

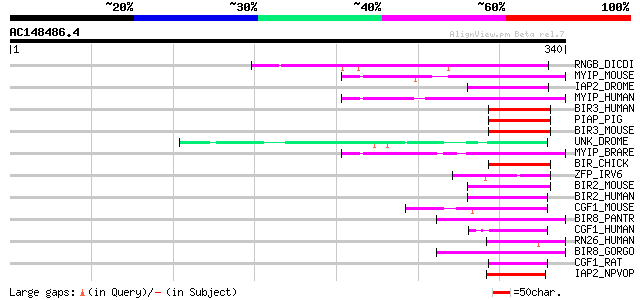
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148486.4 - phase: 0
(340 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
RNGB_DICDI (Q7M3S9) Ring finger protein B (Protein rngB) 72 2e-12
MYIP_MOUSE (Q8BM54) Ubiquitin ligase MYLIP (EC 6.3.2.-) (Myosin ... 53 1e-06
IAP2_DROME (Q24307) Apoptosis 2 inhibitor (Inhibitor of apoptosi... 53 1e-06
MYIP_HUMAN (Q8WY64) Ubiquitin ligase MYLIP (EC 6.3.2.-) (Myosin ... 52 3e-06
BIR3_HUMAN (Q13489) Baculoviral IAP repeat-containing protein 3 ... 52 3e-06
PIAP_PIG (O62640) Putative inhibitor of apoptosis 50 8e-06
BIR3_MOUSE (O08863) Baculoviral IAP repeat-containing protein 3 ... 50 8e-06
UNK_DROME (Q86B79) Unkempt protein 49 1e-05
MYIP_BRARE (Q6TEM9) Ubiquitin ligase MYLIP (EC 6.3.2.-) (Myosin ... 49 2e-05
BIR_CHICK (Q90660) Inhibitor of apoptosis protein (IAP) (Inhibit... 49 2e-05
ZFP_IRV6 (P47732) Zinc finger protein 48 4e-05
BIR2_MOUSE (Q62210) Baculoviral IAP repeat-containing protein 2 ... 48 4e-05
BIR2_HUMAN (Q13490) Baculoviral IAP repeat-containing protein 2 ... 47 5e-05
CGF1_MOUSE (Q8BMJ7) Cell growth regulator with RING finger domai... 47 7e-05
BIR8_PANTR (Q95M72) Baculoviral IAP repeat-containing protein 8 ... 46 2e-04
CGF1_HUMAN (Q99675) Cell growth regulator with RING finger domai... 45 2e-04
RN26_HUMAN (Q9BY78) RING finger protein 26 45 3e-04
BIR8_GORGO (Q95M71) Baculoviral IAP repeat-containing protein 8 ... 44 4e-04
CGF1_RAT (P97587) Cell growth regulator with RING finger domain ... 44 6e-04
IAP2_NPVOP (O10324) Probable apoptosis inhibitor 2 (IAP-2) 44 8e-04
>RNGB_DICDI (Q7M3S9) Ring finger protein B (Protein rngB)
Length = 943
Score = 72.4 bits (176), Expect = 2e-12
Identities = 58/206 (28%), Positives = 100/206 (48%), Gaps = 25/206 (12%)
Query: 149 RELQQQDLEMDRFLKLQGEQLRQTILEKVQATQLQSVSIIEDKVLQKLREKETE--VENI 206
++ QQQ + + L++Q EQL L+ Q +Q Q I + + +KL++++ + ++NI
Sbjct: 729 QQQQQQQQQQQQPLEIQ-EQLTMLQLQLSQLSQQQQNQIDKQQKQEKLQQEQQQQQLKNI 787
Query: 207 NKRNME-------------LEDQMEQLSVEAGAWQQRARYNENMIAALKFNLQQAYLQGR 253
N+ ++ E ++++LS + QQ +N I + L + L G
Sbjct: 788 NRLSISSNSSTLSSKDSFYFESKIQELSNQLKEKQQAITDRDNKIKDFENQLNKYKLIGL 847
Query: 254 DSKEGCGDSEVDDT--------ASCCNGRSLDFHL-LSNENSNMKDLMKCKACRVNEVTM 304
DS + E++ + S + R L+ + L E +KD C C N +
Sbjct: 848 DSMDHYQLLELESSFHNGLKQIGSIKDQRYLNRLVSLEKEKDQLKDQNSCVICASNPPNI 907
Query: 305 VLLPCKHLCLCKDCESKLSFCPLCQS 330
VLLPC+H LC DC SKL+ CP+C+S
Sbjct: 908 VLLPCRHSSLCSDCCSKLTKCPICRS 933
