

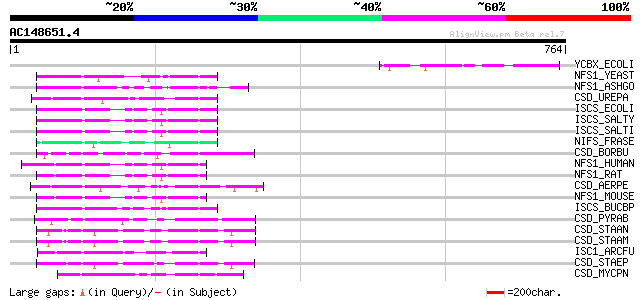
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148651.4 + phase: 0 /pseudo
(764 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
YCBX_ECOLI (P75863) Hypothetical protein ycbX 82 7e-15
NFS1_YEAST (P25374) Cysteine desulfurase, mitochondrial precurso... 65 5e-10
NFS1_ASHGO (O60028) Cysteine desulfurase, mitochondrial precurso... 62 4e-09
CSD_UREPA (Q9PQ36) Probable cysteine desulfurase (EC 2.8.1.7) 62 6e-09
ISCS_ECOLI (P39171) Cysteine desulfurase (EC 2.8.1.7) (ThiI tran... 60 2e-08
ISCS_SALTY (Q8ZN40) Cysteine desulfurase (EC 2.8.1.7) (ThiI tran... 59 6e-08
ISCS_SALTI (Q8Z4N0) Cysteine desulfurase (EC 2.8.1.7) (ThiI tran... 59 6e-08
NIFS_FRASE (Q9Z5X5) Cysteine desulfurase (EC 2.8.1.7) (Nitrogena... 57 1e-07
CSD_BORBU (O51111) Probable cysteine desulfurase (EC 2.8.1.7) 56 3e-07
NFS1_HUMAN (Q9Y697) Cysteine desulfurase, mitochondrial precurso... 56 4e-07
NFS1_RAT (Q99P39) Cysteine desulfurase, mitochondrial precursor ... 55 9e-07
CSD_AERPE (Q9YAB6) Probable cysteine desulfurase (EC 2.8.1.7) 55 9e-07
NFS1_MOUSE (Q9Z1J3) Cysteine desulfurase, mitochondrial precurso... 54 2e-06
ISCS_BUCBP (Q89A19) Cysteine desulfurase (EC 2.8.1.7) 53 3e-06
CSD_PYRAB (Q9V242) Probable cysteine desulfurase (EC 2.8.1.7) 52 6e-06
CSD_STAAN (P99177) Probable cysteine desulfurase (EC 2.8.1.7) 50 2e-05
CSD_STAAM (P63518) Probable cysteine desulfurase (EC 2.8.1.7) 50 2e-05
ISC1_ARCFU (O30052) Probable cysteine desulfurase 1 (EC 2.8.1.7) 49 6e-05
CSD_STAEP (Q8CTA4) Probable cysteine desulfurase (EC 2.8.1.7) 47 2e-04
CSD_MYCPN (P75298) Probable cysteine desulfurase (EC 2.8.1.7) 46 3e-04
>YCBX_ECOLI (P75863) Hypothetical protein ycbX
Length = 369
Score = 81.6 bits (200), Expect = 7e-15
Identities = 74/257 (28%), Positives = 114/257 (43%), Gaps = 43/257 (16%)
Query: 510 NPVEASVQAVGL---LATMGSLKHDREWILKSLSGEILTLKRVPEMGLISSFIDLSQGML 566
+PV+ S++ +GL LA + L DR +++ G +T ++ P+M +
Sbjct: 10 HPVK-SMRGIGLTHALADVSGLAFDRIFMITEPDGTFITARQFPQM------------VR 56
Query: 567 FVESP-----HCKERLQIRLQLDFYDSAIQDI--ELQGQRYKVYSYDNETNSWFSKAIER 619
F SP H + F D A QD E+ G + + N W S R
Sbjct: 57 FTPSPVHDGLHLTAPDGSSAYVRFADFATQDAPTEVWGTHFTARIAPDAINKWLSGFFSR 116
Query: 620 PCTLLRYSGSSHDFVLDRTKDIVTCKDTNSAVSFANEGQFLLVSEESVSDLNKRLCSDVQ 679
L R+ G + R + +SFA+ +LL +E S+ DL +R + V+
Sbjct: 117 EVQL-RWVGPQMTRRVKRHNTV--------PLSFADGYPYLLANEASLRDLQQRCPASVK 167
Query: 680 MDMCETEIEINTNRFRPNLVVSGGRPYDEDGWSDIRIGNKYFRSLGGCNRCQVINLTLNA 739
M+ +FRPNLVVSG ++ED W IRIG+ F + C+RC ++
Sbjct: 168 ME-----------QFRPNLVVSGASAWEEDRWKVIRIGDVVFDVVKPCSRCIFTTVSPEK 216
Query: 740 GQVQKSKEPLATLASYR 756
GQ + EPL TL S+R
Sbjct: 217 GQKHPAGEPLKTLQSFR 233
>NFS1_YEAST (P25374) Cysteine desulfurase, mitochondrial precursor
(EC 2.8.1.7) (tRNA splicing protein SPL1)
Length = 497
Score = 65.5 bits (158), Expect = 5e-10
Identities = 65/257 (25%), Positives = 113/257 (43%), Gaps = 37/257 (14%)
Query: 38 VYLDHAGATLYSELQMESVFKDLTTNVYGNPHSQSDSSA-ATHDIVRDARQQVLDYCNAS 96
+YLD T ++++ K T +YGNPHS + S T+ V +AR V NA
Sbjct: 100 IYLDMQATTPTDPRVLDTMLK-FYTGLYGNPHSNTHSYGWETNTAVENARAHVAKMINAD 158
