

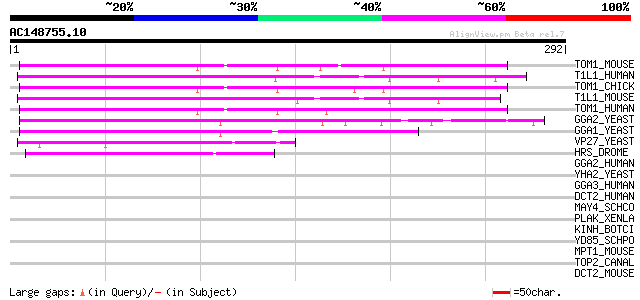
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148755.10 - phase: 0 /pseudo
(292 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TOM1_MOUSE (O88746) Target of Myb protein 1 79 2e-14
T1L1_HUMAN (O75674) TOM1-like 1 protein (Target of myb-like 1 pr... 79 2e-14
TOM1_CHICK (O12940) Target of Myb protein 1 (Tom-1 protein) 78 2e-14
T1L1_MOUSE (Q923U0) TOM1-like 1 protein (Target of myb-like 1 pr... 76 1e-13
TOM1_HUMAN (O60784) Target of Myb protein 1 72 2e-12
GGA2_YEAST (P38817) ADP-ribosylation factor binding protein GGA2... 49 1e-05
GGA1_YEAST (Q06336) ADP-ribosylation factor binding protein GGA1... 49 1e-05
VP27_YEAST (P40343) Vacuolar protein sorting-associated protein ... 48 3e-05
HRS_DROME (Q960X8) Hepatocyte growth factor-regulated tyrosine k... 47 6e-05
GGA2_HUMAN (Q9UJY4) ADP-ribosylation factor binding protein GGA2... 42 0.002
YHA2_YEAST (P38753) Hypothetical 51.2 kDa protein in LAG1-RPL14B... 39 0.020
GGA3_HUMAN (Q9NZ52) ADP-ribosylation factor binding protein GGA3... 37 0.075
DCT2_HUMAN (Q13561) Dynactin complex 50 kDa subunit (50 kDa dyne... 35 0.22
MAY4_SCHCO (P37935) Mating-type protein A-alpha Y4 34 0.49
PLAK_XENLA (P30998) Junction plakoglobin (Desmoplakin III) 33 0.83
KINH_BOTCI (Q86ZC1) Kinesin heavy chain 33 0.83
YD85_SCHPO (Q10410) Hypothetical protein C1F3.05 in chromosome I 33 1.1
MPT1_MOUSE (Q9DBR7) Protein phosphatase 1 regulatory subunit 12A... 33 1.1
TOP2_CANAL (P87078) DNA topoisomerase II (EC 5.99.1.3) 32 1.4
DCT2_MOUSE (Q99KJ8) Dynactin complex 50 kDa subunit (50 kDa dyne... 32 1.4
>TOM1_MOUSE (O88746) Target of Myb protein 1
Length = 492
Score = 78.6 bits (192), Expect = 2e-14
Identities = 69/285 (24%), Positives = 124/285 (43%), Gaps = 30/285 (10%)
Query: 6 VNAATSEKLSEIDWMKNIEISELVARDQRKAKDVVKAIKKR-LGNKNPNAQLYAVMLLEM 64
+ AT L DW N+EI +++ + KD +A+KKR +GNKN + + A+ +LE
Sbjct: 17 IEKATDGSLQSEDWALNMEICDIINETEEGPKDAFRAVKKRIMGNKNFHEVMLALTVLET 76
Query: 65 LMNNIGDHINEQVVRAEVIP-ILVKIVKKKSDLP--VREQIFLLLDATQTSLGGASGKFP 121
+ N G + V + + +LV+ + K++ P V +++ L+ + + +S
Sbjct: 77 CVKNCGHRFHVLVANQDFVENVLVRTILPKNNPPTIVHDKVLNLIQSWADAF-RSSPDLT 135
Query: 122 QYYKAYYDLVSAGVQFPQ------------RAQVVQSNRPSLQPNTTNNVPKR-----EP 164
Y DL G++FP + V S PS Q + ++N +R
Sbjct: 136 GVVAVYEDLRRKGLEFPMTDLDMLSPIHTPQRTVFNSETPSRQNSVSSNTSQRGDLSQHA 195