>MYIP_MOUSE (Q8BM54) Ubiquitin ligase MYLIP (EC 6.3.2.-) (Myosin
regulatory light chain interacting protein) (MIR)
Length = 445
Score = 53.1 bits (126), Expect = 1e-06
Identities = 39/141 (27%), Positives = 61/141 (42%), Gaps = 14/141 (9%)
Query: 204 ENINKRNMELEDQMEQLSVEAGAWQQRARYNENMIAALKFNLQQ---AYLQGRDSKEGCG 260
ENIN + D +++ S E +RA YN ++ + + Q + L+ DS C
Sbjct: 303 ENINLGKKYVFD-IKRTSKEVYDHARRALYNAGVVDLVSRSDQSPPSSPLKSSDSSMSC- 360
Query: 261 DSEVDDTASCCNGRSLD-FHLLSNENSNMKDLMKCKACRVNEVTMVLLPCKHLCLCKDCE 319
S C G S +L + +K+ M C AC E+ PC H C+ C
Sbjct: 361 --------SSCEGLSCQQTRVLQEKLRKLKEAMLCMACCEEEINSTFCPCGHTVCCESCA 412
Query: 320 SKLSFCPLCQSSKFIGMEVYM 340
++L CP+C+S VY+
Sbjct: 413 AQLQSCPVCRSRVEHVQHVYL 433
>IAP2_DROME (Q24307) Apoptosis 2 inhibitor (Inhibitor of apoptosis
2) (dIAP2) (DIAP) (IAP homolog A) (IAP-like protein)
(DILP)
Length = 498
Score = 53.1 bits (126), Expect = 1e-06
Identities = 21/50 (42%), Positives = 28/50 (56%)
Query: 281 LSNENSNMKDLMKCKACRVNEVTMVLLPCKHLCLCKDCESKLSFCPLCQS 330
L EN +KD CK C EV +V LPC HL C C ++ CP+C++
Sbjct: 438 LEEENRQLKDARLCKVCLDEEVGVVFLPCGHLATCNQCAPSVANCPMCRA 487
>MYIP_HUMAN (Q8WY64) Ubiquitin ligase MYLIP (EC 6.3.2.-) (Myosin
regulatory light chain interacting protein) (MIR)
(BM023)
Length = 445
Score = 51.6 bits (122), Expect = 3e-06
Identities = 37/138 (26%), Positives = 57/138 (40%), Gaps = 8/138 (5%)
Query: 204 ENINKRNMELEDQMEQLSVEAGAWQQRARYNENMIAALKFNLQQAYLQGRDSKEGCGDSE 263
ENIN + D +++ S E +RA YN ++ + + Q S SE
Sbjct: 303 ENINLGKKYVFD-IKRTSKEVYDHARRALYNAGVVDLVSRSNQSP------SHSPLKSSE 355
Query: 264 VDDTASCCNGRSLD-FHLLSNENSNMKDLMKCKACRVNEVTMVLLPCKHLCLCKDCESKL 322
S C G S +L + +K+ M C C E+ PC H C+ C ++L
Sbjct: 356 SSMNCSSCEGLSCQQTRVLQEKLRKLKEAMLCMVCCEEEINSTFCPCGHTVCCESCAAQL 415
Query: 323 SFCPLCQSSKFIGMEVYM 340
CP+C+S VY+
Sbjct: 416 QSCPVCRSRVEHVQHVYL 433
>BIR3_HUMAN (Q13489) Baculoviral IAP repeat-containing protein 3
(Inhibitor of apoptosis protein 1) (HIAP1) (HIAP-1)
(C-IAP2) (TNFR2-TRAF signaling complex protein 1) (IAP
homolog C) (Apoptosis inhibitor 2) (API2)
Length = 604
Score = 51.6 bits (122), Expect = 3e-06
Identities = 19/38 (50%), Positives = 26/38 (68%)
Query: 294 CKACRVNEVTMVLLPCKHLCLCKDCESKLSFCPLCQSS 331
CK C EV++V +PC HL +CKDC L CP+C+S+
Sbjct: 557 CKVCMDKEVSIVFIPCGHLVVCKDCAPSLRKCPICRST 594
>PIAP_PIG (O62640) Putative inhibitor of apoptosis
Length = 358
Score = 50.1 bits (118), Expect = 8e-06
Identities = 18/38 (47%), Positives = 25/38 (65%)
Query: 294 CKACRVNEVTMVLLPCKHLCLCKDCESKLSFCPLCQSS 331
