

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148653.1 - phase: 0
(693 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
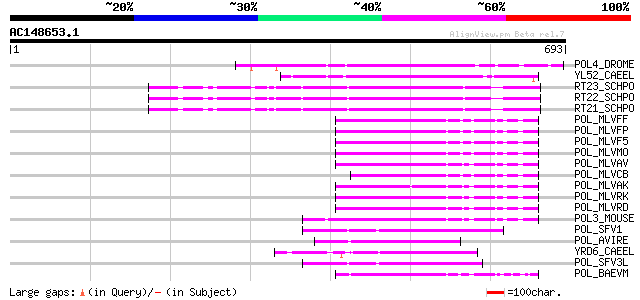
Score E
Sequences producing significant alignments: (bits) Value
POL4_DROME (P10394) Retrovirus-related Pol polyprotein from tran... 117 8e-26
YL52_CAEEL (P34431) Hypothetical protein F44E2.2 in chromosome III 107 8e-23
RT23_SCHPO (Q9UR07) Retrotransposable element Tf2 155 kDa protei... 100 1e-20
RT22_SCHPO (Q9C0R2) Retrotransposable element Tf2 155 kDa protei... 100 1e-20
RT21_SCHPO (Q05654) Retrotransposable element Tf2 155 kDa protei... 100 1e-20
POL_MLVFF (P26809) Pol polyprotein [Contains: Protease (EC 3.4.2... 94 2e-18
POL_MLVFP (P26808) Pol polyprotein [Contains: Protease (EC 3.4.2... 93 2e-18
POL_MLVF5 (P26810) Pol polyprotein [Contains: Protease (EC 3.4.2... 93 2e-18
POL_MLVMO (P03355) Pol polyprotein [Contains: Protease (EC 3.4.2... 92 5e-18
POL_MLVAV (P03356) Pol polyprotein [Contains: Protease (EC 3.4.2... 92 5e-18
POL_MLVCB (P08361) Pol polyprotein [Contains: Reverse transcript... 90 2e-17
POL_MLVAK (P03357) Pol polyprotein [Contains: Reverse transcript... 90 2e-17
POL_MLVRK (P31795) Pol polyprotein [Contains: Protease (EC 3.4.2... 87 1e-16
POL_MLVRD (P11227) Pol polyprotein [Contains: Protease (EC 3.4.2... 87 1e-16
POL3_MOUSE (P11367) Retrovirus-related Pol polyprotein (Endonucl... 87 1e-16
POL_SFV1 (P23074) Pol polyprotein [Contains: Protease (EC 3.4.23... 86 3e-16
POL_AVIRE (P03360) Pol polyprotein [Contains: Reverse transcript... 86 4e-16
YRD6_CAEEL (Q09575) Hypothetical protein K02A2.6 in chromosome II 85 7e-16
POL_SFV3L (P27401) Pol polyprotein [Contains: Protease (EC 3.4.2... 84 1e-15
POL_BAEVM (P10272) Pol polyprotein [Contains: Protease (EC 3.4.2... 80 2e-14
>POL4_DROME (P10394) Retrovirus-related Pol polyprotein from
transposon 412 [Contains: Protease (EC 3.4.23.-); Reverse
transcriptase (EC 2.7.7.49); Endonuclease]
Length = 1237
Score = 117 bits (294), Expect = 8e-26
Identities = 104/424 (24%), Positives = 188/424 (43%), Gaps = 30/424 (7%)
Query: 282 DIKCFLQRQEYPLGASNKD-------KKTLRRLSGNFFLNGDILYKRNFDMVLLRCV--- 331
D+ FLQR E G + KK +S + F N +N + LL V
Sbjct: 828 DLDQFLQRLELQAGIYDISQIKMAPWKKIFEHVSIDKFKNMGNKILKNLKVALLNPVTQI 887
Query: 332 -DKHEADLLMHEIHEGSFGTHSSGPTMAKKMLRAGYYWMTMEADCYKHAKRCYKCQIYAD 390
++ E + ++ +H+ +G T ++ YYW M ++ ++C KCQ
Sbjct: 888 NNEKEKEAILSTLHDDPIQGGHTGITKTLAKVKRHYYWKNMSKYIKEYVRKCQKCQKAKT 947
Query: 391 RIHVPPTALNVLSSP-WPFSMWGIDMIGRIEPKASNGHRFILVAIDYFTKWVEAASYANV 449
H T + + +P F +D IG + PK+ NG+ + + I TK++ A AN
Sbjct: 948 TKHTK-TPMTITETPEHAFDRVVVDTIGPL-PKSENGNEYAVTLICDLTKYLVAIPIANK 1005
Query: 450 TKQVVVKFIKNHIICRYGVPSRIITDNGTNLNNKMMKELCDDFKIEHHNSSPYRPQMNGA 509
+ + V K I I +YG ITD GT N ++ +LC KI++ S+ + Q G
Sbjct: 1006 SAKTVAKAIFESFILKYGPMKTFITDMGTEYKNSIITDLCKYLKIKNITSTAHHHQTVGV 1065
Query: 510 VEAANKNIKKIIQKMVVTYK-DWHEMLPFALHGYRTSIRTSTGATPFSLVYGMEAVLPIE 568
VE +++ + + I+ + T K DW L + ++ + T+ P+ LV+G + LP
Sbjct: 1066 VERSHRTLNEYIRSYISTDKTDWDVWLQYFVYCFNTTQSMVHNYCPYELVFGRTSNLPKH 1125
Query: 569 V-EIPSIRVLMEAELSEAEWVQSRYDQLNLIEEKRMSALCHGQLYQKRMKHAFDKKVHPR 627
++ SI + + ++ + +L + + L + ++++ K +D KV
Sbjct: 1126 FNKLHSIEPIYNID----DYAKESKYRLEVAYARARKLL---EAHKEKNKENYDLKVKDI 1178
Query: 628 EFKEGDLVLKKILTFQPDSRGKWTPNYEGPYVVKKAYSGGAMTLQTMDGEELQRPVNTDA 687
E + GD VL + + K Y GPY ++ +TL T + ++ V+ D
Sbjct: 1179 ELEVGDKVL-----LRNEVGHKLDFKYTGPYKIESIGDNNNITLLT--NKNKKQIVHKDR 1231
Query: 688 VKKY 691
+KK+
Sbjct: 1232 LKKF 1235
>YL52_CAEEL (P34431) Hypothetical protein F44E2.2 in chromosome III
Length = 2186
Score = 107 bits (268), Expect = 8e-23