Query: 97 PEDYKCIFTSGATAALKLVGEAFP---WSCNSNFMYTMENHNSVLGIREYALGQGAAAIA 153
P++ IFTSGAT + +V + P + + T H VL + +G
Sbjct: 159 PKE--IIFTSGATESNNMVLKGVPRFYKKTKKHIITTRTEHKCVLEAARAMMKEGFEVTF 216
Query: 154 VDIEDVHPRIEGEKFPTKISLHQEQRRKVTGLQEEE----PMECNFSGLRFDLDLAKIIK 209
++++D + + L+E E P C S + + ++ I
Sbjct: 217 LNVDD---------------------QGLIDLKELEDAIRPDTCLVSVMAVNNEIGVI-- 253
Query: 210 EDSSKILGASVCKKGRWLVLIDAAKGSATMPPDLSKYPVDFVALSFYKLFGYPTGLGALV 269
K +GA +C+K + DAA+ + D+++ +D +++S +K++G P G+GA+
Sbjct: 254 -QPIKEIGA-ICRKNKIYFHTDAAQAYGKIHIDVNEMNIDLLSISSHKIYG-PKGIGAIY 310
Query: 270 VRNDAAKLLKKSYFSGG 286
VR L+ GG
Sbjct: 311 VRRRPRVRLEPLLSGGG 327
>NFS1_ASHGO (O60028) Cysteine desulfurase, mitochondrial precursor
(EC 2.8.1.7) (tRNA splicing protein SPL1)
Length = 490
Score = 62.4 bits (150), Expect = 4e-09
Identities = 76/297 (25%), Positives = 131/297 (43%), Gaps = 50/297 (16%)
Query: 38 VYLDHAGATLYSELQMESVFKDLTTNVYGNPHSQSDSSA-ATHDIVRDARQQVLDYCNAS 96
+YLD T ++++ K T +YGNPHS + S T V AR+ V D A
Sbjct: 93 IYLDMQATTPTDPRVVDTMLK-FYTGLYGNPHSNTHSYGWETSQEVEKARKNVADVIKAD 151
Query: 97 PEDYKCIFTSGATAA--LKLVGEA-FPWSCNSNFMYTMENHNSVLGIREYALGQGAAAIA 153
P++ IFTSGAT + + L G A F ++ + T H VL +G
Sbjct: 152 PKE--IIFTSGATESNNMALKGVARFYKKRKNHIITTRTEHKCVLEAARSMKDEG----- 204
Query: 154 VDIEDVHPRIEGEKFPTKISLHQEQR--RKVTGLQEEEPMECNFSGLRFDLDLAKIIKED 211
D+ ++ +G +SL + ++ R T L + + ++ + + IKE
Sbjct: 205 FDVTFLNVNEDG-----LVSLEELEQAIRPETSL-------VSVMSVNNEIGVVQPIKE- 251
Query: 212 SSKILGASVCKKGRWLVLIDAAKGSATMPPDLSKYPVDFVALSFYKLFGYPTGLGALVVR 271
+GA +C++ + DAA+ +P D+ + +D +++S +K++G P G+GAL VR
Sbjct: 252 ----IGA-ICRRHKVFFHSDAAQAYGKIPIDVDEMNIDLLSISSHKIYG-PKGIGALYVR 305
Query: 272 NDAAKLLKKSYFSGGTVAASIADIDFIKRREGIEELFEDGTVSFLSIASIRHGFKIL 328
++ + SGG G E F GT+ + + H K++
Sbjct: 306 R-RPRVRMEPLLSGG----------------GQERGFRSGTLPPPLVVGLGHAAKLM 345
>CSD_UREPA (Q9PQ36) Probable cysteine desulfurase (EC 2.8.1.7)
Length = 402
Score = 62.0 bits (149), Expect = 6e-09
Identities = 64/260 (24%), Positives = 113/260 (42%), Gaps = 28/260 (10%)
Query: 31 FNRLQDLVYLDHAGATLYSELQMESVFKDLTTNVYGNPH-SQSDSSAATHDIVRDARQQV 89
F +D++YLD + +L ++ ++++ D T NPH + S+ + TH+I+ + R V
Sbjct: 11 FKNNKDVIYLDSSATSLKPQVVIDAIV-DYYTKYSTNPHNTDSNFTFHTHEIMNETRANV 69
Query: 90 LDYCNASPEDYKCIFTSGATAALKLVGEAFPWSCNSN---FMYTMENHNSVLGIREYALG 146
+ NA+ ++ IFTSGAT +L L+ + +E+ +++L +
Sbjct: 70 AKFINANFDE--IIFTSGATESLNLIANGLRPHLKKGDEIILTYIEHASNLLPWYKLRDD 127
Query: 147 QGAAAIAVDIEDVHPRIEGEKFPTKISLHQEQRRKVTGLQEEEPMECNFSGLRFDLDLAK 206
G I + ++ P++ F IS + KV + N + K
Sbjct: 128 LGVKIIFANQKNQFPQL--SDFLKVIS----PKTKVVSFASGGNLIGNVLDENLIIKNIK 181
Query: 207 IIKEDSSKILGASVCKKGRWLVLIDAAKGSATMPPDLSKYPVDFVALSFYKLFGYPTGLG 266
IK D LV +DA + D+ K DF+ S +KL G PTG+G
Sbjct: 182 KIKPDV--------------LVCVDATQSIQHRMFDVKKCKSDFMVFSAHKLLG-PTGIG 226
Query: 267 ALVVRNDAAKLLKKSYFSGG 286
++ND K ++ + GG
Sbjct: 227 VAYIKNDLIKKMQPLKYGGG 246
>ISCS_ECOLI (P39171) Cysteine desulfurase (EC 2.8.1.7) (ThiI
transpersulfidase) (NifS protein homolog)
Length = 404
Score = 60.1 bits (144), Expect = 2e-08