Query: 165 SPLRRGRVAQKAESNTVPESRIIQKASNVLE-------VLKEVLDAVDAKHPQGARDEFT 217
+PL V +S P I K + LE V+ E+L + + A E
Sbjct: 196 TPLPTPAVL-PGDSPITPTPEQIGKLRSELEMVSGNVRVMSEMLTELVPTQVEPADLELL 254
Query: 218 LDLVEQCSFQKQRVMHLVMASRDERIVSRAIEVNEQLQKVLERHD 262
+L C +QR++ L+ +E++ + +N+ L V RH+
Sbjct: 255 QELNRTCRAMQQRILELIPRISNEQLTEELLMINDNLNNVFLRHE 299
>T1L1_HUMAN (O75674) TOM1-like 1 protein (Target of myb-like 1
protein) (Src activating and signaling molecule protein)
Length = 476
Score = 78.6 bits (192), Expect = 2e-14
Identities = 75/296 (25%), Positives = 137/296 (45%), Gaps = 33/296 (11%)
Query: 5 LVNAATSEKLSEIDWMKNIEISELVARDQRKAKDVVKAIKKRLG-NKNPNAQLYAVMLLE 63
L+ AT + DW + + I +++ Q KD VKA+KKR+ N N + L++
Sbjct: 18 LIEKATFAGVQTEDWGQFMHICDIINTTQDAPKDAVKALKKRISKNYNHKEIQLTLSLID 77
Query: 64 MLMNNIGDHINEQVVRAEVI-PILVKIVKKKSDLPVREQIFLLLDATQTSLGGASG-KFP 121
M + N G +V+ E + LVK++ + +LP+ Q +L S G G
Sbjct: 78 MCVQNCGPSFQSLIVKKEFVKENLVKLLNPRYNLPLDIQNRILNFIKTWSQGFPGGVDVS 137
Query: 122 QYYKAYYDLVSAGVQFP---QRAQVVQSNRPSLQPNTTNNVPKREPSPLRRGRVAQKAES 178
+ + Y DLV GVQFP A+ + + N +VP +P +A K +
Sbjct: 138 EVKEVYLDLVKKGVQFPPSEAEAETARQETAQISSNPPTSVP---TAPALSSVIAPKNST 194
Query: 179 NT-VPESRIIQKASNVLEVLK---EVLDAVDAKHPQGARDEFTLDLVEQC----SFQKQR 230
T VPE I K + L+++K V+ A+ ++ G+ + ++L+++ ++R
Sbjct: 195 VTLVPEQ--IGKLHSELDMVKMNVRVMSAILMENTPGSENHEDIELLQKLYKTGREMQER 252
Query: 231 VMHLVMASRDERIVSRAIEVNEQL--------------QKVLERHDDLLSSKDTTT 272
+M L++ +E + I+VNE L Q++LE++ + + +TT+
Sbjct: 253 IMDLLVVVENEDVTVELIQVNEDLNNAILGYERFTRNQQRILEQNKNQKEATNTTS 308
>TOM1_CHICK (O12940) Target of Myb protein 1 (Tom-1 protein)
Length = 515
Score = 78.2 bits (191), Expect = 2e-14
Identities = 69/284 (24%), Positives = 125/284 (43%), Gaps = 28/284 (9%)
Query: 6 VNAATSEKLSEIDWMKNIEISELVARDQRKAKDVVKAIKKRL-GNKNPNAQLYAVMLLEM 64
+ AT L DW N+EI +++ + KD +AIKKR+ GNKN + + A+ +LE
Sbjct: 17 IERATDGSLRGEDWSLNMEICDIINETEEGPKDAFRAIKKRIVGNKNFHEVMLALTVLET 76
Query: 65 LMNNIGDHINEQVVRAEVI-PILVKIVKKKSDLP--VREQIFLLLDATQTSLGGASGKFP 121
+ N G + V + + +LV+ + K++ P V +++ L+ + + +S
Sbjct: 77 CVKNCGHRFHILVASQDFVESVLVRTILPKNNPPAIVHDKVLTLIQSWADAF-RSSPDLT 135
Query: 122 QYYKAYYDLVSAGVQFPQ------------RAQVVQSNRPSLQPNTTNNVPKREPSPLRR 169
Y DL G++FP R V SN S Q + N P++ S L
Sbjct: 136 GVVAVYEDLRRKGLEFPMTDLDMLSPIHTPRRSVYSSNSQSGQNSPAVNSPQQMESILHP 195
Query: 170 GRVAQKAESNT----VPESRIIQKASNVLE-------VLKEVLDAVDAKHPQGARDEFTL 218