Identities = 80/327 (24%), Positives = 147/327 (44%), Gaps = 14/327 (4%)
Query: 339 LMHEIHEGSFGTHSSGPTMAKKMLRAGYYWMTMEADCYKHAKRCYKCQIYADRIHVPPTA 398
L+ E+HEG H G +M+ +YW M + C KC D + ++
Sbjct: 1468 LLKELHEGMLAGHF-GIKKMWRMVHRKFYWPQMRVCVENCVRTCAKCLCANDHSKLT-SS 1525
Query: 399 LNVLSSPWPFSMWGIDMIGRIEPKASNGHRFILVAIDYFTKWVEAASYANVTKQVVVK-F 457
L +P + D++ + G+R+IL ID FTK+ A + + V+K F
Sbjct: 1526 LTPYRMTFPLEIVACDLMD--VGLSVQGNRYILTIIDLFTKYGTAVPIPDKKAETVLKAF 1583
Query: 458 IKNHIICRYGVPSRIITDNGTNLNNKMMKELCDDFKIEHHNSSPYRPQMNGAVEAANKNI 517
++ I +P +++TD G N + + KIEH + Y + NGAVE NK I
Sbjct: 1584 VERWAIGEGRIPLKLLTDQGKEFVNGLFAQFTHMLKIEHITTKGYNSRANGAVERFNKTI 1643
Query: 518 KKIIQKMVVTYKDWHEMLPFALHGYRTSIRTSTGATPFSLVYGMEAVLPIEVEIPSIRVL 577
I++K +W + + +A++ Y + +TG TP L++G + + P+E+ +
Sbjct: 1644 MHIMKKKTAVPMEWDDQVVYAVYAYNNCVHENTGETPMFLMHGRDVMGPLEMSGEDAVGI 1703
Query: 578 MEAELSEAEWVQSRYDQLNLIEEKRMSALCHGQLYQKRMKHAFDKKVHPREFKEGDLVLK 637
A++ E + + ++ L++ +++ A H Q+ K FD+K ++ + +
Sbjct: 1704 NYADMDEYKHLLTQ----ELLKVQKI-AKEHAMREQESYKSLFDQKYASKKHRFPQPGSR 1758
Query: 638 KILTFQPDSRGKWTP----NYEGPYVV 660
+L + G P + GPY V
Sbjct: 1759 VLLEIPSEKLGAQCPKLVNKWSGPYRV 1785
>RT23_SCHPO (Q9UR07) Retrotransposable element Tf2 155 kDa protein
type 3
Length = 1333
Score = 100 bits (249), Expect = 1e-20
Identities = 117/497 (23%), Positives = 208/497 (41%), Gaps = 45/497 (9%)
Query: 174 IDLRIKNLDIYGDSALVINQITGEWETRHPGLVPYKDYARRLLTFFNKVELHHIPRDENQ 233
++ I+ I D +I +IT E E + L ++ + L FN E+++ P N
Sbjct: 770 LESTIEPFKILTDHRNLIGRITNESEPENKRLARWQLF----LQDFN-FEINYRPGSANH 824
Query: 234 MADALATLSSMYQVNRWNEVPLIHIRHLERPAHVFAIEEVVDDKPWFHDIKCFLQRQEYP 293
+ADAL+ + V+ +P + E + F + + D + + +
Sbjct: 825 IADALSRI-----VDETEPIP----KDSEDNSINFVNQISITDDFKNQVVTEYTNDTKLL 875
Query: 294 LGASNKDKKTLRRLSGNFFLNGDILYKRNFDMVLLRCVDKHEADLLMHEIHEGSFGTHSS 353
+N+DK R+ N L +L D +LL D ++ + HE H
Sbjct: 876 NLLNNEDK----RVEENIQLKDGLLINSK-DQILLPN-DTQLTRTIIKKYHEEGKLIHPG 929
Query: 354 GPTMAKKMLRAGYYWMTMEADCYKHAKRCYKCQIYADRIHVPPTALNVLS-SPWPFSMWG 412
+ +LR + W + ++ + C+ CQI R H P L + S P+
Sbjct: 930 IELLTNIILRR-FTWKGIRKQIQEYVQNCHTCQINKSRNHKPYGPLQPIPPSERPWESLS 988
Query: 413 IDMIGRIEPKASNGHRFILVAIDYFTKW-VEAASYANVTKQVVVKFIKNHIICRYGVPSR 471
+D I + S+G+ + V +D F+K + ++T + + +I +G P
Sbjct: 989 MDFITALPE--SSGYNALFVVVDRFSKMAILVPCTKSITAEQTARMFDQRVIAYFGNPKE 1046
Query: 472 IITDNGTNLNNKMMKELCDDFKIEHHNSSPYRPQMNGAVEAANKNIKKIIQKMVVTYKD- 530
II DN ++ K+ + S PYRPQ +G E N+ ++K+++ + T+ +
Sbjct: 1047 IIADNDHIFTSQTWKDFAHKYNFVMKFSLPYRPQTDGQTERTNQTVEKLLRCVCSTHPNT 1106
Query: 531 WHEMLPFALHGYRTSIRTSTGATPFSLVYGMEAVLPIEVEIPSIRVLMEAELSEAEWV-Q 589
W + + Y +I ++T TPF +V+ L +E+PS + E V Q
Sbjct: 1107 WVDHISLVQQSYNNAIHSATQMTPFEIVHRYSPALS-PLELPSFSDKTDENSQETIQVFQ 1165
Query: 590 SRYDQLNLIEEKRMSALCHGQLYQKRMKHAFDKKVHP-REFKEGDLVL-KKILTFQPDSR 647
+ + LN K MK FD K+ EF+ GDLV+ K+ T
Sbjct: 1166 TVKEHLNTNNIK--------------MKKYFDMKIQEIEEFQPGDLVMVKRTKTGFLHKS 1211
Query: 648 GKWTPNYEGP-YVVKKA 663
K P++ GP YV++K+
Sbjct: 1212 NKLAPSFAGPFYVLQKS 1228
Score = 34.7 bits (78), Expect = 0.87
Identities = 31/110 (28%), Positives = 52/110 (47%), Gaps = 24/110 (21%)
Query: 3 EKTCCALAWAAKRLRHYMISHTTWLISKMDPIKYIFEKPALTGRI-----------ARWQ 51
+K A+ + K RHY L S ++P K + + L GRI ARWQ
Sbjct: 753 DKEMLAIIKSLKHWRHY-------LESTIEPFKILTDHRNLIGRITNESEPENKRLARWQ 805
Query: 52 MLLSE--YDIEYRSQKAIKGSILADHLAHQPIEDYQPIKFDFPDEEIMYL 99
+ L + ++I YR A + +AD L+ + +++ +PI D D I ++
Sbjct: 806 LFLQDFNFEINYRPGSA---NHIADALS-RIVDETEPIPKDSEDNSINFV 851
>RT22_SCHPO (Q9C0R2) Retrotransposable element Tf2 155 kDa protein
type 2
Length = 1333
Score = 100 bits (249), Expect = 1e-20
Identities = 117/497 (23%), Positives = 208/497 (41%), Gaps = 45/497 (9%)
