

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148755.4 + phase: 1 /partial
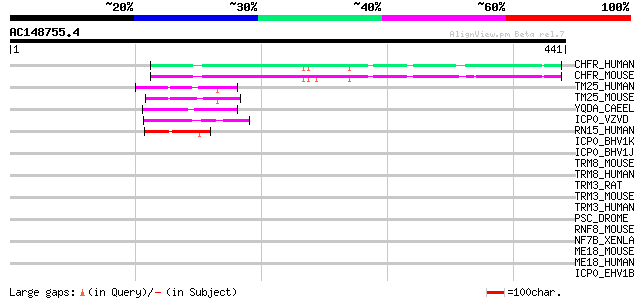
(441 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
CHFR_HUMAN (Q96EP1) Ubiquitin ligase protein CHFR (EC 6.3.2.-) (... 129 2e-29
CHFR_MOUSE (Q810L3) Ubiquitin ligase protein CHFR (EC 6.3.2.-) (... 125 3e-28
TM25_HUMAN (Q14258) Tripartite motif-containing protein 25 (Zinc... 56 2e-07
TM25_MOUSE (Q61510) Tripartite motif-containing protein 25 (Zinc... 50 1e-05
YQDA_CAEEL (Q09268) Hypothetical RING finger protein C32D5.10 in... 47 7e-05
ICP0_VZVD (P09309) Trans-acting transcriptional protein ICP0 45 5e-04
RN15_HUMAN (O00635) RING finger protein 15 (Zinc finger protein ... 44 8e-04
ICP0_BHV1K (P29836) Trans-acting transcriptional protein ICP0 (P... 42 0.002
ICP0_BHV1J (P29128) Trans-acting transcriptional protein ICP0 (P... 42 0.002
TRM8_MOUSE (Q99PJ2) Tripartite motif-containing protein 8 (RING ... 42 0.003
TRM8_HUMAN (Q9BZR9) Tripartite motif-containing protein 8 (RING ... 42 0.003
TRM3_RAT (O70277) Tripartite motif-containing protein 3 (RING fi... 41 0.007
TRM3_MOUSE (Q9R1R2) Tripartite motif-containing protein 3 (RING ... 41 0.007
TRM3_HUMAN (O75382) Tripartite motif-containing protein 3 (RING ... 41 0.007
PSC_DROME (P35820) Posterior sex combs protein 41 0.007
RNF8_MOUSE (Q8VC56) Ubiquitin ligase protein RNF8 (EC 6.3.2.-) (... 40 0.009
NF7B_XENLA (Q92021) Nuclear factor 7, brain (xnf7) (xnf7-B) 40 0.009
ME18_MOUSE (P23798) DNA-binding protein Mel-18 (RING finger prot... 40 0.009
ME18_HUMAN (P35227) DNA-binding protein Mel-18 (RING finger prot... 40 0.009
ICP0_EHV1B (P28990) Trans-acting transcriptional protein ICP0 40 0.009
>CHFR_HUMAN (Q96EP1) Ubiquitin ligase protein CHFR (EC 6.3.2.-)
(Checkpoint with forkhead and RING finger domains
protein)
Length = 664
Score = 129 bits (323), Expect = 2e-29
Identities = 94/378 (24%), Positives = 151/378 (39%), Gaps = 72/378 (19%)
Query: 113 CSICLNIWHDVVTVAPCLHNFCNGCFSEWLRRSQEKRSTVLCPQCRAVVQFVGKNHFLRT 172
C IC ++ HD V++ PC+H FC C+S W+ RS LCP CR V+ + KNH L
Sbjct: 304 CIICQDLLHDCVSLQPCMHTFCAACYSGWMERSS------LCPTCRCPVERICKNHILNN 357
Query: 173 IAEDMVKADSSLRRSHDEVALLDTYASVRSNLVIGSGKKSRKRERANTTMVNQSDGPDHH 232
+ E + RS ++V +D + +++ ++S E ++ + + D