+ ++++ P I+K + LE V+ E+L + + + E
Sbjct: 196 VTLPSGRDTSSNVPITPTQEQIKKLRSELEVVNGNVKVMSEMLTELVPSQAETSDLELLQ 255
Query: 219 DLVEQCSFQKQRVMHLVMASRDERIVSRAIEVNEQLQKVLERHD 262
+L C +QRV+ L+ + E++ + +N+ L V RH+
Sbjct: 256 ELNRTCRAMQQRVLELIPRVQHEQLTEELLLINDNLNNVFLRHE 299
>T1L1_MOUSE (Q923U0) TOM1-like 1 protein (Target of myb-like 1
protein) (Src activating and signaling molecule protein)
Length = 474
Score = 75.9 bits (185), Expect = 1e-13
Identities = 69/267 (25%), Positives = 127/267 (46%), Gaps = 18/267 (6%)
Query: 5 LVNAATSEKLSEIDWMKNIEISELVARDQRKAKDVVKAIKKRLG-NKNPNAQLYAVMLLE 63
L+ AT + DW + + I +++ Q KD VKA+KKR+ N N ++ L++
Sbjct: 18 LIEKATFAGVLTEDWGQFLHICDIINTTQDGPKDAVKALKKRISKNYNHKEIQLSLSLID 77
Query: 64 MLMNNIGDHINEQVVRAEVI-PILVKIVKKKSDLPVREQIFLLLDATQTSLGGASG-KFP 121
M + N G +V+ E I LVK++ + LP+ Q +L S G G
Sbjct: 78 MCVQNCGPSFQSLIVKKEFIKDTLVKLLNPRYTLPLETQNRILNFIKTWSQGFPGGVDVS 137
Query: 122 QYYKAYYDLVSAGVQFPQRAQVVQSNRPS--LQPNTTNNVPKREPSPLRRGRVAQKAES- 178
+ + Y DL+ GVQFP ++ + + + PN +VP +P +A K +
Sbjct: 138 EVKEVYLDLLKKGVQFPPSDGEPETRQEAGQISPNRPTSVP---TAPALSSIIAPKNPTI 194
Query: 179 NTVPESRIIQKASNVLEVLK---EVLDAVDAKHPQGARDEFTLDLVEQC----SFQKQRV 231
+ VPE I K + L+++K +V+ A+ ++ G+ + ++L+ + ++R+
Sbjct: 195 SLVPEQ--IGKLHSELDMVKMNVKVMTAILMENTPGSENHEDIELLRKLYKTGREMQERI 252
Query: 232 MHLVMASRDERIVSRAIEVNEQLQKVL 258
M L++ +E + I+VNE L +
Sbjct: 253 MDLLVVVENEDVTMELIQVNEDLNNAV 279
>TOM1_HUMAN (O60784) Target of Myb protein 1
Length = 492
Score = 72.0 bits (175), Expect = 2e-12
Identities = 65/284 (22%), Positives = 122/284 (42%), Gaps = 28/284 (9%)
Query: 6 VNAATSEKLSEIDWMKNIEISELVARDQRKAKDVVKAIKKRL-GNKNPNAQLYAVMLLEM 64
+ AT L DW N+EI +++ + KD ++A+KKR+ GNKN + + A+ +LE
Sbjct: 17 IEKATDGSLQSEDWALNMEICDIINETEEGPKDALRAVKKRIVGNKNFHEVMLALTVLET 76
Query: 65 LMNNIGDHINEQVVRAEVI-PILVKIVKKKSDLP--VREQIFLLLDATQTSLGGASGKFP 121
+ N G + V + + +LV+ + K++ P V +++ L+ + + +S
Sbjct: 77 CVKNCGHRFHVLVASQDFVESVLVRTILPKNNPPTIVHDKVLNLIQSWADAF-RSSPDLT 135
Query: 122 QYYKAYYDLVSAGVQFPQ------------RAQVVQSNRPSLQPNTTNNVPKREPS---- 165
Y DL G++FP + V S S Q + + ++E S
Sbjct: 136 GVVTIYEDLRRKGLEFPMTDLDMLSPIHTPQRTVFNSETQSGQDSVGTDSSQQEDSGQHA 195
Query: 166 ------PLRRGRVAQKAESNTVPESRI-IQKASNVLEVLKEVLDAVDAKHPQGARDEFTL 218
P+ G + + R ++ S + V+ E+L + + A E
Sbjct: 196 APLPAPPILSGDTPIAPTPEQIGKLRSELEMVSGNVRVMSEMLTELVPTQAEPADLELLQ 255