Query: 174 IDLRIKNLDIYGDSALVINQITGEWETRHPGLVPYKDYARRLLTFFNKVELHHIPRDENQ 233
++ I+ I D +I +IT E E + L ++ + L FN E+++ P N
Sbjct: 770 LESTIEPFKILTDHRNLIGRITNESEPENKRLARWQLF----LQDFN-FEINYRPGSANH 824
Query: 234 MADALATLSSMYQVNRWNEVPLIHIRHLERPAHVFAIEEVVDDKPWFHDIKCFLQRQEYP 293
+ADAL+ + V+ +P + E + F + + D + + +
Sbjct: 825 IADALSRI-----VDETEPIP----KDSEDNSINFVNQISITDDFKNQVVTEYTNDTKLL 875
Query: 294 LGASNKDKKTLRRLSGNFFLNGDILYKRNFDMVLLRCVDKHEADLLMHEIHEGSFGTHSS 353
+N+DK R+ N L +L D +LL D ++ + HE H
Sbjct: 876 NLLNNEDK----RVEENIQLKDGLLINSK-DQILLPN-DTQLTRTIIKKYHEEGKLIHPG 929
Query: 354 GPTMAKKMLRAGYYWMTMEADCYKHAKRCYKCQIYADRIHVPPTALNVLS-SPWPFSMWG 412
+ +LR + W + ++ + C+ CQI R H P L + S P+
Sbjct: 930 IELLTNIILRR-FTWKGIRKQIQEYVQNCHTCQINKSRNHKPYGPLQPIPPSERPWESLS 988
Query: 413 IDMIGRIEPKASNGHRFILVAIDYFTKW-VEAASYANVTKQVVVKFIKNHIICRYGVPSR 471
+D I + S+G+ + V +D F+K + ++T + + +I +G P
Sbjct: 989 MDFITALPE--SSGYNALFVVVDRFSKMAILVPCTKSITAEQTARMFDQRVIAYFGNPKE 1046
Query: 472 IITDNGTNLNNKMMKELCDDFKIEHHNSSPYRPQMNGAVEAANKNIKKIIQKMVVTYKD- 530
II DN ++ K+ + S PYRPQ +G E N+ ++K+++ + T+ +
Sbjct: 1047 IIADNDHIFTSQTWKDFAHKYNFVMKFSLPYRPQTDGQTERTNQTVEKLLRCVCSTHPNT 1106
Query: 531 WHEMLPFALHGYRTSIRTSTGATPFSLVYGMEAVLPIEVEIPSIRVLMEAELSEAEWV-Q 589
W + + Y +I ++T TPF +V+ L +E+PS + E V Q
Sbjct: 1107 WVDHISLVQQSYNNAIHSATQMTPFEIVHRYSPALS-PLELPSFSDKTDENSQETIQVFQ 1165
Query: 590 SRYDQLNLIEEKRMSALCHGQLYQKRMKHAFDKKVHP-REFKEGDLVL-KKILTFQPDSR 647
+ + LN K MK FD K+ EF+ GDLV+ K+ T
Sbjct: 1166 TVKEHLNTNNIK--------------MKKYFDMKIQEIEEFQPGDLVMVKRTKTGFLHKS 1211
Query: 648 GKWTPNYEGP-YVVKKA 663
K P++ GP YV++K+
Sbjct: 1212 NKLAPSFAGPFYVLQKS 1228
Score = 34.7 bits (78), Expect = 0.87
Identities = 31/110 (28%), Positives = 52/110 (47%), Gaps = 24/110 (21%)
Query: 3 EKTCCALAWAAKRLRHYMISHTTWLISKMDPIKYIFEKPALTGRI-----------ARWQ 51
+K A+ + K RHY L S ++P K + + L GRI ARWQ
Sbjct: 753 DKEMLAIIKSLKHWRHY-------LESTIEPFKILTDHRNLIGRITNESEPENKRLARWQ 805
Query: 52 MLLSE--YDIEYRSQKAIKGSILADHLAHQPIEDYQPIKFDFPDEEIMYL 99
+ L + ++I YR A + +AD L+ + +++ +PI D D I ++
Sbjct: 806 LFLQDFNFEINYRPGSA---NHIADALS-RIVDETEPIPKDSEDNSINFV 851
>RT21_SCHPO (Q05654) Retrotransposable element Tf2 155 kDa protein
type 1
Length = 1333
Score = 100 bits (249), Expect = 1e-20
Identities = 117/497 (23%), Positives = 208/497 (41%), Gaps = 45/497 (9%)
Query: 174 IDLRIKNLDIYGDSALVINQITGEWETRHPGLVPYKDYARRLLTFFNKVELHHIPRDENQ 233
++ I+ I D +I +IT E E + L ++ + L FN E+++ P N
Sbjct: 770 LESTIEPFKILTDHRNLIGRITNESEPENKRLARWQLF----LQDFN-FEINYRPGSANH 824
Query: 234 MADALATLSSMYQVNRWNEVPLIHIRHLERPAHVFAIEEVVDDKPWFHDIKCFLQRQEYP 293
+ADAL+ + V+ +P + E + F + + D + + +
Sbjct: 825 IADALSRI-----VDETEPIP----KDSEDNSINFVNQISITDDFKNQVVTEYTNDTKLL 875
Query: 294 LGASNKDKKTLRRLSGNFFLNGDILYKRNFDMVLLRCVDKHEADLLMHEIHEGSFGTHSS 353
+N+DK R+ N L +L D +LL D ++ + HE H
Sbjct: 876 NLLNNEDK----RVEENIQLKDGLLINSK-DQILLPN-DTQLTRTIIKKYHEEGKLIHPG 929
Query: 354 GPTMAKKMLRAGYYWMTMEADCYKHAKRCYKCQIYADRIHVPPTALNVLS-SPWPFSMWG 412
+ +LR + W + ++ + C+ CQI R H P L + S P+
Sbjct: 930 IELLTNIILRR-FTWKGIRKQIQEYVQNCHTCQINKSRNHKPYGPLQPIPPSERPWESLS 988
Query: 413 IDMIGRIEPKASNGHRFILVAIDYFTKW-VEAASYANVTKQVVVKFIKNHIICRYGVPSR 471
+D I + S+G+ + V +D F+K + ++T + + +I +G P
Sbjct: 989 MDFITALPE--SSGYNALFVVVDRFSKMAILVPCTKSITAEQTARMFDQRVIAYFGNPKE 1046
Query: 472 IITDNGTNLNNKMMKELCDDFKIEHHNSSPYRPQMNGAVEAANKNIKKIIQKMVVTYKD- 530
II DN ++ K+ + S PYRPQ +G E N+ ++K+++ + T+ +
Sbjct: 1047 IIADNDHIFTSQTWKDFAHKYNFVMKFSLPYRPQTDGQTERTNQTVEKLLRCVCSTHPNT 1106
Query: 531 WHEMLPFALHGYRTSIRTSTGATPFSLVYGMEAVLPIEVEIPSIRVLMEAELSEAEWV-Q 589
W + + Y +I ++T TPF +V+ L +E+PS + E V Q
Sbjct: 1107 WVDHISLVQQSYNNAIHSATQMTPFEIVHRYSPALS-PLELPSFSDKTDENSQETIQVFQ 1165
