

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
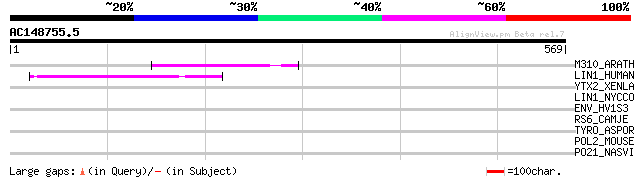
Query= AC148755.5 + phase: 0
(569 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
M310_ARATH (P93295) Hypothetical mitochondrial protein AtMg00310... 88 7e-17
LIN1_HUMAN (P08547) LINE-1 reverse transcriptase homolog 47 1e-04
YTX2_XENLA (P14381) Transposon TX1 hypothetical 149 kDa protein ... 37 0.18
LIN1_NYCCO (P08548) LINE-1 reverse transcriptase homolog 35 0.53
ENV_HV1S3 (P19549) Envelope polyprotein GP160 precursor [Contain... 33 2.0
RS6_CAMJE (Q9ZAH3) 30S ribosomal protein S6 32 3.4
TYRO_ASPOR (Q00234) Tyrosinase (EC 1.14.18.1) (Monophenol monoox... 31 7.6
POL2_MOUSE (P11369) Retrovirus-related Pol polyprotein [Contains... 31 9.9
PO21_NASVI (Q03278) Retrovirus-related Pol polyprotein from type... 31 9.9
>M310_ARATH (P93295) Hypothetical mitochondrial protein AtMg00310
(ORF154)
Length = 154
Score = 87.8 bits (216), Expect = 7e-17
Identities = 54/155 (34%), Positives = 79/155 (50%), Gaps = 14/155 (9%)
Query: 146 SLPVYFLSFFKAPTGIISSIESIFKKKFWGGCEVSRKIAWVKWDSVCLPM-SQGGLGVRK 204
+LPVY +S F+ + + S + +W CE RKI+WV W +C GGLG R
Sbjct: 2 ALPVYAMSCFRLSKLLCKKLTSAMTEFWWSSCENKRKISWVAWQKLCKSKEDDGGLGFRD 61
Query: 205 LGEFNLSLLGKWCWRLLVDKNGLWYRVLKARYGEIGGMVK-EGGRHSSRWWRMICNVRD- 262
LG FN +LL K +R++ + L R+L++RY M++ G S WR I + R+
Sbjct: 62 LGWFNQALLAKQSFRIIHQPHTLLSRLLRSRYFPHSSMMECSVGTRPSYAWRSIIHGREL 121
Query: 263 -GRGLLVGKWFDDNIRRVVGNGRNTFFWYDNWLGD 296
RGLL R +G+G +T W D W+ D
Sbjct: 122 LSRGLL----------RTIGDGIHTKVWLDRWIMD 146
>LIN1_HUMAN (P08547) LINE-1 reverse transcriptase homolog
Length = 1259
Score = 47.0 bits (110), Expect = 1e-04
Identities = 46/202 (22%), Positives = 84/202 (40%), Gaps = 11/202 (5%)
Query: 21 EVRLSHLQFADDTLIIGEKSWLNVRTIRAVLLLLEKVSGLKVNFNKSMVTGVNISTSWLS 80
EV+LS FADD ++ E ++ + + ++ KVSG K+N KS + S
Sbjct: 693 EVKLS--LFADDMIVYLENPIVSAQNLLKLISNFSKVSGYKINVQKSQAFLYTNNRQTES 750
Query: 81 EAAAVLNCKTGLIPFMYLGLPIGGDERKLC--FWKPVLDRISARLSSWNNKYLSSGGRLI 138
+ + L YLG+ + D + L +KP+L+ I + W N S GR+
Sbjct: 751 QIMSELPFTIASKRIKYLGIQLTRDVKDLFKENYKPLLNEIKEDTNKWKNIPCSWVGRIN 810
Query: 139 LLKSVLSSLPVYFLSF--FKAPTGIISSIESIFKKKFWGGCEVSRKIAWVKWDSVCLPMS 196
++K + +Y + K P + +E K W ++K A + ++
Sbjct: 811 IVKMAILPKVIYRFNAIPIKLPMTFFTELEKTTLKFIW-----NQKRAHIAKSTLSQKNK 865
Query: 197 QGGLGVRKLGEFNLSLLGKWCW 218
GG+ + + + + K W
Sbjct: 866 AGGITLPDFKLYYKATVTKTAW 887
>YTX2_XENLA (P14381) Transposon TX1 hypothetical 149 kDa protein
(ORF 2)
Length = 1308
Score = 36.6 bits (83), Expect = 0.18
Identities = 47/231 (20%), Positives = 100/231 (42%), Gaps = 22/231 (9%)
Query: 21 EVRLSHLQFADDTLIIGEKSWLNVRTIRAVLLLLEKVSGLKVNFNKS--MVTGVNISTSW 78