Identities = 67/262 (25%), Positives = 113/262 (42%), Gaps = 45/262 (17%)
Query: 38 VYLDHAGATLYSELQMESVFKDLTTN-VYGNPHSQSDSSA-ATHDIVRDARQQVLDYCNA 95
+YLD++ T E + + +T + +GNP S+S + V AR Q+ D A
Sbjct: 5 IYLDYSATTPVDPRVAEKMMQFMTMDGTFGNPASRSHRFGWQAEEAVDIARNQIADLVGA 64
Query: 96 SPEDYKCIFTSGATAA--LKLVGEA-FPWSCNSNFMYTMENHNSVLGIREYALGQGAAAI 152
P + +FTSGAT + L + G A F + + + H +VL
Sbjct: 65 DPREI--VFTSGATESDNLAIKGAANFYQKKGKHIITSKTEHKAVL-------------- 108
Query: 153 AVDIEDVHPRIEGEKFPTKISLHQEQRRKVTGLQEEEPMECNFSGLRFDLDLAKI----- 207
D ++E E F +++ QR + L+E E + +R D L I
Sbjct: 109 -----DTCRQLEREGF--EVTYLAPQRNGIIDLKELE------AAMRDDTILVSIMHVNN 155
Query: 208 ---IKEDSSKILGASVCKKGRWLVLIDAAKGSATMPPDLSKYPVDFVALSFYKLFGYPTG 264
+ +D + I +C+ + +DA + +P DLS+ VD ++ S +K++G P G
Sbjct: 156 EIGVVQDIAAI--GEMCRARGIIYHVDATQSVGKLPIDLSQLKVDLMSFSGHKIYG-PKG 212
Query: 265 LGALVVRNDAAKLLKKSYFSGG 286
+GAL VR ++ GG
Sbjct: 213 IGALYVRRKPRVRIEAQMHGGG 234
>ISCS_SALTY (Q8ZN40) Cysteine desulfurase (EC 2.8.1.7) (ThiI
transpersulfidase) (NifS protein homolog)
Length = 404
Score = 58.5 bits (140), Expect = 6e-08
Identities = 67/262 (25%), Positives = 112/262 (42%), Gaps = 45/262 (17%)
Query: 38 VYLDHAGATLYSELQMESVFKDLTTN-VYGNPHSQSDSSA-ATHDIVRDARQQVLDYCNA 95
+YLD++ T E + + LT + +GNP S+S + V AR Q+ + A
Sbjct: 5 IYLDYSATTPVDPRVAEKMMQFLTLDGTFGNPASRSHRFGWQAEEAVDIARNQIAELVGA 64
Query: 96 SPEDYKCIFTSGATAA--LKLVGEA-FPWSCNSNFMYTMENHNSVLGIREYALGQGAAAI 152
P + +FTSGAT + L + G A F + + + H +VL
Sbjct: 65 DPREI--VFTSGATESDNLAIKGAANFYQKKGKHIITSKTEHKAVL-------------- 108
Query: 153 AVDIEDVHPRIEGEKFPTKISLHQEQRRKVTGLQEEEPMECNFSGLRFDLDLAKI----- 207
D ++E E F +++ QR + L E E + +R D L I
Sbjct: 109 -----DTCRQLEREGF--EVTYLAPQRNGIIDLNELE------AAMRDDTILVSIMHVNN 155
Query: 208 ---IKEDSSKILGASVCKKGRWLVLIDAAKGSATMPPDLSKYPVDFVALSFYKLFGYPTG 264
+ +D + I +C+ + +DA + +P DLS+ VD ++ S +K++G P G
Sbjct: 156 EIGVVQDIATI--GEMCRARGIIYHVDATQSVGKLPIDLSQLKVDLMSFSGHKIYG-PKG 212
Query: 265 LGALVVRNDAAKLLKKSYFSGG 286
+GAL VR ++ GG
Sbjct: 213 IGALYVRRKPRIRIEAQMHGGG 234
>ISCS_SALTI (Q8Z4N0) Cysteine desulfurase (EC 2.8.1.7) (ThiI
transpersulfidase) (NifS protein homolog)
Length = 404
Score = 58.5 bits (140), Expect = 6e-08
Identities = 67/262 (25%), Positives = 112/262 (42%), Gaps = 45/262 (17%)
Query: 38 VYLDHAGATLYSELQMESVFKDLTTN-VYGNPHSQSDSSA-ATHDIVRDARQQVLDYCNA 95
+YLD++ T E + + LT + +GNP S+S + V AR Q+ + A
Sbjct: 5 IYLDYSATTPVDPRVAEKMMQFLTLDGTFGNPASRSHRFGWQAEEAVDIARNQIAELVGA 64
Query: 96 SPEDYKCIFTSGATAA--LKLVGEA-FPWSCNSNFMYTMENHNSVLGIREYALGQGAAAI 152
P + +FTSGAT + L + G A F + + + H +VL
Sbjct: 65 DPREI--VFTSGATESDNLAIKGAANFYQKKGKHIITSKTEHKAVL-------------- 108
Query: 153 AVDIEDVHPRIEGEKFPTKISLHQEQRRKVTGLQEEEPMECNFSGLRFDLDLAKI----- 207
D ++E E F +++ QR + L E E + +R D L I
Sbjct: 109 -----DTCRQLEREGF--EVTYLAPQRNGIIDLNELE------AAMRDDTILVSIMHVNN 155
Query: 208 ---IKEDSSKILGASVCKKGRWLVLIDAAKGSATMPPDLSKYPVDFVALSFYKLFGYPTG 264
+ +D + I +C+ + +DA + +P DLS+ VD ++ S +K++G P G
Sbjct: 156 EIGVVQDIATI--GEMCRARGIIYHVDATQSVGKLPIDLSQLKVDLMSFSGHKIYG-PKG 212
Query: 265 LGALVVRNDAAKLLKKSYFSGG 286
+GAL VR ++ GG
Sbjct: 213 IGALYVRRKPRIRIEAQMHGGG 234