Query: 590 SRYDQLNLIEEKRMSALCHGQLYQKRMKHAFDKKVHP-REFKEGDLVL-KKILTFQPDSR 647
+ + LN K MK FD K+ EF+ GDLV+ K+ T
Sbjct: 1166 TVKEHLNTNNIK--------------MKKYFDMKIQEIEEFQPGDLVMVKRTKTGFLHKS 1211
Query: 648 GKWTPNYEGP-YVVKKA 663
K P++ GP YV++K+
Sbjct: 1212 NKLAPSFAGPFYVLQKS 1228
Score = 34.7 bits (78), Expect = 0.87
Identities = 31/110 (28%), Positives = 52/110 (47%), Gaps = 24/110 (21%)
Query: 3 EKTCCALAWAAKRLRHYMISHTTWLISKMDPIKYIFEKPALTGRI-----------ARWQ 51
+K A+ + K RHY L S ++P K + + L GRI ARWQ
Sbjct: 753 DKEMLAIIKSLKHWRHY-------LESTIEPFKILTDHRNLIGRITNESEPENKRLARWQ 805
Query: 52 MLLSE--YDIEYRSQKAIKGSILADHLAHQPIEDYQPIKFDFPDEEIMYL 99
+ L + ++I YR A + +AD L+ + +++ +PI D D I ++
Sbjct: 806 LFLQDFNFEINYRPGSA---NHIADALS-RIVDETEPIPKDSEDNSINFV 851
>POL_MLVFF (P26809) Pol polyprotein [Contains: Protease (EC 3.4.23.-);
Reverse transcriptase/ribonuclease H (EC 2.7.7.49) (EC
3.1.26.4) (RT); Integrase (IN)]
Length = 1204
Score = 93.6 bits (231), Expect = 2e-18
Identities = 74/256 (28%), Positives = 123/256 (47%), Gaps = 22/256 (8%)
Query: 407 PFSMWGIDMIGRIEPKASNGHRFILVAIDYFTKWVEAASYANVTKQVVVKFIKNHIICRY 466
P + W ID ++P G++++LV ID F+ WVEA T +VV K + I R+
Sbjct: 914 PGTHWEIDFT-EVKP-GLYGYKYLLVFIDTFSGWVEAFPTKKETAKVVTKKLLEEIFPRF 971
Query: 467 GVPSRIITDNGTNLNNKMMKELCDDFKIEHHNSSPYRPQMNGAVEAANKNIKKIIQKMVV 526
G+P + TDNG +K+ + + D ++ YRPQ +G VE N+ IK+ + K+ +
Sbjct: 972 GMPQVLGTDNGPAFVSKVSQTVADLLGVDWKLHCAYRPQSSGQVERMNRTIKETLTKLTL 1031
Query: 527 T--YKDWHEMLPFALHGYRTSIRTSTGATPFSLVYGMEAVLPIEVEIPSIRVLMEAELSE 584
+DW +LP AL+ R + G TP+ ++YG P V P + A+++
Sbjct: 1032 ATGSRDWVLLLPLALYRARNT-PGPHGLTPYEILYGAP---PPLVNFPDPDM---AKVTH 1084
Query: 585 AEWVQSRYDQLNLIEEKRMSALCHGQLYQKRMKHAFDKKVHPREFKEGDLVLKKILTFQP 644
+Q+ L L++ + L YQ+++ D+ V P F+ GD V +
Sbjct: 1085 NPSLQAHLQALYLVQHEVWRPL--AAAYQEQL----DRPVVPHPFRVGDTV-----WVRR 1133
Query: 645 DSRGKWTPNYEGPYVV 660
P ++GPY V
Sbjct: 1134 HQTKNLEPRWKGPYTV 1149
>POL_MLVFP (P26808) Pol polyprotein [Contains: Protease (EC 3.4.23.-);
Reverse transcriptase/ribonuclease H (EC 2.7.7.49) (EC
3.1.26.4) (RT); Integrase (IN)]
Length = 1204
Score = 93.2 bits (230), Expect = 2e-18
Identities = 73/256 (28%), Positives = 123/256 (47%), Gaps = 22/256 (8%)
Query: 407 PFSMWGIDMIGRIEPKASNGHRFILVAIDYFTKWVEAASYANVTKQVVVKFIKNHIICRY 466
P + W ID ++P G++++LV +D F+ WVEA T +VV K + I R+
Sbjct: 914 PGTHWEIDFT-EVKP-GLYGYKYLLVFVDTFSGWVEAFPTKKETAKVVTKKLLEEIFPRF 971
Query: 467 GVPSRIITDNGTNLNNKMMKELCDDFKIEHHNSSPYRPQMNGAVEAANKNIKKIIQKMVV 526
G+P + TDNG +K+ + + D ++ YRPQ +G VE N+ IK+ + K+ +
Sbjct: 972 GMPQVLGTDNGPAFVSKVSQTVADLLGVDWKLHCAYRPQSSGQVERMNRTIKETLTKLTL 1031
Query: 527 T--YKDWHEMLPFALHGYRTSIRTSTGATPFSLVYGMEAVLPIEVEIPSIRVLMEAELSE 584
+DW +LP AL+ R + G TP+ ++YG P V P + A+++
Sbjct: 1032 ATGSRDWVLLLPLALYRARNT-PGPHGLTPYEILYGAP---PPLVNFPDPDM---AKVTH 1084
Query: 585 AEWVQSRYDQLNLIEEKRMSALCHGQLYQKRMKHAFDKKVHPREFKEGDLVLKKILTFQP 644
+Q+ L L++ + L YQ+++ D+ V P F+ GD V +
Sbjct: 1085 NPSLQAHLQALYLVQHEVWRPL--AAAYQEQL----DRPVVPHPFRVGDTV-----WVRR 1133
Query: 645 DSRGKWTPNYEGPYVV 660
P ++GPY V
Sbjct: 1134 HQTKNLEPRWKGPYTV 1149
>POL_MLVF5 (P26810) Pol polyprotein [Contains: Protease (EC 3.4.23.-);
Reverse transcriptase/ribonuclease H (EC 2.7.7.49) (EC
3.1.26.4) (RT); Integrase (IN)]
Length = 1204
Score = 93.2 bits (230), Expect = 2e-18
Identities = 73/256 (28%), Positives = 123/256 (47%), Gaps = 22/256 (8%)
Query: 407 PFSMWGIDMIGRIEPKASNGHRFILVAIDYFTKWVEAASYANVTKQVVVKFIKNHIICRY 466
P + W ID ++P G++++LV +D F+ WVEA T +VV K + I R+
Sbjct: 914 PGTHWEIDFT-EVKP-GLYGYKYLLVFVDTFSGWVEAFPTKKETAKVVTKKLLEEIFPRF 971
Query: 467 GVPSRIITDNGTNLNNKMMKELCDDFKIEHHNSSPYRPQMNGAVEAANKNIKKIIQKMVV 526
G+P + TDNG +K+ + + D ++ YRPQ +G VE N+ IK+ + K+ +