Query: 219 DLVEQCSFQKQRVMHLVMASRDERIVSRAIEVNEQLQKVLERHD 262
+L C +QRV+ L+ +E++ + VN+ L V RH+
Sbjct: 256 ELNRTCRAMQQRVLELIPQIANEQLTEELLIVNDNLNNVFLRHE 299
>GGA2_YEAST (P38817) ADP-ribosylation factor binding protein GGA2
(Golgi-localized, gamma ear-containing, ARF-binding
protein 2)
Length = 585
Score = 49.3 bits (116), Expect = 1e-05
Identities = 76/316 (24%), Positives = 134/316 (42%), Gaps = 48/316 (15%)
Query: 6 VNAATSEKLSEIDWMKNIEISELVARDQRKA-KDVVKAIKKRLGNKNPNAQLYAVMLLEM 64
+ A L+E D N++I++ + Q A +D A+ K + N+ + ++A+ LL++
Sbjct: 30 IQRACRMSLAEPDLALNLDIADYINEKQGAAPRDAAIALAKLINNRESHVAIFALSLLDV 89
Query: 65 LMNNIGDHINEQVVRAEVIPILVKIVKKKSDLPVREQIFLLLDAT----QTSLGGASGKF 120
L+ N G + Q+ R E + LVK L + L+L A QT +S K
Sbjct: 90 LVKNCGYPFHLQISRKEFLNELVKRFPGHPPLRYSKIQRLILTAIEEWYQTICKHSSYKN 149
Query: 121 PQ-YYKAYYDLVS-AGVQFPQRAQV-VQSNRPSLQPNTTNNVPKRE-------------- 163
Y + + L+ G FP+ ++ + +PS Q T + + K +
Sbjct: 150 DMGYIRDMHRLLKYKGYAFPKISESDLAVLKPSNQLKTASEIQKEQEIAQAAKLEELIRR 209
Query: 164 --PSPLRRGRVAQK-----AESNTVPESRIIQKASNVL----EVLKEVLDAVDAKHPQGA 212
P LR K E N V + I N L ++L E+L++ D+ Q
Sbjct: 210 GKPEDLREANKLMKIMAGFKEDNAVQAKQAISSELNKLKRKADLLNEMLESPDS---QNW 266
Query: 213 RDEFTLDL-----VEQCSFQKQRVMHLVMASRDERIVSRAIEVNEQLQKVLERHDDLLSS 267
+E T +L V Q FQK + D+ +V ++ N+ + ++LE+ +LL +
Sbjct: 267 DNETTQELHSALKVAQPKFQK----IIEEEQEDDALVQDLLKFNDTVNQLLEKF-NLLKN 321
Query: 268 KDTTTVN--HFDHEEA 281
D+ + H H A
Sbjct: 322 GDSNAASQIHPSHVSA 337
>GGA1_YEAST (Q06336) ADP-ribosylation factor binding protein GGA1
(Golgi-localized, gamma ear-containing, ARF-binding
protein 1)
Length = 557
Score = 48.9 bits (115), Expect = 1e-05
Identities = 52/217 (23%), Positives = 96/217 (43%), Gaps = 10/217 (4%)
Query: 6 VNAATSEKLSEIDWMKNIEISELVARDQRKA-KDVVKAIKKRLGNKNPNAQLYAVMLLEM 64
+ A L E D N+++++ + Q ++ V AI+K + N + A ++A+ LL++
Sbjct: 26 IQRACRSTLPEPDLGLNLDVADYINSKQGATPREAVLAIEKLVNNGDTQAAVFALSLLDV 85
Query: 65 LMNNIGDHINEQVVRAEVIPILVKIVKKKSDLPVREQIFLLLDAT----QTSLGGASGKF 120
L+ N G I+ Q+ R E + LVK ++ L + ++L+A QT AS K
Sbjct: 86 LVKNCGYSIHLQISRKEFLNDLVKRFPEQPPLRYSKVQQMILEAIEEWYQTICKHASYKD 145
Query: 121 P-QYYKAYYDLVS-AGVQFPQRAQVVQSNRPSLQPNTTNNVPKREPSPLRRGRVAQKAES 178
QY + L+ G FP +V N L+PN P R + A+ E
Sbjct: 146 DLQYINDMHKLLKYKGYTFP---KVGSENLAVLRPNDQLRTPSELQEEQERAQAAKLEEL 202
Query: 179 NTVPESRIIQKASNVLEVLKEVLDAVDAKHPQGARDE 215
+ +++A+ +++++ D Q +E