CK C EV++V +PC HL +CKDC L CP+C+ +
Sbjct: 311 CKVCMDKEVSIVFIPCGHLVVCKDCAPSLRKCPICRGT 348
>BIR3_MOUSE (O08863) Baculoviral IAP repeat-containing protein 3
(Inhibitor of apoptosis protein 1) (MIAP1) (MIAP-1)
Length = 600
Score = 50.1 bits (118), Expect = 8e-06
Identities = 18/38 (47%), Positives = 25/38 (65%)
Query: 294 CKACRVNEVTMVLLPCKHLCLCKDCESKLSFCPLCQSS 331
CK C EV++V +PC HL +CKDC L CP+C+ +
Sbjct: 553 CKVCMDREVSIVFIPCGHLVVCKDCAPSLRKCPICRGT 590
>UNK_DROME (Q86B79) Unkempt protein
Length = 599
Score = 49.3 bits (116), Expect = 1e-05
Identities = 62/239 (25%), Positives = 92/239 (37%), Gaps = 48/239 (20%)
Query: 105 SQISSVDFLQPRSVSTGLGLSLDNTRLASTGDSALLSLIGDDIDRELQQQDLEMDRFLKL 164
+Q S F R +S G G D ++ + + L+ I DDI+ L
Sbjct: 387 TQNDSSLFFPSRIISPGFG---DGLSISPSVRISELNTIRDDINSSSVGNSL-------- 435
Query: 165 QGEQLRQTILEKVQATQLQSVSIIEDKVLQKL-REKETEVENINKRNMELEDQMEQLSV- 222
T+ A LQS+ + L ++ E T+ I+K N ED +L +
Sbjct: 436 ----FENTLNTAKNAFSLQSLQSQNNSDLGRITNELLTKNAQIHKLNGRFEDMACKLKIA 491
Query: 223 ---------EAGAWQQR---ARYNENMIAALKFNLQQAYLQGRDSKEGCGDSEVDDTASC 270
EA W++R A+ N+ A L+ +L L+ SK EVD
Sbjct: 492 ELHRDKAKQEAQEWKERYDLAQIQLNLPAELR-DLSIQKLKQLQSKLRTDLEEVDK---- 546
Query: 271 CNGRSLDFHLLSNENSNMKDLMKCKACRVNEVTMVLLPCKHLCLCKDCESKLSFCPLCQ 329
+L EN+ KC C N T+ L PC HL +C C ++ CP CQ
Sbjct: 547 ---------VLYLENAK-----KCMKCEENNRTVTLEPCNHLSICNTCAESVTECPYCQ 591
>MYIP_BRARE (Q6TEM9) Ubiquitin ligase MYLIP (EC 6.3.2.-) (Myosin
regulatory light chain interacting protein) (MIR)
Length = 472
Score = 48.9 bits (115), Expect = 2e-05
Identities = 34/137 (24%), Positives = 59/137 (42%), Gaps = 9/137 (6%)
Query: 204 ENINKRNMELEDQMEQLSVEAGAWQQRARYNENMIAALKFNLQQAYLQGRDSKEGCGDSE 263
ENIN + D + + S E + +RA YN ++ + ++ S+E G
Sbjct: 303 ENINLGKKYVFD-IRRTSKEVYDYARRALYNAGIVDMMARPGERTPSNRSPSREQEGAL- 360
Query: 264 VDDTASCCNGRSLDFHLLSNENSNMKDLMKCKACRVNEVTMVLLPCKHLCLCKDCESKLS 323
D C R LL + +++ + C C E+ PC H+ C++C ++L
Sbjct: 361 --DCGGCQQSR-----LLQEKLQKLREALLCMLCCEEEIDAAFCPCGHMVCCQNCAAQLQ 413
Query: 324 FCPLCQSSKFIGMEVYM 340
CP+C+S VY+
Sbjct: 414 SCPVCRSEVEHVQHVYL 430
>BIR_CHICK (Q90660) Inhibitor of apoptosis protein (IAP) (Inhibitor
of T cell apoptosis protein)
Length = 611
Score = 48.5 bits (114), Expect = 2e-05
Identities = 17/38 (44%), Positives = 25/38 (65%)
Query: 294 CKACRVNEVTMVLLPCKHLCLCKDCESKLSFCPLCQSS 331
CK C EV++V +PC HL +CK+C L CP+C+ +
Sbjct: 564 CKVCMDKEVSIVFIPCGHLVVCKECAPSLRKCPICRGT 601
>ZFP_IRV6 (P47732) Zinc finger protein
Length = 208
Score = 47.8 bits (112), Expect = 4e-05