Sbjct: 972 GMPQVLGTDNGPAFVSKVSQTVADLLGVDWKLHCAYRPQSSGQVERMNRTIKETLTKLTL 1031
Query: 527 T--YKDWHEMLPFALHGYRTSIRTSTGATPFSLVYGMEAVLPIEVEIPSIRVLMEAELSE 584
+DW +LP AL+ R + G TP+ ++YG P V P + A+++
Sbjct: 1032 ATGSRDWVLLLPLALYRARNT-PGPHGLTPYEILYGAP---PPLVNFPDPDM---AKVTH 1084
Query: 585 AEWVQSRYDQLNLIEEKRMSALCHGQLYQKRMKHAFDKKVHPREFKEGDLVLKKILTFQP 644
+Q+ L L++ + L YQ+++ D+ V P F+ GD V +
Sbjct: 1085 NPSLQAHLQALYLVQHEVWRPL--AAAYQEQL----DRPVVPHPFRVGDTV-----WVRR 1133
Query: 645 DSRGKWTPNYEGPYVV 660
P ++GPY V
Sbjct: 1134 HQTKNLEPRWKGPYTV 1149
>POL_MLVMO (P03355) Pol polyprotein [Contains: Protease (EC 3.4.23.-);
Reverse transcriptase/ribonuclease H (EC 2.7.7.49) (EC
3.1.26.4) (RT); Integrase (IN)]
Length = 1199
Score = 92.0 bits (227), Expect = 5e-18
Identities = 73/256 (28%), Positives = 122/256 (47%), Gaps = 22/256 (8%)
Query: 407 PFSMWGIDMIGRIEPKASNGHRFILVAIDYFTKWVEAASYANVTKQVVVKFIKNHIICRY 466
P + W ID I+P G++++LV ID F+ W+EA T +VV K + I R+
Sbjct: 909 PGTHWEIDFT-EIKP-GLYGYKYLLVFIDTFSGWIEAFPTKKETAKVVTKKLLEEIFPRF 966
Query: 467 GVPSRIITDNGTNLNNKMMKELCDDFKIEHHNSSPYRPQMNGAVEAANKNIKKIIQKMVV 526
G+P + TDNG +K+ + + D I+ YRPQ +G VE N+ IK+ + K+ +
Sbjct: 967 GMPQVLGTDNGPAFVSKVSQTVADLLGIDWKLHCAYRPQSSGQVERMNRTIKETLTKLTL 1026
Query: 527 T--YKDWHEMLPFALHGYRTSIRTSTGATPFSLVYGMEAVLPIEVEIPSIRVLMEAELSE 584
+DW +LP AL+ R + G TP+ ++YG P V P + ++
Sbjct: 1027 ATGSRDWVLLLPLALYRARNT-PGPHGLTPYEILYGAP---PPLVNFPDPDM---TRVTN 1079
Query: 585 AEWVQSRYDQLNLIEEKRMSALCHGQLYQKRMKHAFDKKVHPREFKEGDLVLKKILTFQP 644
+ +Q+ L L++ + L YQ+++ D+ V P ++ GD V +
Sbjct: 1080 SPSLQAHLQALYLVQHEVWRPL--AAAYQEQL----DRPVVPHPYRVGDTV-----WVRR 1128
Query: 645 DSRGKWTPNYEGPYVV 660
P ++GPY V
Sbjct: 1129 HQTKNLEPRWKGPYTV 1144
>POL_MLVAV (P03356) Pol polyprotein [Contains: Protease (EC 3.4.23.-);
Reverse transcriptase/ribonuclease H (EC 2.7.7.49) (EC
3.1.26.4) (RT); Integrase (IN)]
Length = 1196
Score = 92.0 bits (227), Expect = 5e-18
Identities = 71/256 (27%), Positives = 123/256 (47%), Gaps = 22/256 (8%)
Query: 407 PFSMWGIDMIGRIEPKASNGHRFILVAIDYFTKWVEAASYANVTKQVVVKFIKNHIICRY 466
P S W ID ++P G++++LV +D F+ WVEA T +VV K + I R+
Sbjct: 909 PGSHWEIDFT-EVKP-GLYGYKYLLVFVDTFSGWVEAFPTKRETARVVSKKLLEEIFPRF 966
Query: 467 GVPSRIITDNGTNLNNKMMKELCDDFKIEHHNSSPYRPQMNGAVEAANKNIKKIIQKMVV 526
G+P + +DNG +++ + + D I+ YRPQ +G VE N+ IK+ + K+ +
Sbjct: 967 GMPQVLGSDNGPAFTSQVSQSVADLLGIDWKLHCAYRPQSSGQVERMNRTIKETLTKLTL 1026
Query: 527 T--YKDWHEMLPFALHGYRTSIRTSTGATPFSLVYGMEAVLPIEVEIPSIRVLMEAELSE 584
+DW +LP AL+ R + G TP+ ++YG L + P + +EL+
Sbjct: 1027 AAGTRDWVLLLPLALYRARNT-PGPHGLTPYEILYGAPPPL-VNFHDPDM-----SELTN 1079
Query: 585 AEWVQSRYDQLNLIEEKRMSALCHGQLYQKRMKHAFDKKVHPREFKEGDLVLKKILTFQP 644
+ +Q+ L ++ + L + Y+ ++ D+ V P F+ GD V +
Sbjct: 1080 SPSLQAHLQALQTVQREIWKPL--AEAYRDQL----DQPVIPHPFRIGDSV-----WVRR 1128
Query: 645 DSRGKWTPNYEGPYVV 660
P ++GPY V
Sbjct: 1129 HQTKNLEPRWKGPYTV 1144
>POL_MLVCB (P08361) Pol polyprotein [Contains: Reverse
transcriptase/ribonuclease H (EC 2.7.7.49) (EC 3.1.26.4)
(RT); Integrase (IN)] (Fragment)
Length = 282
Score = 89.7 bits (221), Expect = 2e-17
Identities = 66/237 (27%), Positives = 114/237 (47%), Gaps = 20/237 (8%)
Query: 426 GHRFILVAIDYFTKWVEAASYANVTKQVVVKFIKNHIICRYGVPSRIITDNGTNLNNKMM 485
G++++LV +D F+ W+EA T +VV K + I R+G+P + TDNG +K+
Sbjct: 12 GYKYLLVFVDTFSGWIEAFPTKKETAKVVTKKLLEEIFPRFGMPQVLGTDNGPAFVSKVS 71
Query: 486 KELCDDFKIEHHNSSPYRPQMNGAVEAANKNIKKIIQKMVVT--YKDWHEMLPFALHGYR 543
+ + D I+ YRPQ +G VE N+ IK+ + K+ + +DW +LP AL+ R
Sbjct: 72 QTVADLLGIDWKLHCAYRPQSSGQVERMNRTIKETLTKLTLATGSRDWVLLLPLALYRAR 131
Query: 544 TSIRTSTGATPFSLVYGMEAVLPIEVEIPSIRVLMEAELSEAEWVQSRYDQLNLIEEKRM 603
+ G TP+ ++YG P V P + ++ + +Q+ L L++ +