Sbjct: 358 LVEAYLIQHPDKSRSEEDVQSMDARNKITQDMLQPKVRRSFSDEEGSSEDLLELSDVDSE 417
Query: 233 ----------CSQC------------------------------------VTVVGGFQCD 246
C QC T V + C
Sbjct: 418 SSDISQPYVVCRQCPEYRRQAAQPPHCPAPEGEPGAPQALGDAPSTSVSLTTAVQDYVCP 477
Query: 247 PSTIHLQCQECGGMMPSRTGLG------VPQYCLGCDRPFCGAYWSALGVTGSGSYPVCS 300
H C C MP R PQ C C +PFC YW G T +G Y +
Sbjct: 478 LQGSHALCTCCFQPMPDRRAEREQDPRVAPQQCAVCLQPFCHLYW---GCTRTGCYGCLA 534
Query: 301 PDTLRPISEHTISGIPLVAHEKNLHEQNITESCIRQMGRTLQDVISEWIAKLNNREIDTT 360
P + + + G+ N +E +I ++ + G T +++++E + L
Sbjct: 535 PFCELNLGDKCLDGVL----NNNSYESDILKNYLATRGLTWKNMLTESLVALQRG----- 585
Query: 361 RLMLNHAEMMTAGTLVCSDCYQKLVSFFLYWFRISTPKHLLPPEASTREDCWYGYACRTQ 420
+ L +T T++C C + Y +R + P LP ++R DC++G CRTQ
Sbjct: 586 -VFLLSDYRVTGDTVLCYCCGLRSFRELTYQYRQNIPASELPVAVTSRPDCYWGRNCRTQ 644
Query: 421 HRSEEHARKRNHVCRPTR 438
++ HA K NH+C TR
Sbjct: 645 VKA-HHAMKFNHICEQTR 661
>CHFR_MOUSE (Q810L3) Ubiquitin ligase protein CHFR (EC 6.3.2.-)
(Checkpoint with forkhead and RING finger domains
protein)
Length = 664
Score = 125 bits (313), Expect = 3e-28
Identities = 95/379 (25%), Positives = 152/379 (40%), Gaps = 73/379 (19%)
Query: 113 CSICLNIWHDVVTVAPCLHNFCNGCFSEWLRRSQEKRSTVLCPQCRAVVQFVGKNHFLRT 172
C IC ++ HD V++ PC+H FC C+S W+ RS LCP CR V+ + KNH L
Sbjct: 303 CIICQDLLHDCVSLQPCMHTFCAACYSGWMERSS------LCPTCRCPVERICKNHILNN 356
Query: 173 IAEDMVKADSSLRRSHDEVALLDTYASVRSNLVIGSGKKSRKRERANTTMVNQSDGPDHH 232
+ E + RS ++V +D + +++ ++S E ++ + + D
Sbjct: 357 LVEAYLIQHPDKSRSEEDVRSMDARNKITQDMLQPKVRRSFSDEEGSSEDLLELSDVDSE 416
Query: 233 ----------CSQC----------------------VTVVGG---------------FQC 245
C QC +GG + C
Sbjct: 417 SSDISQPYIVCRQCPEYRRQAVQSLPCPVPESELGATLALGGEAPSTSASLPTAAPDYMC 476
Query: 246 DPSTIHLQCQECGGMMPSRTGLG------VPQYCLGCDRPFCGAYWSALGVTGSGSYPVC 299
H C C MP R PQ C C +PFC YW G T +G +
Sbjct: 477 PLQGSHAICTCCFQPMPDRRAEREQDPRVAPQQCAVCLQPFCHLYW---GCTRTGCFGCL 533
Query: 300 SPDTLRPISEHTISGIPLVAHEKNLHEQNITESCIRQMGRTLQDVISEWIAKLNNREIDT 359
+P + + + G+ N +E +I ++ + G T + V++E + L
Sbjct: 534 APFCELNLGDKCLDGVL----NNNNYESDILKNYLATRGLTWKSVLTESLLALQRGVF-- 587
Query: 360 TRLMLNHAEMMTAGTLVCSDCYQKLVSFFLYWFRISTPKHLLPPEASTREDCWYGYACRT 419
ML+ + T T++C C + Y +R + P LP ++R DC++G CRT
Sbjct: 588 ---MLSDYRI-TGNTVLCYCCGLRSFRELTYQYRQNIPASELPVTVTSRPDCYWGRNCRT 643
Query: 420 QHRSEEHARKRNHVCRPTR 438