++R+ +ADD +++ + +++ + + S ++N++KS ++ G ++ +
Sbjct: 686 DMRVVLSAYADDVILVAQ-DLVDLERAQECQEVYAAASSARINWSKSSGLLEG-SLKVDF 743
Query: 79 LSEAAAVLNCKTGLIPFMYLGLPIGGDERKLCFWKPVLDR-ISARLSSWNN--KYLSSGG 135
L A ++ ++ +I YLG+ + +E + L+ + RL W K LS G
Sbjct: 744 LPPAFRDISWESKIIK--YLGVYLSAEEYPVSQNFIELEECVLTRLGKWKGFAKVLSMRG 801
Query: 136 RLILLKSVLSSLPVYFLSFFKAPTGIISSIESIFKKKFWGGCEVSRKIAWVKWDSVCLPM 195
R +++ +++S Y L I+ I+ W G WV LP+
Sbjct: 802 RALVINQLVASQIWYRLICLSPTQEFIAKIQRRLLDFLWIGKH------WVSAGVSSLPL 855
Query: 196 SQGGLGV----RKLGEFNLSLLGKWCWRLLVDKNGLWYRVLKARYGEIGGM 242
+GG GV ++ F L + ++ L D + W + + Y ++ M
Sbjct: 856 KEGGQGVVCIRSQVHTFRLQQIQRY---LYADPSPQWCTLASSFYRQVRNM 903
>LIN1_NYCCO (P08548) LINE-1 reverse transcriptase homolog
Length = 1260
Score = 35.0 bits (79), Expect = 0.53
Identities = 41/165 (24%), Positives = 73/165 (43%), Gaps = 20/165 (12%)
Query: 21 EVRLSHLQFADDTLIIGEKSWLNVRTIRAVLLLLEKVSGLKVNFNKSMV---TGVNISTS 77
E++LS FADD ++ E + + + V+ VSG K+N +KS+ T N +
Sbjct: 693 EIKLS--LFADDMIVYLENTRDSTTKLLEVIKEYSNVSGYKINTHKSVAFIYTNNNQAEK 750
Query: 78 WLSEAAAVLNCKTGLIP--FMYLGLPIGGDERKLC--FWKPVLDRISARLSSWNNKYLSS 133
+ ++ ++P YLG+ + D + L ++ + I+ ++ W N S
Sbjct: 751 TVKDSIPFT-----VVPKKMKYLGVYLTKDVKDLYKENYETLRKEIAEDVNKWKNIPCSW 805
Query: 134 GGRLILLKSVLSSLPVYFLSF----FKAPTGIISSIESIFKKKFW 174
GR+ ++K +S LP +F KAP +E I W
Sbjct: 806 LGRINIVK--MSILPKAIYNFNAIPIKAPLSYFKDLEKIILHFIW 848
>ENV_HV1S3 (P19549) Envelope polyprotein GP160 precursor [Contains:
Exterior membrane glycoprotein (GP120); Transmembrane
glycoprotein (GP41)]
Length = 852
Score = 33.1 bits (74), Expect = 2.0
Identities = 52/207 (25%), Positives = 78/207 (37%), Gaps = 41/207 (19%)
Query: 178 EVSRKIAWVKWD-------SVCLPMSQGGLGVRKLGEFNLSLLGKWC--WRLLVDKNGLW 228
++ + W++W+ S+ + + ++ E L L KW W N LW
Sbjct: 617 KIWNNMTWMEWEREIDNYTSLIYTLLEESQNQQEKNEQELLELDKWASLWNWFSITNWLW 676
Query: 229 YRVLKARYGEIGGMVKEGGRHSSRWWRMICNVRDGRGLLV-------------------- 268
Y ++ +GG++ G R ++ VR G L
Sbjct: 677 Y--IRIFIMIVGGLI--GLRIIFAVLSIVNRVRQGYSPLSFQTLIPAQRGPDRPEGIEEG 732
Query: 269 -GKWFDDNIRRVVGNGRNTFFWYDNWLGDVPLKFQFSRLFDLAVNKECSVEDMVRAGWEE 327
G+ D R+V NG FW D L + L F + RL DL + VE + R GWE
Sbjct: 733 GGERDRDRSTRLV-NGFLALFWDD--LRSLCL-FSYHRLTDLLLIVARIVELLGRRGWEV 788
Query: 328 GGGGWVWRRRLLAWEEEGVRECVNLLN 354
W LL W +E V+LLN
Sbjct: 789 LK---YWWNLLLYWSQELKNSAVSLLN 812
>RS6_CAMJE (Q9ZAH3) 30S ribosomal protein S6
Length = 125
Score = 32.3 bits (72), Expect = 3.4
Identities = 20/63 (31%), Positives = 32/63 (50%), Gaps = 4/63 (6%)
Query: 108 KLCFWKPVLDRISARLSSWNNKYLSSGGRLILLKSVLSSLPVYFLSFFKAPTGIISSIES 167
KL F K VL + SA + + + G R + K YF+ +FKAPT +I+ +E
Sbjct: 22 KLEFVKEVLTKNSAEIET----VVPMGTRKLAYKIKKYERGTYFVIYFKAPTNLIAELER 77