Sbjct: 203 LRSGKPDDLKEANKLMKIMAGFKDDTKVAVKQAINNE 239
>VP27_YEAST (P40343) Vacuolar protein sorting-associated protein
VPS27
Length = 622
Score = 47.8 bits (112), Expect = 3e-05
Identities = 39/151 (25%), Positives = 76/151 (49%), Gaps = 8/151 (5%)
Query: 5 LVNAATSEKL--SEIDWMKNIEISELVARDQRKAKDVVKAIKKRLGN--KNPNAQLYAVM 60
L+ ATSE + ++D +EIS+++ + KD ++ IKKR+ N NPN QL +
Sbjct: 12 LIEQATSESIPNGDLDLPIALEISDVLRSRRVNPKDSMRCIKKRILNTADNPNTQLSSWK 71
Query: 61 LLEMLMNNIGDHINEQVVRAEVIPILVKIV-KKKSDLPVREQIFLLLDATQTSLGGASGK 119
L + + N G +++ E + + ++ ++ S+ + E + +L + S +
Sbjct: 72 LTNICVKNGGTPFIKEICSREFMDTMEHVILREDSNEELSELVKTILYELYVAFKNDS-Q 130
Query: 120 FPQYYKAYYDLVSAGVQFPQRAQVVQSNRPS 150
K Y L+S G++FP++ + SN P+
Sbjct: 131 LNYVAKVYDKLISRGIKFPEK--LTLSNSPT 159
>HRS_DROME (Q960X8) Hepatocyte growth factor-regulated tyrosine
kinase substrate
Length = 760
Score = 47.0 bits (110), Expect = 6e-05
Identities = 32/131 (24%), Positives = 62/131 (46%), Gaps = 1/131 (0%)
Query: 9 ATSEKLSEIDWMKNIEISELVARDQRKAKDVVKAIKKRLGNKNPNAQLYAVMLLEMLMNN 68
ATS E DW + I + + + K+ AIKK++ + NP++ Y++++LE ++ N
Sbjct: 13 ATSHLRLEPDWPSILLICDEINQKDVTPKNAFAAIKKKMNSPNPHSSCYSLLVLESIVKN 72
Query: 69 IGDHINEQVVRAEVIPILVKIVKKKSDLPVREQIFLLLDATQTSLGGASGKFPQYYKAYY 128
G ++E+V E + ++ VR+++ L+ T +S K+
Sbjct: 73 CGAPVHEEVFTKENCEMFSSFLESTPHENVRQKMLELVQ-TWAYAFRSSDKYQAIKDTMT 131
Query: 129 DLVSAGVQFPQ 139
L + G FP+
Sbjct: 132 ILKAKGHTFPE 142
>GGA2_HUMAN (Q9UJY4) ADP-ribosylation factor binding protein GGA2
(Golgi-localized, gamma ear-containing, ARF-binding
protein 2) (Gamma-adaptin related protein 2) (Vear) (VHS
domain and ear domain of gamma-adaptin)
Length = 613
Score = 42.0 bits (97), Expect = 0.002
Identities = 23/88 (26%), Positives = 40/88 (45%)
Query: 6 VNAATSEKLSEIDWMKNIEISELVARDQRKAKDVVKAIKKRLGNKNPNAQLYAVMLLEML 65
+N AT +SE DW E V D + ++ + LYA+ +LEM
Sbjct: 30 LNKATDPSMSEQDWSAIQNFCEQVNTDPNGPTHAPWLLAHKIQSPQEKEALYALTVLEMC 89
Query: 66 MNNIGDHINEQVVRAEVIPILVKIVKKK 93
MN+ G+ + +V + + L+K++ K
Sbjct: 90 MNHCGEKFHSEVAKFRFLNELIKVLSPK 117
>YHA2_YEAST (P38753) Hypothetical 51.2 kDa protein in LAG1-RPL14B
intergenic region
Length = 452
Score = 38.5 bits (88), Expect = 0.020
Identities = 21/87 (24%), Positives = 42/87 (48%), Gaps = 1/87 (1%)
Query: 9 ATSEKLSEIDWMKNIEISELVARD-QRKAKDVVKAIKKRLGNKNPNAQLYAVMLLEMLMN 67
AT KL +W +++ +LV D + ++V+ I+KRL ++ N L + L L