Sbjct: 132 NT-PGPHGLTPYEILYGAP---PPLVNFPDPDM---TRVTNSPSLQAHLQALYLVQHEVW 184
Query: 604 SALCHGQLYQKRMKHAFDKKVHPREFKEGDLVLKKILTFQPDSRGKWTPNYEGPYVV 660
L YQ+++ D+ V P ++ GD V + P ++GPY V
Sbjct: 185 RPL--AAAYQEQL----DRPVVPHPYRVGDTV-----WVRRHQTKNLEPRWKGPYTV 230
>POL_MLVAK (P03357) Pol polyprotein [Contains: Reverse
transcriptase/ribonuclease H (EC 2.7.7.49) (EC 3.1.26.4)
(RT); Integrase (IN)] (Fragment)
Length = 843
Score = 89.7 bits (221), Expect = 2e-17
Identities = 71/256 (27%), Positives = 125/256 (48%), Gaps = 23/256 (8%)
Query: 407 PFSMWGIDMIGRIEPKASNGHRFILVAIDYFTKWVEAASYANVTKQVVVKFIKNHIICRY 466
P S W ID ++P G++++LV +D F+ WVEA T +VV K + I R+
Sbjct: 557 PGSHWEIDFT-EVKP-GLYGYKYLLVFVDTFSGWVEAFPTKRETARVVSKKLLEEIFPRF 614
Query: 467 GVPSRIITDNGTNLNNKMMKELCDDFKIEHHNSSPYRPQMNGAVEAANKNIKKIIQKMVV 526
G+P + +DNG +++ + + D I+ + + YRPQ +G VE N+ IK+ + K+ +
Sbjct: 615 GMPQVLGSDNGPAFTSQVSQSVADLLGIDKLHCA-YRPQSSGQVERMNRTIKETLTKLTL 673
Query: 527 T--YKDWHEMLPFALHGYRTSIRTSTGATPFSLVYGMEAVLPIEVEIPSIRVLMEAELSE 584
+DW +LP AL+ R + G TP+ ++YG L + P + +EL+
Sbjct: 674 AAGTRDWVLLLPLALYRARNT-PGPHGLTPYEILYGAPPPL-VNFHDPDM-----SELTN 726
Query: 585 AEWVQSRYDQLNLIEEKRMSALCHGQLYQKRMKHAFDKKVHPREFKEGDLVLKKILTFQP 644
+ +Q+ L ++ + L + Y+ ++ D+ V P F+ GD V +
Sbjct: 727 SPSLQAHLQALQTVQREIWKPL--AEAYRDQL----DQPVIPHPFRIGDSV-----WVRR 775
Query: 645 DSRGKWTPNYEGPYVV 660
P ++GPY V
Sbjct: 776 HQTKNLEPRWKGPYTV 791
>POL_MLVRK (P31795) Pol polyprotein [Contains: Protease (EC
3.4.23.-); Reverse transcriptase/ribonuclease H (EC
2.7.7.49) (EC 3.1.26.4) (RT); Integrase (IN)] (Fragment)
Length = 581
Score = 87.4 bits (215), Expect = 1e-16
Identities = 68/256 (26%), Positives = 121/256 (46%), Gaps = 22/256 (8%)
Query: 407 PFSMWGIDMIGRIEPKASNGHRFILVAIDYFTKWVEAASYANVTKQVVVKFIKNHIICRY 466
P + W ID ++P G++++LV +D F+ WVEA + T ++V K + I R+
Sbjct: 294 PGTHWEIDFT-EVKP-GLYGYKYLLVFVDTFSGWVEAFPTKHETAKIVTKKLLEEIFPRF 351
Query: 467 GVPSRIITDNGTNLNNKMMKELCDDFKIEHHNSSPYRPQMNGAVEAANKNIKKIIQKMVV 526
G+P + TDNG +++ + + I+ YRPQ +G VE N+ IK+ + K+ +
Sbjct: 352 GMPQVLGTDNGPAFVSQVSQSVAKLLGIDWKLHCAYRPQSSGQVERMNRTIKETLTKLTL 411
Query: 527 T--YKDWHEMLPFALHGYRTSIRTSTGATPFSLVYGMEAVLPIEVEIPSIRVLMEAELSE 584
+DW +LP AL+ R + G TP+ ++YG L + P + ++ +
Sbjct: 412 ATGTRDWVLLLPLALYRARNT-PGPHGLTPYEILYGAPPPL-VNFHDPEM-----SKFTN 464
Query: 585 AEWVQSRYDQLNLIEEKRMSALCHGQLYQKRMKHAFDKKVHPREFKEGDLVLKKILTFQP 644
+ +Q+ L ++ + L YQ ++ D+ V P F+ GD V +
Sbjct: 465 SPSLQAHLQALQAVQREVWKPL--AAAYQDQL----DQPVIPHPFRVGDTV-----WVRR 513
Query: 645 DSRGKWTPNYEGPYVV 660
P ++GPY V
Sbjct: 514 HQTKNLEPRWKGPYTV 529
>POL_MLVRD (P11227) Pol polyprotein [Contains: Protease (EC 3.4.23.-);
Reverse transcriptase/ribonuclease H (EC 2.7.7.49) (EC
3.1.26.4) (RT); Integrase (IN)]
Length = 1196
Score = 87.4 bits (215), Expect = 1e-16
Identities = 68/256 (26%), Positives = 121/256 (46%), Gaps = 22/256 (8%)
Query: 407 PFSMWGIDMIGRIEPKASNGHRFILVAIDYFTKWVEAASYANVTKQVVVKFIKNHIICRY 466
P + W ID ++P G++++LV +D F+ WVEA + T ++V K + I R+
Sbjct: 909 PGTHWEIDFT-EVKP-GLYGYKYLLVFVDTFSGWVEAFPTKHETAKIVTKKLLEEIFPRF 966
Query: 467 GVPSRIITDNGTNLNNKMMKELCDDFKIEHHNSSPYRPQMNGAVEAANKNIKKIIQKMVV 526
G+P + TDNG +++ + + I+ YRPQ +G VE N+ IK+ + K+ +
Sbjct: 967 GMPQVLGTDNGPAFVSQVSQSVAKLLGIDWKLHCAYRPQSSGQVERMNRTIKETLTKLTL 1026
Query: 527 T--YKDWHEMLPFALHGYRTSIRTSTGATPFSLVYGMEAVLPIEVEIPSIRVLMEAELSE 584
+DW +LP AL+ R + G TP+ ++YG L + P + ++ +
Sbjct: 1027 ATGTRDWVLLLPLALYRARNT-PGPHGLTPYEILYGAPPPL-VNFHDPEM-----SKFTN 1079
Query: 585 AEWVQSRYDQLNLIEEKRMSALCHGQLYQKRMKHAFDKKVHPREFKEGDLVLKKILTFQP 644
+ +Q+ L ++ + L YQ ++ D+ V P F+ GD V +
Sbjct: 1080 SPSLQAHLQALQAVQREVWKPL--AAAYQDQL----DQPVIPHPFRVGDTV-----WVRR 1128
Query: 645 DSRGKWTPNYEGPYVV 660
P ++GPY V
