSVLRIEPWGYLDMKGIMYSCKKSDLEKS------KLLQCEKHLKDLEAQSEKVG 84
Query: 288 GVCT-IVHVNDLKDSPGPGKWELR-QATKQALQLFQDNYPEFVAKQVFINVPWWYLAVNR 345
CT + V D+++ W+ +Q+ +DNYPE + + IN P + + +
Sbjct: 85 KPCTGLTVVFDMENVGSKHMWKPGLDMYLYLVQVLEDNYPEMMKRLFVINAPTLFPVLYK 144
Query: 346 MISPFLTQRTKSK-FVFAGPSKSTETLLSYIAPEQLPVKYGGLSKDGE------------ 392
++ P L++ K+K FV G K +TLL YI E+LP GG +G+
Sbjct: 145 LVKPLLSEDMKNKIFVLGGDYK--DTLLEYIDAEELPAYLGGTKSEGDEKCSELICHGGE 202
Query: 393 ------FGNSD---SVTEITIRPASKHTVEFPV-TEKCLLSWEVRVIGWEVRYGAEFVPS 442
N+D ++ IT+ K VE+ + E + WE + ++ +G
Sbjct: 203 VPKEFYLENTDDFETMETITVGSGDKIYVEYEIENENTYIKWEYKTEEHDIGFGLFRKNG 262
Query: 443 NEGSYTVIVQKARKVASSEEAVLCNSFKINEPGKVVLTIDNTSS 486
+E V +++ L S K +PG L DN+ S
Sbjct: 263 DEWEEVVPIER----TDCSIMTLDGSHKCKDPGTYALCFDNSFS 302
>YJO5_YEAST (P47008) Hypothetical 34.4 kDa protein in IDS2-MPI2
intergenic region
Length = 294
Score = 66.6 bits (161), Expect = 1e-10
Identities = 52/195 (26%), Positives = 93/195 (47%), Gaps = 19/195 (9%)
Query: 175 DETSDVILLKFLRARDFKVKEAFTMIKNTILWRKEFG-IEELMDEKLGDELEKVVYMHGF 233
++ +D + K +A F+ + + + WR+EF + E EL+ V + F
Sbjct: 54 EKIADRLTYKLCKAYQFEYSTIVQNLIDILNWRREFNPLSCAYKEVHNTELQNVGILT-F 112
Query: 234 DKEGHP----VCYNIYGEFQNKELYNKTFSDEEKRHNFLKWRIQFLEKSIRNLDFNHGGV 289
D G V +N+YG+ K+ + F + +K F+++RI +EK + LDF
Sbjct: 113 DANGDANKKAVTWNLYGQLVKKK---ELFQNVDK---FVRYRIGLMEKGLSLLDFTSSDN 166
Query: 290 CTIVHVNDLKDSPGPGKW----ELRQATKQALQLFQDNYPEFVAKQVFINVPWWYLAVNR 345
+ V+D K G W +++ +K + +FQ YPE + + F+NVP + V
Sbjct: 167 NYMTQVHDYK---GVSVWRMDSDIKNCSKTVIGIFQKYYPELLYAKYFVNVPTVFGWVYD 223
Query: 346 MISPFLTQRTKSKFV 360
+I F+ + T+ KFV
Sbjct: 224 LIKKFVDETTRKKFV 238
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.313 0.131 0.377
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 61,509,801
Number of Sequences: 164201
Number of extensions: 2724131
Number of successful extensions: 7662
Number of sequences better than 10.0: 126
Number of HSP's better than 10.0 without gapping: 32
Number of HSP's successfully gapped in prelim test: 98
Number of HSP's that attempted gapping in prelim test: 7479
Number of HSP's gapped (non-prelim): 209
length of query: 503
length of database: 59,974,054
effective HSP length: 114
effective length of query: 389
effective length of database: 41,255,140
effective search space: 16048249460
effective search space used: 16048249460
T: 11
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 68 (30.8 bits)
Medicago: description of AC148755.7