>NIFS_FRASE (Q9Z5X5) Cysteine desulfurase (EC 2.8.1.7) (Nitrogenase
metalloclusters biosynthesis protein nifS)
Length = 419
Score = 57.4 bits (137), Expect = 1e-07
Identities = 65/263 (24%), Positives = 101/263 (37%), Gaps = 40/263 (15%)
Query: 37 LVYLDHAGATLYSELQMESVFKDLTTNVYGNPHSQSDSSAATHDIVRDARQQVLDYCNAS 96
++YLDH AT + ++ V L T + NP S+ IV AR++V A
Sbjct: 1 MIYLDH-NATTPVDPRVVEVMLPLFTERFANPASKHAPGQEATRIVERARREVASLAGAR 59
Query: 97 PEDYKCIFTSGATAALKL-----VGEAFPWSCNSNFMYTMENHNSVLGIREYALGQGAAA 151
P + +FTSGAT A L + A P + H +VLG + A+ G+A
Sbjct: 60 PREI--VFTSGATEAANLAIGGVLATAEPG--RRRILVGATEHPAVLGAADAAVSAGSAV 115
Query: 152 IAVDIEDVHPRIEGEKFPTKISLHQEQRRKVTGLQEEEPMECNFSGLRFDLDLAKIIKED 211
A +G +I++H + V L+ R D+A +
Sbjct: 116 SA----------DGTTAAERIAVHPDGTVDVDDLRR-----------RLGPDVALVAVMA 154
Query: 212 SSKILGA--------SVCKKGRWLVLIDAAKGSATMPPDLSKYPVDFVALSFYKLFGYPT 263
++ GA + D + +P + + VD S +KL+G P
Sbjct: 155 ANNETGAVNDPRPLVEAARSAGRCCCADVTQALGRVPVEFERTGVDLAVSSAHKLYG-PK 213
Query: 264 GLGALVVRNDAAKLLKKSYFSGG 286
G+GAL+ DA L GG
Sbjct: 214 GVGALIASKDAWSRLAPRVHGGG 236
>CSD_BORBU (O51111) Probable cysteine desulfurase (EC 2.8.1.7)
Length = 422
Score = 56.2 bits (134), Expect = 3e-07
Identities = 70/311 (22%), Positives = 131/311 (41%), Gaps = 39/311 (12%)
Query: 37 LVYLDHAGAT------LYSELQMESVFKDLTTNVYGNPHSQSDSSAATHDIVRDARQQVL 90
++Y D+A + +YS ++ +++ NV+ + H + S+ + R+ V
Sbjct: 32 IIYFDNAATSQKPKNVIYSNVEY---YENYNANVHRSGHKFAIQSSIK---IEKTRELVK 85
Query: 91 DYCNASPEDYKCIFTSGATAALKLVGEAFPWSCNSNFMYTMENHNSVLGIREYALGQGAA 150
++ NA IFTSG T + + +F +S Y + +L E+
Sbjct: 86 NFINAESAK-NIIFTSGTTDGINTIASSFFYS-----KYFKKKDEIILTTLEHNSNLLPW 139
Query: 151 AIAVDIEDVHPRIEGEKFPTKISLHQEQRRKVTGLQEEEPMECNFSGLRFDL----DLAK 206
++ ++ +I+ KF + E+ K L E+ + SG+ L DL
Sbjct: 140 VNLANLANL--KIKLAKFNEMGIITPEEIEK---LITEKTKLISISGINNTLGTINDLES 194
Query: 207 IIKEDSSKILGASVCKKGRWLVLIDAAKGSATMPPDLSKYPVDFVALSFYKLFGYPTGLG 266
I K + KK + +DAA+ + + D+ K DF+ S +K+ PTG+G
Sbjct: 195 IGK----------IAKKYNICLFVDAAQMAPHIKIDVKKIGCDFLVFSGHKMLA-PTGIG 243
Query: 267 ALVVRNDAAKLLKKSYFSGGTVAASIADIDFIK-RREGIEELFEDGTVSFLSIASIRHGF 325
L + N+ A+ L S G TV + + IK + FE GT + I +
Sbjct: 244 ILYISNNMAEKLHSSKLGGNTVEEIFIENEKIKFKASDSPNKFESGTPNIAGIIGLEEAI 303
Query: 326 KILNSLTVSAI 336
K ++++++ I
Sbjct: 304 KYIDNISMDFI 314
>NFS1_HUMAN (Q9Y697) Cysteine desulfurase, mitochondrial precursor
(EC 2.8.1.7) (HUSSY-08)
Length = 457
Score = 55.8 bits (133), Expect = 4e-07
Identities = 65/267 (24%), Positives = 115/267 (42%), Gaps = 45/267 (16%)
Query: 17 PNAARTIDQIRATEFNRLQDLVYLDHAGATLYSELQMESVFKDLTTNVYGNPHSQSDSSA 76
P +A D A E + +Y+D T ++++ L N YGNPHS++ +
Sbjct: 38 PQSAVPADTTAAPEVGPVLRPLYMDVQATTPLDPRVLDAMLPYLI-NYYGNPHSRTHAYG 96
Query: 77 ATHDIVRD-ARQQVLDYCNASPEDYKCIFTSGATAA--LKLVGEA-FPWSCNSNFMYTME 132
+ + ARQQV A P + IFTSGAT + + + G A F S + + T
Sbjct: 97 WESEAAMERARQQVASLIGADPREI--IFTSGATESNNIAIKGVARFYRSRKKHLITTQT 154
Query: 133 NHNSVLGIREYALGQGAAAIAVDIEDVHPRIEGEKFPTKISLHQEQRRKVTGLQEEEPME 192
H VL D +E E F +++ Q+ + L+E E
Sbjct: 155 EHKCVL-------------------DSCRSLEAEGF--QVTYLPVQKSGIIDLKELE--- 190