Sbjct: 1129 HQTKNLEPRWKGPYTV 1144
>POL3_MOUSE (P11367) Retrovirus-related Pol polyprotein
(Endonuclease) (Fragment)
Length = 390
Score = 87.4 bits (215), Expect = 1e-16
Identities = 75/298 (25%), Positives = 137/298 (45%), Gaps = 25/298 (8%)
Query: 366 YYWMTMEADCYKHAKRCYKC-QIYADRIHVPPTALNVLSSPWPFSMWGIDMIGRIEPKAS 424
YY + + ++ A+ C C Q+ A + + A + + W ID ++P
Sbjct: 78 YYMLNKDKILHEVAESCQACVQVNASKTKI--RAGTRVRGHRLGTHWEIDFT-EVKP-GL 133
Query: 425 NGHRFILVAIDYFTKWVEAASYANVTKQVVVKFIKNHIICRYGVPSRIITDNGTNLNNKM 484
G++++LV +D F+ WVEA + T ++V K + I R+G+P + TDNG +++
Sbjct: 134 YGYKYLLVFVDTFSGWVEAFPTKHETAKIVTKKLLEEIFPRFGMPQVLGTDNGPAFVSQV 193
Query: 485 MKELCDDFKIEHHNSSPYRPQMNGAVEAANKNIKKIIQKMVVT--YKDWHEMLPFALHGY 542
+ + I+ YRPQ +G VE N+ IK+ + K+ + +DW +LP AL+
Sbjct: 194 SQSVAKLLGIDWKLHCAYRPQSSGQVERMNRTIKETLTKLTLATGTRDWVLLLPLALYRA 253
Query: 543 RTSIRTSTGATPFSLVYGMEAVLPIEVEIPSIRVLMEAELSEAEWVQSRYDQLNLIEEKR 602
R + G TP+ ++YG L + P + ++ + + +Q+ L ++ +
Sbjct: 254 RNT-PGPHGLTPYEILYGAPPPL-VNFHDPEM-----SKFTNSPSLQAHLQALQAVQREV 306
Query: 603 MSALCHGQLYQKRMKHAFDKKVHPREFKEGDLVLKKILTFQPDSRGKWTPNYEGPYVV 660
L YQ ++ D+ V P F+ GD V + P ++GPY V
Sbjct: 307 WKPL--AAAYQDQL----DQPVIPHPFRVGDTV-----WVRRHQTKNLEPRWKGPYTV 353
>POL_SFV1 (P23074) Pol polyprotein [Contains: Protease (EC 3.4.23.-);
Reverse transcriptase/ribonuclease H (EC 2.7.7.49) (EC
3.1.26.4) (RT); Integrase (IN)]
Length = 1161
Score = 86.3 bits (212), Expect = 3e-16
Identities = 63/253 (24%), Positives = 116/253 (44%), Gaps = 7/253 (2%)
Query: 366 YYWMTMEADCYKHAKRCYKCQIYADRIHVPPTALNVLSSPWPFSMWGIDMIGRIEPKASN 425
Y+W + D K ++C +C + P L + PF + ID IG + P SN
Sbjct: 842 YWWPNLRKDVVKSIRQCKQCLVTNATNLTSPPILRPVKPLKPFDKFYIDYIGPLPP--SN 899
Query: 426 GHRFILVAIDYFTKWVEAASYANVTKQVVVKFIKNHIICRYGVPSRIITDNGTNLNNKMM 485
G+ +LV +D T +V + VK + +++ +P + +D G +
Sbjct: 900 GYLHVLVVVDSMTGFVWLYPTKAPSTSATVKAL--NMLTSIAIPKVLHSDQGAAFTSSTF 957
Query: 486 KELCDDFKIEHHNSSPYRPQMNGAVEAANKNIKKIIQKMVV-TYKDWHEMLPFALHGYRT 544
+ + I+ S+PY PQ +G VE N +IK+++ K+++ W+++LP
Sbjct: 958 ADWAKEKGIQLEFSTPYHPQSSGKVERKNSDIKRLLTKLLIGRPAKWYDLLPVVQLALNN 1017
Query: 545 SIRTSTGATPFSLVYGMEAVLPIEVEIPSIRVLMEAELSEAEWVQSRYDQ-LNLIEEKRM 603
S S+ TP L++G+++ P ++ + E ELS + ++S Q + R
Sbjct: 1018 SYSPSSKYTPHQLLFGVDSNTPF-ANSDTLDLSREEELSLLQEIRSSLHQPTSPPASSRS 1076
Query: 604 SALCHGQLYQKRM 616
+ GQL Q+R+
Sbjct: 1077 WSPSVGQLVQERV 1089
>POL_AVIRE (P03360) Pol polyprotein [Contains: Reverse
transcriptase/ribonuclease H (EC 2.7.7.49) (EC 3.1.26.4)
(RT); Integrase (IN)] (Fragment)
Length = 473
Score = 85.5 bits (210), Expect = 4e-16
Identities = 56/183 (30%), Positives = 87/183 (46%), Gaps = 3/183 (1%)
Query: 381 RCYKCQIYADRIHVPPTALNV-LSSPWPFSMWGIDMIGRIEPKASNGHRFILVAIDYFTK 439
RC C R LN + P W +D I K G++++LV +D F+
Sbjct: 161 RCVACAQVNPRAAPVEKGLNSRIRGAAPGEHWEVDFTEMITAKG--GYKYLLVLVDTFSG 218
Query: 440 WVEAASYANVTKQVVVKFIKNHIICRYGVPSRIITDNGTNLNNKMMKELCDDFKIEHHNS 499
WVEA T QVV+K + II R+G+P +I +DNG K+ ++LC+ +
Sbjct: 219 WVEAYPAKRETSQVVIKHLILDIIPRFGLPVQIGSDNGPAFVAKVTQQLCEALNVSWKLH 278
Query: 500 SPYRPQMNGAVEAANKNIKKIIQKMVVTYKDWHEMLPFALHGYRTSIRTSTGATPFSLVY 559
YRPQ +G VE N+ +KK I K+ + + P + T G +PF ++Y
Sbjct: 279 CAYRPQSSGQVERMNRTLKKAIAKLEDRDRRGLGLPPPSGFAPGTVYPGREGLSPFEILY 338
Query: 560 GME 562
G++
Sbjct: 339 GLK 341
>YRD6_CAEEL (Q09575) Hypothetical protein K02A2.6 in chromosome II
Length = 1268
Score = 84.7 bits (208), Expect = 7e-16
Identities = 67/260 (25%), Positives = 119/260 (45%), Gaps = 24/260 (9%)
Query: 331 VDKHEADLLMHEIHEGSFGTHSSGPTMAKKMLRAGYYWMTMEADCYKHAKRCYKCQIYAD 390
V K +++ ++HEG G K+ R+ +W +++D + C CQ +
Sbjct: 778 VPKSLQKIVLKQLHEGH-----PGIVQMKQKARSFVFWRGLDSDIENMVRHCNNCQENSK 832