Q ++ HA K NH+C TR
Sbjct: 644 QVKA-HHAMKFNHICEQTR 661
>TM25_HUMAN (Q14258) Tripartite motif-containing protein 25 (Zinc
finger protein 147) (Estrogen responsive finger protein)
(Efp)
Length = 630
Score = 56.2 bits (134), Expect = 2e-07
Identities = 33/85 (38%), Positives = 45/85 (52%), Gaps = 9/85 (10%)
Query: 101 LLKICVDVDHAKCSICLNIWHDVVTVAPCLHNFCNGCFSE-WLRRSQEKRSTVLCPQCRA 159
+ ++C + CSICL + + VT PC HNFC C +E W + S LCPQCRA
Sbjct: 1 MAELCPLAEELSCSICLEPFKEPVTT-PCGHNFCGSCLNETWAVQG----SPYLCPQCRA 55
Query: 160 VVQF---VGKNHFLRTIAEDMVKAD 181
V Q + KN L + E ++AD
Sbjct: 56 VYQARPQLHKNTVLCNVVEQFLQAD 80
>TM25_MOUSE (Q61510) Tripartite motif-containing protein 25 (Zinc
finger protein 147) (Estrogen responsive finger protein)
(Efp)
Length = 634
Score = 50.1 bits (118), Expect = 1e-05
Identities = 27/79 (34%), Positives = 41/79 (51%), Gaps = 9/79 (11%)
Query: 109 DHAKCSICLNIWHDVVTVAPCLHNFCNGCFSE-WLRRSQEKRSTVLCPQCRAVVQF---V 164
+ CS+CL ++ + VT PC HNFC C E W+ + R CPQCR V Q +
Sbjct: 9 EELSCSVCLELFKEPVTT-PCGHNFCTSCLDETWVVQGPPYR----CPQCRKVYQVRPQL 63
Query: 165 GKNHFLRTIAEDMVKADSS 183
KN + + E ++A+ +
Sbjct: 64 QKNTVMCAVVEQFLQAEQA 82
>YQDA_CAEEL (Q09268) Hypothetical RING finger protein C32D5.10 in
chromosome II
Length = 610
Score = 47.4 bits (111), Expect = 7e-05
Identities = 22/76 (28%), Positives = 34/76 (43%), Gaps = 4/76 (5%)
Query: 106 VDVDHAKCSICLNIWHDVVTVAPCLHNFCNGCFSEWLRRSQEKRSTVLCPQCRAVVQFVG 165
+D +CS+C N D +++ C H FC C WL K S CP C+ V F+
Sbjct: 34 IDEPEEECSVCKNEIIDTTSLSDCCHEFCYDCIVGWL----TKGSGPFCPMCKTPVSFIQ 89
Query: 166 KNHFLRTIAEDMVKAD 181
+ I +K++
Sbjct: 90 RKGTDEKITVQQIKSE 105
>ICP0_VZVD (P09309) Trans-acting transcriptional protein ICP0
Length = 467
Score = 44.7 bits (104), Expect = 5e-04
Identities = 26/84 (30%), Positives = 35/84 (40%), Gaps = 11/84 (13%)
Query: 107 DVDHAKCSICLNIWHDVVTVAPCLHNFCNGCFSEWLRRSQEKRSTVLCPQCRAVVQFVGK 166
D C+IC++ D+ PCLH+FC C W S V CP CR VQ +
Sbjct: 13 DASDNTCTICMSTVSDLGKTMPCLHDFCFVCIRAWTSTS------VQCPLCRCPVQSI-- 64
Query: 167 NHFLRTIAEDMVKADSSLRRSHDE 190
L I D + + S D+
Sbjct: 65 ---LHKIVSDTSYKEYEVHPSDDD 85
>RN15_HUMAN (O00635) RING finger protein 15 (Zinc finger protein
RoRet) (Tripartite motif-containing protein 38)
Length = 465
Score = 43.9 bits (102), Expect = 8e-04