Query: 193 CNFSGLRFDLDLAKI--------IKEDSSKILGASVCKKGRWLVLIDAAKGSATMPPDLS 244
+ ++ D L + +K+ ++I +C + DAA+ +P D++
Sbjct: 191 ---AAIQPDTSLVSVMTVNNEIGVKQPIAEI--GRICSSRKVYFHTDAAQAVGKIPLDVN 245
Query: 245 KYPVDFVALSFYKLFGYPTGLGALVVR 271
+D +++S +K++G P G+GA+ +R
Sbjct: 246 DMKIDLMSISGHKIYG-PKGVGAIYIR 271
>NFS1_RAT (Q99P39) Cysteine desulfurase, mitochondrial precursor (EC
2.8.1.7)
Length = 451
Score = 54.7 bits (130), Expect = 9e-07
Identities = 61/246 (24%), Positives = 108/246 (43%), Gaps = 45/246 (18%)
Query: 38 VYLDHAGATLYSELQMESVFKDLTTNVYGNPHSQSDSSAATHDIVRD-ARQQVLDYCNAS 96
+Y+D T ++++ L N YGNPHS++ + + + ARQQV A
Sbjct: 53 LYMDVRATTPLDPRVLDAMLPYLV-NYYGNPHSRTHAYGWESEAAMERARQQVASLIGAD 111
Query: 97 PEDYKCIFTSGATAA--LKLVGEA-FPWSCNSNFMYTMENHNSVLGIREYALGQGAAAIA 153
P + IFTSGAT + + + G A F S + + T H VL
Sbjct: 112 PREI--IFTSGATESNNIAIKGVARFYRSRKKHLVTTQTEHKCVL--------------- 154
Query: 154 VDIEDVHPRIEGEKFPTKISLHQEQRRKVTGLQEEEPMECNFSGLRFDLDLAKI------ 207
D +E E F +++ Q+ + L+E E + ++ D L +
Sbjct: 155 ----DSCRSLEAEGF--RVTYLPVQKSGIIDLKELE------AAIQPDTSLVSVMTVNNE 202
Query: 208 --IKEDSSKILGASVCKKGRWLVLIDAAKGSATMPPDLSKYPVDFVALSFYKLFGYPTGL 265
+K+ ++I +C + DAA+ +P D++ +D +++S +KL+G P G+
Sbjct: 203 IGVKQPIAEI--GQICSSRKLYFHTDAAQAVGKIPLDVNDMKIDLMSISGHKLYG-PKGV 259
Query: 266 GALVVR 271
GA+ +R
Sbjct: 260 GAIYIR 265
>CSD_AERPE (Q9YAB6) Probable cysteine desulfurase (EC 2.8.1.7)
Length = 411
Score = 54.7 bits (130), Expect = 9e-07
Identities = 83/348 (23%), Positives = 143/348 (40%), Gaps = 64/348 (18%)
Query: 29 TEFNRLQ-DLVYLDHAGATLYSELQMESVFKDLTTNVYGNPHSQSDS-SAATHDIVRDAR 86
+EF L+ +VYLD+A +TL +E++ ++ + Y N H S DA
Sbjct: 11 SEFPELERGIVYLDNAASTLKPRRVVEAM-REFSYRSYANVHRGVHRLSMEASKAYEDAH 69
Query: 87 QQVLDYCNASPEDYKCIFTSGATAALKLVGEAFPWSC---NSNFMYTMENHNSVL--GIR 141
+ V S ++ +FT T A++L ++ + T +H+S L +R
Sbjct: 70 EVVARLVGGSWDEV--VFTRNTTEAMQLAALTLAYNGLLRGGEVLVTAADHHSTLLPWVR 127
Query: 142 EYALGQGAAAIAVDIEDVHPRIEGEKFPTKISLH----QEQRRKVTGLQEEEPMECNFSG 197
LG G A I ++G P L ++ R G N +G
Sbjct: 128 AARLGGGRARILP--------LDGRGVPRWDLLEDYITEDTRAVAVGHVS------NVTG 173
Query: 198 LRFDLDLAKIIKEDSSKILGASVCKKGRWLVLIDAAKGSATMPPDLSKYPVDFVALSFYK 257
+ ++ I+K ++K +GA LV++D+A+G +P D + VD A S +K
Sbjct: 174 VVAPVE--DIVK--AAKKVGA--------LVVLDSAQGVPHLPVDFRRMGVDMAAFSGHK 221
Query: 258 LFGYPTGLGALVVRNDAAKLLKKSYFSGGTVAASIADIDFIKRREGIEEL--------FE 309
+ G PTG+G L R D + L+ GGTV+ ++ + G E+ FE
Sbjct: 222 MLG-PTGIGVLWARRDLLEELEPPLGGGGTVSR-------VRLQGGSVEIEWEEPPWKFE 273
Query: 310 DGTVSFLSIASIRHGFKILNSLTVSAISR--------TYNISCPVYEE 349
GT + + +L + + ++R T + P+Y E
Sbjct: 274 AGTPPIIEAVGLAEAANMLMEIGMEHVARHEDELTSHTMKLLEPLYSE 321
>NFS1_MOUSE (Q9Z1J3) Cysteine desulfurase, mitochondrial precursor
(EC 2.8.1.7) (m-Nfs1)
Length = 451
Score = 53.9 bits (128), Expect = 2e-06
Identities = 61/246 (24%), Positives = 108/246 (43%), Gaps = 45/246 (18%)
Query: 38 VYLDHAGATLYSELQMESVFKDLTTNVYGNPHSQSDSSAATHDIVRD-ARQQVLDYCNAS 96
+Y+D T ++++ L N YGNPHS++ + + + ARQQV A
Sbjct: 53 LYMDVQATTPLDPRVLDAMLPYLV-NYYGNPHSRTHAYGWESEAAMERARQQVASLIGAD 111
Query: 97 PEDYKCIFTSGATAA--LKLVGEA-FPWSCNSNFMYTMENHNSVLGIREYALGQGAAAIA 153