Sbjct: 15 ATDPKLRSDNWQYILDVCDLVKEDPEDNGQEVMSLIEKRLEQQDANVILRTLSLTVSLAE 74
Query: 68 NIGDHINEQVVRAEVIPILVKIVKKKS 94
N G + +++ +L +++ S
Sbjct: 75 NCGSRLRQEISSKNFTSLLYALIESHS 101
>GGA3_HUMAN (Q9NZ52) ADP-ribosylation factor binding protein GGA3
(Golgi-localized, gamma ear-containing, ARF-binding
protein 3)
Length = 723
Score = 36.6 bits (83), Expect = 0.075
Identities = 61/293 (20%), Positives = 115/293 (38%), Gaps = 35/293 (11%)
Query: 6 VNAATSEKLSEIDWMKNIEISELVARDQRKAKDVVKAIKKRLGNKNPNAQLYAVMLLEML 65
+N AT+ + DW I + + ++ + V+ + ++ + L A+ +LE
Sbjct: 13 LNKATNPSNRQEDWEYIIGFCDQINKELEGPQIAVRLLAHKIQSPQEWEALQALTVLEAC 72
Query: 66 MNNIGDHINEQVVRAEVIPILVKIVKKK------SDLPVREQIFLLLDATQTSLGGASGK 119
M N G + +V + + L+K+V K S+ + I LL T A K
Sbjct: 73 MKNCGRRFHNEVGKFRFLNELIKVVSPKYLGDRVSEKVKTKVIELLYSWTMALPEEAKIK 132
Query: 120 FPQYYKAYYDLVSAG-VQFPQRAQVVQSNRPSLQPNTTNNV-PKREPSPLRRGRVAQK-- 175
AY+ L G VQ V ++ PS P N V E S L + K
Sbjct: 133 -----DAYHMLKRQGIVQSDPPIPVDRTLIPSPPPRPKNPVFDDEEKSKLLAKLLKSKNP 187
Query: 176 ---AESNTVPESRI------IQKASNVLEVLKEVLDAV----------DAKHPQGARDEF 216
E+N + +S + IQK + L L+EV + V + E
Sbjct: 188 DDLQEANKLIKSMVKEDEARIQKVTKRLHTLEEVNNNVRLLSEMLLHYSQEDSSDGDREL 247
Query: 217 TLDLVEQCSFQKQRVMHLVMASRD-ERIVSRAIEVNEQLQKVLERHDDLLSSK 268
+L +QC +++ + L + D + + ++ ++ L +V+ + ++ +
Sbjct: 248 MKELFDQCENKRRTLFKLASETEDNDNSLGDILQASDNLSRVINSYKTIIEGQ 300
>DCT2_HUMAN (Q13561) Dynactin complex 50 kDa subunit (50 kDa
dynein-associated polypeptide) (p50 dynamitin) (DCTN-50)
(Dynactin 2)
Length = 400
Score = 35.0 bits (79), Expect = 0.22
Identities = 62/248 (25%), Positives = 105/248 (42%), Gaps = 24/248 (9%)
Query: 40 VKAIKKRLGNKNPNAQLYAVMLLEMLMNNIGDHINEQVVRAEVIPIL---VKIVKKKSDL 96
V+ IK + +L V+L + L + +Q+V + + +L I D
Sbjct: 116 VEKIKTTVKESATEEKLTPVLLAKQLAA-----LKQQLVASHLEKLLGPDAAINLTDPDG 170
Query: 97 PVREQIFLLLDATQTSLGGASGKF----PQYYKAYYDLVSAGVQ--FPQRAQVVQSNRPS 150
+ +++ L L+AT+ S GG+ GK P Y+L S Q F Q A+V + +
Sbjct: 171 ALAKRLLLQLEATKNSKGGSGGKTTGTPPDSSLVTYELHSRPEQDKFSQAAKVAELEKRL 230
Query: 151 LQPNTTNNVPKREPSPLRRG-RVAQKAESNTVPESRIIQKASNVLEV----LKEVLDAVD 205
+ T + +PL G + A E+ + ++++ VL+ L+ VL V+
Sbjct: 231 TELETAVRCDQDAQNPLSAGLQGACLMETVELLQAKVSALDLAVLDQVEARLQSVLGKVN 290
Query: 206 --AKHPQGARDEFTLDLVEQCSFQKQRVMHLVMASRDERIVSRAIEVNEQLQKVLERHDD 263
AKH D T V Q QR + AS +V R + + +QL + +
Sbjct: 291 EIAKHKASVEDADTQSKVHQLYETIQRWSPI--ASTLPELVQRLVTI-KQLHEQAMQFGQ 347