Identities = 19/55 (34%), Positives = 35/55 (63%), Gaps = 4/55 (7%)
Query: 108 VDHAKCSICLNIWHDVVTVAPCLHNFCNGCFSEWLRRSQEKR---STVLCPQCRA 159
++ A CSICL++ + V++ C H++C+ C +++ + +K+ T CPQCRA
Sbjct: 11 MEEATCSICLSLMTNPVSIN-CGHSYCHLCITDFFKNPSQKQLRQETFCCPQCRA 64
>ICP0_BHV1K (P29836) Trans-acting transcriptional protein ICP0 (P135
protein) (IER 2.9/ER2.6)
Length = 676
Score = 42.4 bits (98), Expect = 0.002
Identities = 22/50 (44%), Positives = 24/50 (48%), Gaps = 6/50 (12%)
Query: 113 CSICLNIWHDVVTVAPCLHNFCNGCFSEWLRRSQEKRSTVLCPQCRAVVQ 162
C ICL+ PCLH FC C WL E R T CP C+A VQ
Sbjct: 13 CCICLDAITGAARALPCLHAFCLACIRRWL----EGRPT--CPLCKAPVQ 56
>ICP0_BHV1J (P29128) Trans-acting transcriptional protein ICP0 (P135
protein) (IER 2.9/ER2.6)
Length = 676
Score = 42.4 bits (98), Expect = 0.002
Identities = 22/50 (44%), Positives = 24/50 (48%), Gaps = 6/50 (12%)
Query: 113 CSICLNIWHDVVTVAPCLHNFCNGCFSEWLRRSQEKRSTVLCPQCRAVVQ 162
C ICL+ PCLH FC C WL E R T CP C+A VQ
Sbjct: 13 CCICLDAITGAARALPCLHAFCLACIRRWL----EGRPT--CPLCKAPVQ 56
>TRM8_MOUSE (Q99PJ2) Tripartite motif-containing protein 8 (RING
finger protein 27) (Glioblastoma-expressed RING finger
protein)
Length = 551
Score = 42.0 bits (97), Expect = 0.003
Identities = 20/46 (43%), Positives = 27/46 (58%), Gaps = 6/46 (13%)
Query: 113 CSICLNIWHDVVTVAPCLHNFCNGCFSE-WLRRSQEKRSTVLCPQC 157
C ICL+++ + V + PC HNFC GC E W + S V CP+C
Sbjct: 15 CPICLHVFVEPVQL-PCKHNFCRGCIGEAWAKDS----GLVRCPEC 55
>TRM8_HUMAN (Q9BZR9) Tripartite motif-containing protein 8 (RING
finger protein 27) (Glioblastoma-expressed RING finger
protein)
Length = 551
Score = 42.0 bits (97), Expect = 0.003
Identities = 20/46 (43%), Positives = 27/46 (58%), Gaps = 6/46 (13%)
Query: 113 CSICLNIWHDVVTVAPCLHNFCNGCFSE-WLRRSQEKRSTVLCPQC 157
C ICL+++ + V + PC HNFC GC E W + S V CP+C
Sbjct: 15 CPICLHVFVEPVQL-PCKHNFCRGCIGEAWAKDS----GLVRCPEC 55
>TRM3_RAT (O70277) Tripartite motif-containing protein 3 (RING
finger protein 22) (Brain-expressed RING finger protein)
Length = 744
Score = 40.8 bits (94), Expect = 0.007
Identities = 26/91 (28%), Positives = 38/91 (41%), Gaps = 11/91 (12%)
Query: 106 VDVDHAKCSICLNIWHDVVTVAPCLHNFCNGCFSEWLRRSQEKRSTVLCPQCRAV----- 160
+D CSICL+ + V PCLH FC C ++ + T+ CP CR
Sbjct: 15 MDKQFLVCSICLDRYR-CPKVLPCLHTFCERCLQNYI---PPQSLTLSCPVCRQTSILPE 70
Query: 161 --VQFVGKNHFLRTIAEDMVKADSSLRRSHD 189