P + IFTSGAT + + + G A F S + + T H VL
Sbjct: 112 PREI--IFTSGATESNNIAIKGVARFYRSRKKHLVTTQTEHKCVL--------------- 154
Query: 154 VDIEDVHPRIEGEKFPTKISLHQEQRRKVTGLQEEEPMECNFSGLRFDLDLAKI------ 207
D +E E F +++ Q+ + L+E E + ++ D L +
Sbjct: 155 ----DSCRSLEAEGF--RVTYLPVQKSGIIDLKELE------AAIQPDTSLVSVMTVNNE 202
Query: 208 --IKEDSSKILGASVCKKGRWLVLIDAAKGSATMPPDLSKYPVDFVALSFYKLFGYPTGL 265
+K+ ++I +C + DAA+ +P D++ +D +++S +KL+G P G+
Sbjct: 203 IGVKQPIAEI--RQICSSRKVYFHTDAAQAVGKIPLDVNDMKIDLMSISGHKLYG-PKGV 259
Query: 266 GALVVR 271
GA+ +R
Sbjct: 260 GAIYIR 265
>ISCS_BUCBP (Q89A19) Cysteine desulfurase (EC 2.8.1.7)
Length = 404
Score = 52.8 bits (125), Expect = 3e-06
Identities = 67/254 (26%), Positives = 109/254 (42%), Gaps = 29/254 (11%)
Query: 38 VYLDHAGATLYSELQMESVFKDLTTN-VYGNPHSQSDSSA-ATHDIVRDARQQVLDYCNA 95
+YLD+A T M+ + LT +GNP S+S + V AR Q+ + +A
Sbjct: 5 IYLDYAATTPVEFEVMKEMMNYLTLEGEFGNPASRSHKFGWKAEEAVDIARNQIAELIHA 64
Query: 96 SPEDYKCIFTSGATAA--LKLVGEAFPWSCNSNFMYTMEN-HNSVLGIREYALGQGAAAI 152
+ IFTSGAT + L + G A + N + T H + L Y +G
Sbjct: 65 DSREI--IFTSGATESNNLAIKGIAEFYKKKGNHIITCSTEHKATLDTCRYLENKG---- 118
Query: 153 AVDIEDVHPRIEGEKFPTKISLHQEQRRKVTGLQEEEPMECNFSGLRFDLDLAKIIKEDS 212
DI ++P G I QE+ +K T L S + + ++ I +D
Sbjct: 119 -FDITYLNPLQNGT---INIYELQEKIQKNTIL---------VSIMHVNNEIGVI--QDI 163
Query: 213 SKILGASVCKKGRWLVLIDAAKGSATMPPDLSKYPVDFVALSFYKLFGYPTGLGALVVRN 272
KI + +C+ L +DAA+ + +L + +D ++ S +K++G P G+G L +R
Sbjct: 164 HKI--SKICQSNNILFHVDAAQSIGKININLKQLKIDLMSFSAHKIYG-PKGIGGLYIRR 220
Query: 273 DAAKLLKKSYFSGG 286
L GG
Sbjct: 221 KPRVRLSAQIHGGG 234
>CSD_PYRAB (Q9V242) Probable cysteine desulfurase (EC 2.8.1.7)
Length = 401
Score = 52.0 bits (123), Expect = 6e-06
Identities = 67/319 (21%), Positives = 130/319 (40%), Gaps = 54/319 (16%)
Query: 35 QDLVYLDHAGATLYSELQMESV---FKDLTTNVYGNPHSQSDSSAATHDIVRDARQQVLD 91
Q+++Y D+ +L + +E++ + NV+ H S + ++ ++R+ V D
Sbjct: 15 QEVIYFDNTATSLTPKPVIEAMDEYYLRYRANVHRGVHRLSQMATQKYE---ESRKVVAD 71
Query: 92 YCNASPEDYKCIFTSGATAALKLV--GEAFPWSCNSNFMYT-MENHNSVLGIREYALGQG 148
+ NA ++ FT + +L LV G + + T E+H+++L + A +G
Sbjct: 72 FINAEFDEIA--FTKNTSESLNLVALGLEHLFKKGDKIVTTPYEHHSNLLPWQRLAKKKG 129
Query: 149 AAAIAV--------DIEDVHPRIEGEKFPTKISLHQEQRRKVTGLQEEEPMECNFSGLRF 200
+ D+ D +I+G K+ Q + + E E
Sbjct: 130 LKLEFIEGDDEGNLDLADAEKKIKG----AKLVAVQHVSNALGVIHEVE----------- 174
Query: 201 DLDLAKIIKEDSSKILGASVCKKGRWLVLIDAAKGSATMPPDLSKYPVDFVALSFYKLFG 260
+L K++KE+ + + ++DAA+ M D+ K DF+A S +K
Sbjct: 175 --ELGKMVKEEGA-------------IFVVDAAQSVGHMEVDVKKLKADFLAFSGHKGPM 219
Query: 261 YPTGLGALVVRNDAAKLLKKSYFSGGTVAASIADIDF-IKRREGIEELFEDGTVSFLSIA 319
PTG+G L + + + + GGT I D++ + E FE GT +
Sbjct: 220 GPTGIGVLYINKEFFDVFEPPLIGGGT----IEDVELCCYKLTEPPERFEAGTPNIGGAI 275
Query: 320 SIRHGFKILNSLTVSAISR 338
+ G K + + + I +
Sbjct: 276 GLAAGIKYIEKIGIDKIEK 294
>CSD_STAAN (P99177) Probable cysteine desulfurase (EC 2.8.1.7)
Length = 413
Score = 50.4 bits (119), Expect = 2e-05
Identities = 70/314 (22%), Positives = 131/314 (41%), Gaps = 46/314 (14%)
Query: 37 LVYLDHAGATLYSELQ----MESVFKDLTTNVYGNPHSQSDSSAATHDIVRDARQQVLDY 92
L YLD AT + +Q +E +K +NV+ H+ S AT D +AR+ V +