Query: 391 RIHVPPTALNVLSSPWPF--SMWG---IDMIGRIEPKASNGHRFILVAIDYFTKWVEAAS 445
V P +PWP + W ID G + NG ++LV +D TK+ E
Sbjct: 833 MPRVVPL------NPWPVPEAPWKRIHIDFAGPL-----NGC-YLLVVVDAKTKYAEVKL 880
Query: 446 YANVTKQVVVKFIKNHIICRYGVPSRIITDNGTNLNNKMMKELCDDFKIEHHNSSPYRPQ 505
+++ + ++ I +G P II+DNGT L + + ++C IEH S+ Y P+
Sbjct: 881 TRSISAVTTIDLLEE-IFSIHGYPETIISDNGTQLTSHLFAQMCQSHGIEHKTSAVYYPR 939
Query: 506 MNGAVEAANKNIKKIIQKMVVTYKDWHEMLPFALHGYRTSIRTS-TGATPFSLVYGMEAV 564
NGA E +K+ I K+ ++L L YR + ++ G+TP +G +
Sbjct: 940 SNGAAERFVDTLKRGIAKIKGEGSVNQQILNKFLISYRNTPHSALNGSTPAECHFGRKIR 999
Query: 565 LPIEVEIPSIRVLMEAELSE 584
+ + +P+ RVL +L++
Sbjct: 1000 TTMSLLMPTDRVLKVPKLTQ 1019
>POL_SFV3L (P27401) Pol polyprotein [Contains: Protease (EC 3.4.23.-);
Reverse transcriptase/ribonuclease H (EC 2.7.7.49) (EC
3.1.26.4) (RT); Integrase (IN)]
Length = 1157
Score = 84.0 bits (206), Expect = 1e-15
Identities = 59/226 (26%), Positives = 104/226 (45%), Gaps = 6/226 (2%)
Query: 366 YYWMTMEADCYKHAKRCYKCQIYADRIHVPPTALNVLSSPWPFSMWGIDMIGRIEPKASN 425
Y+W + D K ++C +C + P L PF + ID IG + P SN
Sbjct: 844 YWWPNLRKDVVKVIRQCKQCLVTNAATLAAPPILRPERPVKPFDKFFIDYIGPLPP--SN 901
Query: 426 GHRFILVAIDYFTKWVEAASYANVTKQVVVKFIKNHIICRYGVPSRIITDNGTNLNNKMM 485
G+ +LV +D T +V + VK + +++ VP I +D G +
Sbjct: 902 GYLHVLVVVDSMTGFVWLYPTKAPSTSATVKAL--NMLTSIAVPKVIHSDQGAAFTSATF 959
Query: 486 KELCDDFKIEHHNSSPYRPQMNGAVEAANKNIKKIIQKMVV-TYKDWHEMLPFALHGYRT 544
+ + I+ S+PY PQ +G VE N +IK+++ K++V W+++LP
Sbjct: 960 ADWAKNKGIQLEFSTPYHPQSSGKVERKNSDIKRLLTKLLVGRPAKWYDLLPVVQLALNN 1019
Query: 545 SIRTSTGATPFSLVYGMEAVLPIEVEIPSIRVLMEAELSEAEWVQS 590
S S+ TP L++G+++ P ++ + E ELS + ++S
Sbjct: 1020 SYSPSSKYTPHQLLFGIDSNTPF-ANSDTLDLSREEELSLLQEIRS 1064
>POL_BAEVM (P10272) Pol polyprotein [Contains: Protease (EC 3.4.23.-);
Reverse transcriptase/ribonuclease H (EC 2.7.7.49) (EC
3.1.26.4) (RT); Integrase (IN)]
Length = 1189
Score = 80.1 bits (196), Expect = 2e-14
Identities = 72/256 (28%), Positives = 120/256 (46%), Gaps = 25/256 (9%)
Query: 407 PFSMWGIDMIGRIEPKASNGHRFILVAIDYFTKWVEAASYANVTKQVVVKFIKNHIICRY 466
P W ID ++P + G++++LV +D F+ WVEA T +V K I I R+
Sbjct: 904 PGVYWEIDFT-EVKPHYA-GYKYLLVFVDTFSGWVEAFPTRQETAHIVAKKILEEIFPRF 961
Query: 467 GVPSRIITDNGTNLNNKMMKELCDDFKIEHHNSSPYRPQMNGAVEAANKNIKKIIQKMVV 526
G+P I +DNG +++ + L I YRPQ +G VE N+ IK+ + K+ +
Sbjct: 962 GLPKVIGSDNGPAFVSQVSQGLARILGINWKLHCAYRPQSSGQVERMNRTIKETLTKLTL 1021
Query: 527 T--YKDWHEMLPFALHGYRTSIRTSTGATPFSLVYGMEAVLPIEVEIPSIRVLMEAELSE 584
KDW +L AL R + G TP+ ++YG P+ + S + +
Sbjct: 1022 ETGLKDWRRLLSLALLRARNT-PNRFGLTPYEILYG--GPPPLSTLLNSF-----SPSNS 1073
Query: 585 AEWVQSRYDQLNLIEEKRMSALCHGQLYQKRMKHAFDKKVHPREFKEGDLVLKKILTFQP 644
+Q+R L ++ + + L +LY + + HP F+ GD V + +
Sbjct: 1074 KTDLQARLKGLQAVQAQIWAPL--AELY----RPGHSQTSHP--FQVGDSVYVR----RH 1121
Query: 645 DSRGKWTPNYEGPYVV 660
S+G P ++GPY+V
Sbjct: 1122 RSQG-LEPRWKGPYIV 1136
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.322 0.138 0.428
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 86,980,042
Number of Sequences: 164201
Number of extensions: 3819215
Number of successful extensions: 7898
Number of sequences better than 10.0: 120
Number of HSP's better than 10.0 without gapping: 109
Number of HSP's successfully gapped in prelim test: 11
Number of HSP's that attempted gapping in prelim test: 7754
Number of HSP's gapped (non-prelim): 133
length of query: 693
length of database: 59,974,054
effective HSP length: 117
effective length of query: 576
effective length of database: 40,762,537
effective search space: 23479221312
effective search space used: 23479221312
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 69 (31.2 bits)
Medicago: description of AC148653.1