V + N F+ ++ E M +A D
Sbjct: 71 QGVSALQNNFFISSLMEAMQQAPDGAHDPED 101
>TRM3_MOUSE (Q9R1R2) Tripartite motif-containing protein 3 (RING
finger protein 22) (RING finger protein HAC1)
Length = 744
Score = 40.8 bits (94), Expect = 0.007
Identities = 26/91 (28%), Positives = 38/91 (41%), Gaps = 11/91 (12%)
Query: 106 VDVDHAKCSICLNIWHDVVTVAPCLHNFCNGCFSEWLRRSQEKRSTVLCPQCRAV----- 160
+D CSICL+ + V PCLH FC C ++ + T+ CP CR
Sbjct: 15 MDKQFLVCSICLDRYR-CPKVLPCLHTFCERCLQNYI---PPQSLTLSCPVCRQTSILPE 70
Query: 161 --VQFVGKNHFLRTIAEDMVKADSSLRRSHD 189
V + N F+ ++ E M +A D
Sbjct: 71 QGVSALQNNFFISSLMEAMQQAPEGAHDPED 101
>TRM3_HUMAN (O75382) Tripartite motif-containing protein 3 (RING
finger protein 22) (Brain-expressed RING finger protein)
Length = 744
Score = 40.8 bits (94), Expect = 0.007
Identities = 26/91 (28%), Positives = 38/91 (41%), Gaps = 11/91 (12%)
Query: 106 VDVDHAKCSICLNIWHDVVTVAPCLHNFCNGCFSEWLRRSQEKRSTVLCPQCRAV----- 160
+D CSICL+ + V PCLH FC C ++ + T+ CP CR
Sbjct: 15 MDKQFLVCSICLDRYQ-CPKVLPCLHTFCERCLQNYI---PAQSLTLSCPVCRQTSILPE 70
Query: 161 --VQFVGKNHFLRTIAEDMVKADSSLRRSHD 189
V + N F+ ++ E M +A D
Sbjct: 71 QGVSALQNNFFISSLMEAMQQAPDGAHDPED 101
>PSC_DROME (P35820) Posterior sex combs protein
Length = 1603
Score = 40.8 bits (94), Expect = 0.007
Identities = 20/76 (26%), Positives = 34/76 (44%), Gaps = 16/76 (21%)
Query: 110 HAKCSICLNIWHDVVTVAPCLHNFCNGCFSEWLRRSQEKRSTVLCPQCRAVVQFVGKNHF 169
H C +C + T+ CLH+FC+ C LR+ + CP+C V+ N
Sbjct: 262 HIICHLCQGYLINATTIVECLHSFCHSCLINHLRKER------FCPRCEMVINNAKPN-- 313
Query: 170 LRTIAEDMVKADSSLR 185
+K+D++L+
Sbjct: 314 --------IKSDTTLQ 321
>RNF8_MOUSE (Q8VC56) Ubiquitin ligase protein RNF8 (EC 6.3.2.-)
(RING finger protein 8) (AIP37) (ActA-interacting
protein 37) (LaXp180)
Length = 488
Score = 40.4 bits (93), Expect = 0.009
Identities = 18/51 (35%), Positives = 29/51 (56%), Gaps = 7/51 (13%)
Query: 112 KCSICLNIWHDVVTVAPCLHNFCNGCFSEWLRRSQEKRSTVLCPQCRAVVQ 162
+C IC + + VT+ C H+FC+ C +EW++R E CP CR ++
Sbjct: 405 QCIICSEYFIEAVTLN-CAHSFCSFCINEWMKRKVE------CPICRKDIE 448
>NF7B_XENLA (Q92021) Nuclear factor 7, brain (xnf7) (xnf7-B)
Length = 609
Score = 40.4 bits (93), Expect = 0.009
Identities = 18/46 (39%), Positives = 28/46 (60%), Gaps = 5/46 (10%)
Query: 113 CSICLNIWHDVVTVAPCLHNFCNGCFSEWLRRSQEKRSTVLCPQCR 158