Query: 168 IFK 170
+ +
Sbjct: 78 VLR 80
>TYRO_ASPOR (Q00234) Tyrosinase (EC 1.14.18.1) (Monophenol
monooxygenase)
Length = 539
Score = 31.2 bits (69), Expect = 7.6
Identities = 17/61 (27%), Positives = 31/61 (49%), Gaps = 3/61 (4%)
Query: 342 EEEGVRECVNLLNNIVLQDNIQDKWRWLLDPILGYTVKGTYCFLTNMEGQVTSGVGFDVW 401
E + R+ + LN L D + + WRW+ DP T + ++ +L + G+ G G++
Sbjct: 19 ETKAERKSIRDLNEEEL-DKLIEAWRWIQDP--ARTGEDSFFYLAGLHGEPFRGAGYNNS 75
Query: 402 H 402
H
Sbjct: 76 H 76
>POL2_MOUSE (P11369) Retrovirus-related Pol polyprotein [Contains:
Reverse transcriptase (EC 2.7.7.49); Endonuclease]
Length = 1300
Score = 30.8 bits (68), Expect = 9.9
Identities = 36/158 (22%), Positives = 62/158 (38%), Gaps = 6/158 (3%)
Query: 21 EVRLSHLQFADDTLIIGEKSWLNVRTIRAVLLLLEKVSGLKVNFNKSMVTGVNISTSWLS 80
EV++S L ADD ++ + R + ++ +V G K+N NKSM +
Sbjct: 720 EVKISLL--ADDMIVYISDPKNSTRELLNLINSFGEVVGYKINSNKSMAFLYTKNKQAEK 777
Query: 81 EAAAVLNCKTGLIPFMYLGLPIGGDERKLC--FWKPVLDRISARLSSWNNKYLSSGGRLI 138
E YLG+ + + + L +K + I L W + S GR+
Sbjct: 778 EIRETTPFSIVTNNIKYLGVTLTKEVKDLYDKNFKSLKKEIKEDLRRWKDLPCSWIGRIN 837
Query: 139 LLKSVLSSLPVYFLSF--FKAPTGIISSIESIFKKKFW 174
++K + +Y + K PT + +E K W
Sbjct: 838 IVKMAILPKAIYRFNAIPIKIPTQFFNELEGAICKFVW 875
>PO21_NASVI (Q03278) Retrovirus-related Pol polyprotein from type I
retrotransposable element R2 [Contains: Reverse
transcriptase (EC 2.7.7.49); Endonuclease] (Fragment)
Length = 1025
Score = 30.8 bits (68), Expect = 9.9
Identities = 24/87 (27%), Positives = 41/87 (46%), Gaps = 16/87 (18%)
Query: 379 KGTYCFLTNMEGQVTSGVGFDVWHRRVPSKVSLFAWRLLQDRVATRTNLVRRYVLQAADN 438
+ +Y ++ ++ + + V H R+ + +L R+ + R RTN +
Sbjct: 799 EASYSWIRDIHVAIPASVWIKYHHTRINALPTLM--RMSRGR---RTN---------GNA 844
Query: 439 FCVGGCGIPETVDHLFIQC--TSFGRV 463
C GCG+PET+ H+ QC T GRV
Sbjct: 845 LCRAGCGLPETLYHVVQQCPRTHGGRV 871
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.326 0.142 0.483
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 71,875,818
Number of Sequences: 164201
Number of extensions: 3146825
Number of successful extensions: 7621
Number of sequences better than 10.0: 9
Number of HSP's better than 10.0 without gapping: 1
Number of HSP's successfully gapped in prelim test: 8
Number of HSP's that attempted gapping in prelim test: 7611
Number of HSP's gapped (non-prelim): 16
length of query: 569
length of database: 59,974,054
effective HSP length: 116
effective length of query: 453
effective length of database: 40,926,738
effective search space: 18539812314
effective search space used: 18539812314
T: 11
A: 40
X1: 15 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.6 bits)
S2: 68 (30.8 bits)
Medicago: description of AC148755.5