Query: 264 LLSSKDTT 271
LL+ DTT
Sbjct: 348 LLTHLDTT 355
>MAY4_SCHCO (P37935) Mating-type protein A-alpha Y4
Length = 928
Score = 33.9 bits (76), Expect = 0.49
Identities = 20/70 (28%), Positives = 39/70 (55%), Gaps = 3/70 (4%)
Query: 119 KFPQYYKAYYDLVSAGVQFPQRAQV--VQSNRPSLQPNTTNNVPKREPSPLRRGRVAQKA 176
K P+ + + +L A +P +Q V S +PS Q ++++ P+++ L +GR + +
Sbjct: 382 KSPRAFDSRAELAFAQAAYPSPSQYAYVHSRKPSAQKPSSDS-PRKDFGQLGKGRPSSNS 440
Query: 177 ESNTVPESRI 186
S+TVP R+
Sbjct: 441 TSSTVPPHRV 450
>PLAK_XENLA (P30998) Junction plakoglobin (Desmoplakin III)
Length = 738
Score = 33.1 bits (74), Expect = 0.83
Identities = 19/65 (29%), Positives = 31/65 (47%), Gaps = 3/65 (4%)
Query: 220 LVEQCSFQKQRVMHLVMASRDERIVSRAIEVNEQLQKVLERHDDLLSSKDTTTVNHFDHE 279
L E K +MHL+ D + +RAI +L K+L D ++ +K + VN +
Sbjct: 112 LAEPSQMLKSAIMHLINYQDDAELATRAI---PELTKLLNDEDPMVVNKASMIVNQLSKK 168
Query: 280 EAEEE 284
EA +
Sbjct: 169 EASRK 173
>KINH_BOTCI (Q86ZC1) Kinesin heavy chain
Length = 880
Score = 33.1 bits (74), Expect = 0.83
Identities = 20/78 (25%), Positives = 40/78 (50%), Gaps = 9/78 (11%)
Query: 197 LKEVLDAVDAKHPQGAR---------DEFTLDLVEQCSFQKQRVMHLVMASRDERIVSRA 247
+K+V+D +D+ H Q + +E LVE +Q + + +S ++ V R
Sbjct: 577 IKKVIDHIDSLHEQSSAGEAIPPDEFEELKAKLVETQGIVRQAELSMFGSSSNDANVKRR 636
Query: 248 IEVNEQLQKVLERHDDLL 265
E+ ++LQ + + ++DLL
Sbjct: 637 EELEQRLQVLEQEYEDLL 654
>YD85_SCHPO (Q10410) Hypothetical protein C1F3.05 in chromosome I
Length = 510
Score = 32.7 bits (73), Expect = 1.1
Identities = 20/82 (24%), Positives = 41/82 (49%), Gaps = 1/82 (1%)
Query: 3 AELVNAATSEKLSEIDWMKNIEISELVARDQRKA-KDVVKAIKKRLGNKNPNAQLYAVML 61
++ ++ AT + E + NIEI++L+ + ++ I KR+ + NP A+ L
Sbjct: 8 SKYIDKATDQFNLEPNLALNIEIADLINEKKGNTPREAALLILKRVNSANPTVSYLALHL 67
Query: 62 LEMLMNNIGDHINEQVVRAEVI 83
L++ + N G + Q+ E +
Sbjct: 68 LDICVKNCGYPFHFQIASEEFL 89
>MPT1_MOUSE (Q9DBR7) Protein phosphatase 1 regulatory subunit 12A
(Myosin phosphatase targeting subunit 1) (Myosin
phosphatase target subunit 1)
Length = 1004
Score = 32.7 bits (73), Expect = 1.1
Identities = 25/78 (32%), Positives = 41/78 (52%), Gaps = 8/78 (10%)
Query: 214 DEFTLDLVEQCSFQKQRVMHLVMASRDER---IVSRAIEVNEQLQKVLERHDDLL--SSK 268
DE L +E+ +KQ ++H RD++ I S A N Q QK + + L+ K
Sbjct: 273 DEDILGYLEELQ-KKQTLLH--SEKRDKKSPLIESTANMENNQPQKAFKNKETLIIEPEK 329
Query: 269 DTTTVNHFDHEEAEEEEE 286
+ + + +HE+A+EEEE
Sbjct: 330 NASRIESLEHEKADEEEE 347
