

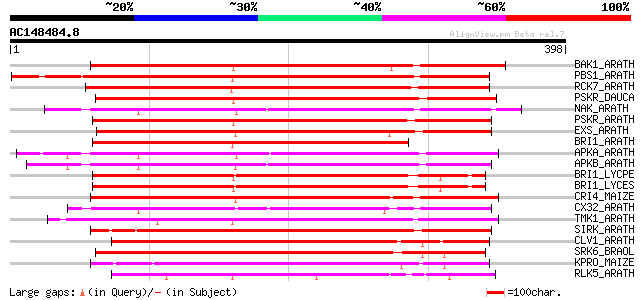
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148484.8 + phase: 0
(398 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
BAK1_ARATH (Q94F62) BRASSINOSTEROID INSENSITIVE 1-associated rec... 271 2e-72
PBS1_ARATH (Q9FE20) Serine/threonine-protein kinase PBS1 (EC 2.7... 263 6e-70
RCK7_ARATH (Q9LQQ8) Putative serine/threonine-protein kinase RLC... 259 6e-69
PSKR_DAUCA (Q8LPB4) Phytosulfokine receptor precursor (EC 2.7.1.... 251 2e-66
NAK_ARATH (P43293) Probable serine/threonine-protein kinase NAK ... 248 3e-65
PSKR_ARATH (Q9ZVR7) Putative phytosulfokine receptor precursor (... 246 6e-65
EXS_ARATH (Q9LYN8) Leucine-rich repeat receptor protein kinase E... 238 2e-62
BRI1_ARATH (O22476) BRASSINOSTEROID INSENSITIVE 1 precursor (EC ... 238 2e-62
APKA_ARATH (Q06548) Protein kinase APK1A, chloroplast precursor ... 238 2e-62
APKB_ARATH (P46573) Protein kinase APK1B, chloroplast precursor ... 236 1e-61
BRI1_LYCPE (Q8L899) Systemin receptor SR160 precursor (EC 2.7.1.... 230 4e-60
BRI1_LYCES (Q8GUQ5) Brassinosteroid LRR receptor kinase precurso... 230 4e-60
CRI4_MAIZE (O24585) Putative receptor protein kinase CRINKLY4 pr... 226 6e-59
CX32_ARATH (P27450) Probable serine/threonine-protein kinase Cx3... 222 1e-57
TMK1_ARATH (P43298) Putative receptor protein kinase TMK1 precur... 219 7e-57
SIRK_ARATH (O64483) Senescence-induced receptor-like serine/thre... 217 4e-56
CLV1_ARATH (Q9SYQ8) Receptor protein kinase CLAVATA1 precursor (... 211 3e-54
SRK6_BRAOL (Q09092) Putative serine/threonine-protein kinase rec... 209 8e-54
KPRO_MAIZE (P17801) Putative receptor protein kinase ZmPK1 precu... 190 5e-48
RLK5_ARATH (P47735) Receptor-like protein kinase 5 precursor (EC... 181 4e-45
>BAK1_ARATH (Q94F62) BRASSINOSTEROID INSENSITIVE 1-associated
receptor kinase 1 precursor (EC 2.7.1.37)
(BRI1-associated receptor kinase 1) (Somatic
embryogenesis receptor-like kinase 3)
Length = 615
Score = 271 bits (693), Expect = 2e-72
Identities = 148/303 (48%), Positives = 202/303 (65%), Gaps = 11/303 (3%)
Query: 59 FTYEELASATKGFANENIIGQGGFGYVHKGILPTGKEIAVKSLKAGSGQG-EREFQAEID 117
F+ EL A+ F+N+NI+G+GGFG V+KG L G +AVK LK QG E +FQ E++
Sbjct: 277 FSLRELQVASDNFSNKNILGRGGFGKVYKGRLADGTLVAVKRLKEERTQGGELQFQTEVE 336
Query: 118 IISRVHHRHLVSLVGYCVSGGQRMLVYEFVPNKTLEYHLHGK--GVPTMDWPTRMRIALG 175
+IS HR+L+ L G+C++ +R+LVY ++ N ++ L + P +DWP R RIALG
Sbjct: 337 MISMAVHRNLLRLRGFCMTPTERLLVYPYMANGSVASCLRERPESQPPLDWPKRQRIALG 396
Query: 176 SARGLAYLHEDCSPRIIHRDIKAANVLIDDSFEAKVADFGLAKLTTDTNTHVSTRVMGTF 235
SARGLAYLH+ C P+IIHRD+KAAN+L+D+ FEA V DFGLAKL +THV+T V GT
Sbjct: 397 SARGLAYLHDHCDPKIIHRDVKAANILLDEEFEAVVGDFGLAKLMDYKDTHVTTAVRGTI 456
Query: 236 GYMAPEYASSGKLTEKSDVFSFGVMLLELLTGKRPLD---LTNAMDESLVDWARPLLSRA 292