Identities = 22/62 (35%), Positives = 37/62 (59%), Gaps = 3/62 (4%)
Query: 272 NGRSLDFHLLSNENSNMKD--LMKCKACRVNEVTMVLLPCKHLCLCKDCESKLSFCPLCQ 329
+ + LD + ++NS D ++ CK C N++T VL+PC H C +C KL CP+C+
Sbjct: 139 HNQDLDHNQDLDQNSTTSDCDVLTCKICFTNKITKVLIPCGH-SSCYECVFKLQTCPICK 197
Query: 330 SS 331
++
Sbjct: 198 NN 199
>BIR2_MOUSE (Q62210) Baculoviral IAP repeat-containing protein 2
(Inhibitor of apoptosis protein 2) (MIAP2) (MIAP-2)
Length = 612
Score = 47.8 bits (112), Expect = 4e-05
Identities = 17/51 (33%), Positives = 30/51 (58%)
Query: 281 LSNENSNMKDLMKCKACRVNEVTMVLLPCKHLCLCKDCESKLSFCPLCQSS 331
L + +++ CK C EV++V +PC HL +C++C L CP+C+ +
Sbjct: 552 LEEQLRRLQEERTCKVCMDREVSIVFIPCGHLVVCQECAPSLRKCPICRGT 602
>BIR2_HUMAN (Q13490) Baculoviral IAP repeat-containing protein 2
(Inhibitor of apoptosis protein 2) (HIAP2) (HIAP-2)
(C-IAP1) (TNFR2-TRAF signaling complex protein 2) (IAP
homolog B)
Length = 618
Score = 47.4 bits (111), Expect = 5e-05
Identities = 17/49 (34%), Positives = 29/49 (58%)
Query: 281 LSNENSNMKDLMKCKACRVNEVTMVLLPCKHLCLCKDCESKLSFCPLCQ 329
L + +++ CK C EV++V +PC HL +C++C L CP+C+
Sbjct: 558 LEEQLRRLQEERTCKVCMDKEVSVVFIPCGHLVVCQECAPSLRKCPICR 606
>CGF1_MOUSE (Q8BMJ7) Cell growth regulator with RING finger domain 1
(Cell growth regulatory gene 19 protein)
Length = 332
Score = 47.0 bits (110), Expect = 7e-05
Identities = 26/94 (27%), Positives = 44/94 (46%), Gaps = 14/94 (14%)
Query: 243 FNLQQAYLQGRDSKEGCGDSEVDDTASCCNGRSLDFHLLS------NENSNMKDLMK-CK 295
++L+Q ++ +S D +D RS++ LL N+ +++ + C
Sbjct: 223 YDLKQLFMSANNSPPPSNDESPED-------RSVEQSLLEKVGLAGNDGDPVEESSRDCV 275
Query: 296 ACRVNEVTMVLLPCKHLCLCKDCESKLSFCPLCQ 329
C+ V VLLPC+H CLC C CP+C+
Sbjct: 276 VCQNGGVNWVLLPCRHACLCDSCVRYFKQCPMCR 309
>BIR8_PANTR (Q95M72) Baculoviral IAP repeat-containing protein 8
(Inhibitor of apoptosis-like protein 2) (IAP-like
protein 2) (ILP-2)
Length = 236
Score = 45.8 bits (107), Expect = 2e-04
Identities = 23/80 (28%), Positives = 36/80 (44%), Gaps = 1/80 (1%)
Query: 262 SEVDDTASCCNGRSLDFHLLSNEN-SNMKDLMKCKACRVNEVTMVLLPCKHLCLCKDCES 320
++ D T + N SL + E ++D CK C + +V +PC HL CK C
Sbjct: 156 AQKDTTENESNQTSLQREISPEEPLRRLQDEKLCKICMDRHIAVVFIPCGHLVTCKQCAE 215
Query: 321 KLSFCPLCQSSKFIGMEVYM 340
+ CP+C + V+M
Sbjct: 216 AVDRCPMCSAVIDFKQRVFM 235
>CGF1_HUMAN (Q99675) Cell growth regulator with RING finger domain 1
(Cell growth regulatory gene 19 protein)
Length = 332
Score = 45.4 bits (106), Expect = 2e-04
Identities = 22/48 (45%), Positives = 26/48 (53%), Gaps = 5/48 (10%)