Sbjct: 27 LAYLDST-ATSQTPMQVLNVLEDYYKRYNSNVHRGVHTLG--SLAT-DGYENARETVRRF 82
Query: 93 CNASPEDYKCIFTSGATAALKLV----GEAFPWSCNSNFMYTMENHNSVLGIREYALGQG 148
NA + + IFT G TA++ LV G+A + + ME+H +++ ++ A +
Sbjct: 83 INAKYFE-EIIFTRGTTASINLVAHSYGDANVEEGDEIVVTEMEHHANIVPWQQLAKRKN 141
Query: 149 AAAIAVDIEDVHPRIEGEKFPTKISLHQEQRRKVTGLQEEEPMECNFSGLRFDLDLAKII 208
A ++ + +GE I + K+ + + S + ++ K I
Sbjct: 142 AT-----LKFIPMTADGELNIEDIKQTINDKTKIVAI-------AHISNVLGTINDVKTI 189
Query: 209 KEDSSKILGASVCKKGRWLVLIDAAKGSATMPPDLSKYPVDFVALSFYKLFGYPTGLGAL 268
A + + ++ +D A+ + M D+ + DF + S +K+ G PTG+G L
Sbjct: 190 ---------AEIAHQHGAIISVDGAQAAPHMKLDMQEMNADFYSFSGHKMLG-PTGIGVL 239
Query: 269 VVRNDAAKLLKKSYFSGGTVAASIADIDFIKRREG----IEELFEDGTVSFLSIASIRHG 324
+ + + ++ F G IDF+ + + + FE GT +
Sbjct: 240 FGKRELLQKMEPIEFGGDM-------IDFVSKYDATWADLPTKFEAGTPLIAQAIGLAEA 292
Query: 325 FKILNSLTVSAISR 338
+ L + AI +
Sbjct: 293 IRYLERIGFDAIHK 306
>CSD_STAAM (P63518) Probable cysteine desulfurase (EC 2.8.1.7)
Length = 413
Score = 50.4 bits (119), Expect = 2e-05
Identities = 70/314 (22%), Positives = 131/314 (41%), Gaps = 46/314 (14%)
Query: 37 LVYLDHAGATLYSELQ----MESVFKDLTTNVYGNPHSQSDSSAATHDIVRDARQQVLDY 92
L YLD AT + +Q +E +K +NV+ H+ S AT D +AR+ V +
Sbjct: 27 LAYLDST-ATSQTPMQVLNVLEDYYKRYNSNVHRGVHTLG--SLAT-DGYENARETVRRF 82
Query: 93 CNASPEDYKCIFTSGATAALKLV----GEAFPWSCNSNFMYTMENHNSVLGIREYALGQG 148
NA + + IFT G TA++ LV G+A + + ME+H +++ ++ A +
Sbjct: 83 INAKYFE-EIIFTRGTTASINLVAHSYGDANVEEGDEIVVTEMEHHANIVPWQQLAKRKN 141
Query: 149 AAAIAVDIEDVHPRIEGEKFPTKISLHQEQRRKVTGLQEEEPMECNFSGLRFDLDLAKII 208
A ++ + +GE I + K+ + + S + ++ K I
Sbjct: 142 AT-----LKFIPMTADGELNIEDIKQTINDKTKIVAI-------AHISNVLGTINDVKTI 189
Query: 209 KEDSSKILGASVCKKGRWLVLIDAAKGSATMPPDLSKYPVDFVALSFYKLFGYPTGLGAL 268
A + + ++ +D A+ + M D+ + DF + S +K+ G PTG+G L
Sbjct: 190 ---------AEIAHQHGAIISVDGAQAAPHMKLDMQEMNADFYSFSGHKMLG-PTGIGVL 239
Query: 269 VVRNDAAKLLKKSYFSGGTVAASIADIDFIKRREG----IEELFEDGTVSFLSIASIRHG 324
+ + + ++ F G IDF+ + + + FE GT +
Sbjct: 240 FGKRELLQKMEPIEFGGDM-------IDFVSKYDATWADLPTKFEAGTPLIAQAIGLAEA 292
Query: 325 FKILNSLTVSAISR 338
+ L + AI +
Sbjct: 293 IRYLERIGFDAIHK 306
>ISC1_ARCFU (O30052) Probable cysteine desulfurase 1 (EC 2.8.1.7)
Length = 382
Score = 48.5 bits (114), Expect = 6e-05
Identities = 56/236 (23%), Positives = 99/236 (41%), Gaps = 29/236 (12%)
Query: 39 YLDHAGATLYSELQMESVFKDLTTNVYGNPHSQSDSSAATHDIVRDARQQVLDYCNASPE 98
Y D+A E +E++ +T + +GNP S +++ +AR ++ + N
Sbjct: 3 YFDYASTKPVDERVVEAMLPYMTEH-FGNPSSVHSYGFKAREVIEEARGKISELINGGGG 61
Query: 99 DYKCIFTSGATAA--LKLVGEAFPWS-CNSNFMYTMENHNSVLGIREYALGQGAAAIAVD 155
D +FTSGAT + L L+G A + + + + H SV+ +Y QG +
Sbjct: 62 D--IVFTSGATESNNLALIGYAMRNARKGKHVLVSSIEHMSVINPAKYLQKQGFEVEFIP 119
Query: 156 IEDVHPRIEGEKFPTKISLHQEQRRKVTGLQEEEPMECNFSGLRFDLDLAKIIKEDSSKI 215
+ D + ++ E KI E+ + + ++ + IKE S I
Sbjct: 120 V-DKYGTVDLEFIEEKI--------------REDTILVSVQHANNEIGTIQPIKEISEII 164
Query: 216 LGASVCKKGRWLVLIDAAKGSATMPPDLSKYPVDFVALSFYKLFGYPTGLGALVVR 271
GR + +DA + D+ K D + +S ++G P G+GAL VR
Sbjct: 165 -------NGRAALHVDATASLGQIEVDVEKIGADMLTISSNDIYG-PKGVGALWVR 212