C +C+ ++ D V VA C HNFC C + ++ E +S+ CP+CR
Sbjct: 145 CPLCVELFKDPVMVA-CGHNFCRSC----IDKAWEGQSSFACPECR 185
>ME18_MOUSE (P23798) DNA-binding protein Mel-18 (RING finger protein
110) (Zinc finger protein 144)
Length = 342
Score = 40.4 bits (93), Expect = 0.009
Identities = 24/82 (29%), Positives = 34/82 (41%), Gaps = 9/82 (10%)
Query: 100 TLLKICVDVDHAKCSICLNIWHDVVTVAPCLHNFCNGCFSEWLRRSQEKRSTVLCPQCRA 159
T +KI H C++C + D T+ CLH+FC C +L ++ CP C
Sbjct: 5 TRIKITELNPHLMCALCGGYFIDATTIVECLHSFCKTCIVRYLETNK------YCPMCDV 58
Query: 160 VVQFVGKNHFLRTIAEDMVKAD 181
V K L +I D D
Sbjct: 59 QVH---KTRPLLSIRSDKTLQD 77
>ME18_HUMAN (P35227) DNA-binding protein Mel-18 (RING finger protein
110) (Zinc finger protein 144)
Length = 344
Score = 40.4 bits (93), Expect = 0.009
Identities = 24/82 (29%), Positives = 34/82 (41%), Gaps = 9/82 (10%)
Query: 100 TLLKICVDVDHAKCSICLNIWHDVVTVAPCLHNFCNGCFSEWLRRSQEKRSTVLCPQCRA 159
T +KI H C++C + D T+ CLH+FC C +L ++ CP C
Sbjct: 5 TRIKITELNPHLMCALCGGYFIDATTIVECLHSFCKTCIVRYLETNK------YCPMCDV 58
Query: 160 VVQFVGKNHFLRTIAEDMVKAD 181
V K L +I D D
Sbjct: 59 QVH---KTRPLLSIRSDKTLQD 77
>ICP0_EHV1B (P28990) Trans-acting transcriptional protein ICP0
Length = 532
Score = 40.4 bits (93), Expect = 0.009
Identities = 18/53 (33%), Positives = 26/53 (48%), Gaps = 6/53 (11%)
Query: 112 KCSICLNIWHDVVTVAPCLHNFCNGCFSEWLRRSQEKRSTVLCPQCRAVVQFV 164
+C ICL + PCLH FC C + W+R++ CP C+ V+ V
Sbjct: 7 RCPICLEDPSNYSMALPCLHAFCYVCITRWIRQNP------TCPLCKVPVESV 53
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.321 0.134 0.429
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 53,715,960
Number of Sequences: 164201
Number of extensions: 2278607
Number of successful extensions: 5241
Number of sequences better than 10.0: 155
Number of HSP's better than 10.0 without gapping: 32
Number of HSP's successfully gapped in prelim test: 123
Number of HSP's that attempted gapping in prelim test: 5153
Number of HSP's gapped (non-prelim): 181
length of query: 441
length of database: 59,974,054
effective HSP length: 113
effective length of query: 328
effective length of database: 41,419,341
effective search space: 13585543848
effective search space used: 13585543848
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 67 (30.4 bits)
Medicago: description of AC148755.4