Query: 282 SNENSNMKDLMKCKACRVNEVTMVLLPCKHLCLCKDCESKLSFCPLCQ 329
S ENS KD C C+ V VLLPC+H CLC C CP+C+
Sbjct: 267 SEENS--KD---CVVCQNGTVNWVLLPCRHTCLCDGCVKYFQQCPMCR 309
>RN26_HUMAN (Q9BY78) RING finger protein 26
Length = 433
Score = 45.1 bits (105), Expect = 3e-04
Identities = 21/55 (38%), Positives = 29/55 (52%), Gaps = 7/55 (12%)
Query: 293 KCKACRVNEVTMVLLPCKHLCLCKDCESKL-------SFCPLCQSSKFIGMEVYM 340
KC C+ T++LLPC+HLCLC+ C L CPLC+ + VY+
Sbjct: 379 KCVICQDQSKTVLLLPCRHLCLCQACTEILMRHPVYHRNCPLCRRGILQTLNVYL 433
>BIR8_GORGO (Q95M71) Baculoviral IAP repeat-containing protein 8
(Inhibitor of apoptosis-like protein 2) (IAP-like
protein 2) (ILP-2)
Length = 236
Score = 44.3 bits (103), Expect = 4e-04
Identities = 22/80 (27%), Positives = 36/80 (44%), Gaps = 1/80 (1%)
Query: 262 SEVDDTASCCNGRSLDFHLLSNEN-SNMKDLMKCKACRVNEVTMVLLPCKHLCLCKDCES 320
++ D T + N SL + E +++ CK C + +V +PC HL CK C
Sbjct: 156 AQKDTTENESNQTSLQREISPEEPLRRLQEEKLCKICMDRHIAVVFIPCGHLVTCKQCAE 215
Query: 321 KLSFCPLCQSSKFIGMEVYM 340
+ CP+C + V+M
Sbjct: 216 AVDRCPMCNAVIDFKQRVFM 235
>CGF1_RAT (P97587) Cell growth regulator with RING finger domain 1
(Cell growth regulatory gene 19 protein)
Length = 332
Score = 43.9 bits (102), Expect = 6e-04
Identities = 16/36 (44%), Positives = 20/36 (55%)
Query: 294 CKACRVNEVTMVLLPCKHLCLCKDCESKLSFCPLCQ 329
C C+ V VLLPC+H CLC C CP+C+
Sbjct: 274 CVVCQNGGVNWVLLPCRHACLCDSCVCYFKQCPMCR 309
>IAP2_NPVOP (O10324) Probable apoptosis inhibitor 2 (IAP-2)
Length = 236
Score = 43.5 bits (101), Expect = 8e-04
Identities = 15/36 (41%), Positives = 23/36 (63%)
Query: 293 KCKACRVNEVTMVLLPCKHLCLCKDCESKLSFCPLC 328
+CK C VNE ++ LPC+HL +C +C + C +C
Sbjct: 188 ECKVCFVNEKSVCFLPCRHLVVCAECSPRCKRCCVC 223
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.317 0.133 0.386
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 39,019,528
Number of Sequences: 164201
Number of extensions: 1673026
Number of successful extensions: 8629
Number of sequences better than 10.0: 320
Number of HSP's better than 10.0 without gapping: 78
Number of HSP's successfully gapped in prelim test: 247
Number of HSP's that attempted gapping in prelim test: 7892
Number of HSP's gapped (non-prelim): 890
length of query: 340
length of database: 59,974,054
effective HSP length: 111
effective length of query: 229
effective length of database: 41,747,743
effective search space: 9560233147
effective search space used: 9560233147
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 66 (30.0 bits)
Medicago: description of AC148486.4