>CSD_STAEP (Q8CTA4) Probable cysteine desulfurase (EC 2.8.1.7)
Length = 413
Score = 47.0 bits (110), Expect = 2e-04
Identities = 68/310 (21%), Positives = 127/310 (40%), Gaps = 42/310 (13%)
Query: 37 LVYLDHAGATLYSELQMESVFKDLTTNVYGNPHS--QSDSSAATHDIVRDARQQVLDYCN 94
L YLD AT + +Q+ +V D N H + S AT D +AR+ V + N
Sbjct: 27 LAYLDST-ATSQTPVQVLNVLDDYYKRYNSNVHRGVHTLGSLAT-DGYENARETVRRFIN 84
Query: 95 ASPEDYKCIFTSGATAALKLV----GEAFPWSCNSNFMYTMENHNSVLGIREYALGQGAA 150
A + + IFT G TA++ +V G+A + + ME+H +++ ++ A + A
Sbjct: 85 AKYFE-EIIFTRGTTASINIVAHSYGDANISEGDEIVVTEMEHHANIVPWQQLAKRKNAT 143
Query: 151 AIAVDIEDVHPRIEGEKFPTKISLHQEQRRKVTGLQEEEPMECNFSGLRFDLDLAKIIKE 210
++ + +GE I + K+ + + + ++ +AKI E
Sbjct: 144 -----LKFIPMTKDGELQLDDIKATINDKTKIVAIAHVSNVLGTINDVK---TIAKIAHE 195
Query: 211 DSSKILGASVCKKGRWLVLIDAAKGSATMPPDLSKYPVDFVALSFYKLFGYPTGLGALVV 270
+ ++ +D A+ + M D+ DF + S +K+ G PTG+G L
Sbjct: 196 HGA-------------VISVDGAQSAPHMALDMQDIDADFYSFSGHKMLG-PTGIGVLYG 241
Query: 271 RNDAAKLLKKSYFSGGTVAASIADIDFIKRREG----IEELFEDGTVSFLSIASIRHGFK 326
+ + + ++ F G IDF+ + + + FE GT +
Sbjct: 242 KRELLQNMEPVEFGGDM-------IDFVSKYDSTWADLPTKFEAGTPLIAQAIGLAEAIH 294
Query: 327 ILNSLTVSAI 336
+ +L +AI
Sbjct: 295 YIENLGFNAI 304
>CSD_MYCPN (P75298) Probable cysteine desulfurase (EC 2.8.1.7)
Length = 408
Score = 46.2 bits (108), Expect = 3e-04
Identities = 62/258 (24%), Positives = 108/258 (41%), Gaps = 27/258 (10%)
Query: 67 NPHSQS-DSSAATHDIVRDARQQVLDYCNASPEDYKCIFTSGATAALKLVGEAF-PWSCN 124
NPH+++ D + I+ + RQ V D+ N + + IFTS AT ++ L PW
Sbjct: 53 NPHNKTPDLNNQIIAIIAETRQLVADWFNVTA--LEIIFTSSATESINLFAHGLKPWIKP 110
Query: 125 SNFMYTMENHNSVLGIREYALGQGAAAIAVDIEDVHPRIEGEKFPTKISLHQEQRRKVTG 184
+ + + +S + AL + A V +E + + F + I+ + KV
Sbjct: 111 GDEIVLKGDEHSANVLPWVALAKQTKARLVWVEKQANQSLEDTFKSLIN----PKTKVVA 166
Query: 185 LQEEEPMECNFSGLRFDL-DLAKIIKEDSSKILGASVCKKGRWLVLIDAAKGSATMPPDL 243
+ N G D +A +K+ + K V +DA + D+
Sbjct: 167 ITATS----NLFGNSIDFAQIADYLKQVNPKAF-----------VAVDAVQTVQHKQIDI 211
Query: 244 SKYPVDFVALSFYKLFGYPTGLGALVVRNDAAKLLKKSYFSGGTVAASIADIDFIKRREG 303
+K +DF+A S +K +G PTGLG ++ L+ GG + I D D +
Sbjct: 212 AKTQIDFLAFSTHKFYG-PTGLGVAYIKKSLQPQLQPLKL-GGDIFTQI-DPDNTIHFKS 268
Query: 304 IEELFEDGTVSFLSIASI 321
FE GT + ++I ++
Sbjct: 269 TPLKFEAGTPNIMAIYAL 286
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.322 0.138 0.415
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 87,990,548
Number of Sequences: 164201
Number of extensions: 3719965
Number of successful extensions: 7663
Number of sequences better than 10.0: 96
Number of HSP's better than 10.0 without gapping: 12
Number of HSP's successfully gapped in prelim test: 84
Number of HSP's that attempted gapping in prelim test: 7596
Number of HSP's gapped (non-prelim): 136
length of query: 764
length of database: 59,974,054
effective HSP length: 118
effective length of query: 646
effective length of database: 40,598,336
effective search space: 26226525056
effective search space used: 26226525056
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 70 (31.6 bits)
Medicago: description of AC148651.4