>TOP2_CANAL (P87078) DNA topoisomerase II (EC 5.99.1.3)
Length = 1461
Score = 32.3 bits (72), Expect = 1.4
Identities = 29/112 (25%), Positives = 56/112 (49%), Gaps = 12/112 (10%)
Query: 187 IQKASNVLEVLKEVLDAVDAKHPQGARDEFTLDLVEQCSFQKQRVMHLVMASRDERIVSR 246
I+K ++VLE++K+ V ++ Q +D T +L Q ++ + M E+ +S
Sbjct: 1043 IKKYNDVLEIIKDFY-YVRLEYYQKRKDYMTDNLQNQLLMLSEQARFIKMII--EKQLSV 1099
Query: 247 AIEVNEQLQKVLERHD---------DLLSSKDTTTVNHFDHEEAEEEEEPEQ 289
A + +QL +LE H+ + SS++ T + D EE +E+E ++
Sbjct: 1100 ANKKKKQLVALLEEHNFTKFSKDGKPIKSSEELLTGDDADEEEETQEQEGDE 1151
>DCT2_MOUSE (Q99KJ8) Dynactin complex 50 kDa subunit (50 kDa
dynein-associated polypeptide) (p50 dynamitin) (DCTN-50)
(Dynactin 2) (Growth cone membrane protein 23-48K)
(GMP23-48K)
Length = 401
Score = 32.3 bits (72), Expect = 1.4
Identities = 50/191 (26%), Positives = 84/191 (43%), Gaps = 17/191 (8%)
Query: 95 DLPVREQIFLLLDATQTSLGGASGKF-----PQYYKAYYDLVSAGVQ--FPQRAQVVQSN 147
D + +++ L L+AT++S G + GK P Y+L S Q F Q A+V +
Sbjct: 169 DGALAKRLLLQLEATKSSKGSSGGKATAGAPPDSSLVTYELHSRPEQDKFSQAAKVAELE 228
Query: 148 RPSLQPNTTNNVPKREPSPLRRG-RVAQKAESNTVPESRIIQKASNVLEV----LKEVLD 202
+ + T + +PL G + A E+ + ++++ VL+ L+ VL
Sbjct: 229 KRLTELEATVRCDQDAQNPLSAGLQGACLMETVELLQAKVSALDLAVLDQVEARLQSVLG 288
Query: 203 AVD--AKHPQGARDEFTLDLVEQCSFQKQRVMHLVMASRDERIVSRAIEVNEQLQKVLER 260
V+ AKH D T + V Q QR +AS +V R + + +QL + +
Sbjct: 289 KVNEIAKHKASVEDADTQNKVHQLYETIQR--WSPVASTLPELVQRLVTI-KQLHEQAMQ 345
Query: 261 HDDLLSSKDTT 271
LL+ DTT
Sbjct: 346 FGQLLTHLDTT 356
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.314 0.130 0.348
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 30,518,001
Number of Sequences: 164201
Number of extensions: 1191432
Number of successful extensions: 4011
Number of sequences better than 10.0: 53
Number of HSP's better than 10.0 without gapping: 14
Number of HSP's successfully gapped in prelim test: 39
Number of HSP's that attempted gapping in prelim test: 3979
Number of HSP's gapped (non-prelim): 61
length of query: 292
length of database: 59,974,054
effective HSP length: 109
effective length of query: 183
effective length of database: 42,076,145
effective search space: 7699934535
effective search space used: 7699934535
T: 11
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (22.0 bits)
S2: 65 (29.6 bits)
Medicago: description of AC148755.10