G++APEY S+GK +EK+DVF +GVMLLEL+TG+R D L N D L+DW + LL
Sbjct: 457 GHIAPEYLSTGKSSEKTDVFGYGVMLLELITGQRAFDLARLANDDDVMLLDWVKGLL--- 513
Query: 293 LEEDGNFAELVDPFLEGNYDHQEMIRLAACAASSIRHSAKKRSKMSQIVRALEGDVSLED 352
++ LVD L+GNY +E+ +L A + S +R KMS++VR LEGD E
Sbjct: 514 --KEKKLEALVDVDLQGNYKDEEVEQLIQVALLCTQSSPMERPKMSEVVRMLEGDGLAER 571
Query: 353 LKE 355
+E
Sbjct: 572 WEE 574
>PBS1_ARATH (Q9FE20) Serine/threonine-protein kinase PBS1 (EC
2.7.1.37) (AvrPphB susceptible protein 1)
Length = 456
Score = 263 bits (672), Expect = 6e-70
Identities = 147/348 (42%), Positives = 217/348 (62%), Gaps = 14/348 (4%)
Query: 2 DHVVRVQQNPNGASGVWGAPHPLMNSGEMSSNYSYGMGPPGSMQSSPGLSLTLKGGTFTY 61
+H + Q P ++ + G P + GE S+ + G + GL + TF +
Sbjct: 22 NHGQKKQSQPTVSNNISGLP----SGGEKLSSKTNGGSKRELLLPRDGLG-QIAAHTFAF 76
Query: 62 EELASATKGFANENIIGQGGFGYVHKGILP-TGKEIAVKSLKAGSGQGEREFQAEIDIIS 120
ELA+AT F + +G+GGFG V+KG L TG+ +AVK L QG REF E+ ++S
Sbjct: 77 RELAAATMNFHPDTFLGEGGFGRVYKGRLDSTGQVVAVKQLDRNGLQGNREFLVEVLMLS 136
Query: 121 RVHHRHLVSLVGYCVSGGQRMLVYEFVPNKTLEYHLHG--KGVPTMDWPTRMRIALGSAR 178
+HH +LV+L+GYC G QR+LVYEF+P +LE HLH +DW RM+IA G+A+
Sbjct: 137 LLHHPNLVNLIGYCADGDQRLLVYEFMPLGSLEDHLHDLPPDKEALDWNMRMKIAAGAAK 196
Query: 179 GLAYLHEDCSPRIIHRDIKAANVLIDDSFEAKVADFGLAKL-TTDTNTHVSTRVMGTFGY 237
GL +LH+ +P +I+RD K++N+L+D+ F K++DFGLAKL T +HVSTRVMGT+GY
Sbjct: 197 GLEFLHDKANPPVIYRDFKSSNILLDEGFHPKLSDFGLAKLGPTGDKSHVSTRVMGTYGY 256
Query: 238 MAPEYASSGKLTEKSDVFSFGVMLLELLTGKRPLDLTNAM-DESLVDWARPLLSRALEED 296
APEYA +G+LT KSDV+SFGV+ LEL+TG++ +D +++LV WARPL + +
Sbjct: 257 CAPEYAMTGQLTVKSDVYSFGVVFLELITGRKAIDSEMPHGEQNLVAWARPLFN----DR 312
Query: 297 GNFAELVDPFLEGNYDHQEMIRLAACAASSIRHSAKKRSKMSQIVRAL 344
F +L DP L+G + + + + A A+ I+ A R ++ +V AL
Sbjct: 313 RKFIKLADPRLKGRFPTRALYQALAVASMCIQEQAATRPLIADVVTAL 360
>RCK7_ARATH (Q9LQQ8) Putative serine/threonine-protein kinase
RLCKVII (EC 2.7.1.37)
Length = 423
Score = 259 bits (663), Expect = 6e-69
Identities = 134/295 (45%), Positives = 197/295 (66%), Gaps = 9/295 (3%)
Query: 55 KGGTFTYEELASATKGFANENIIGQGGFGYVHKGILPT-GKEIAVKSLKAGSGQGEREFQ 113
K TFT++ELA AT F ++ +G+GGFG V KG + + +A+K L QG REF
Sbjct: 87 KAQTFTFQELAEATGNFRSDCFLGEGGFGKVFKGTIEKLDQVVAIKQLDRNGVQGIREFV 146
Query: 114 AEIDIISRVHHRHLVSLVGYCVSGGQRMLVYEFVPNKTLEYHLH--GKGVPTMDWPTRMR 171
E+ +S H +LV L+G+C G QR+LVYE++P +LE HLH G +DW TRM+
Sbjct: 147 VEVLTLSLADHPNLVKLIGFCAEGDQRLLVYEYMPQGSLEDHLHVLPSGKKPLDWNTRMK 206
Query: 172 IALGSARGLAYLHEDCSPRIIHRDIKAANVLIDDSFEAKVADFGLAKL-TTDTNTHVSTR 230
IA G+ARGL YLH+ +P +I+RD+K +N+L+ + ++ K++DFGLAK+ + THVSTR
Sbjct: 207 IAAGAARGLEYLHDRMTPPVIYRDLKCSNILLGEDYQPKLSDFGLAKVGPSGDKTHVSTR 266
Query: 231 VMGTFGYMAPEYASSGKLTEKSDVFSFGVMLLELLTGKRPLDLTNA-MDESLVDWARPLL 289
VMGT+GY AP+YA +G+LT KSD++SFGV+LLEL+TG++ +D T D++LV WARPL
Sbjct: 267 VMGTYGYCAPDYAMTGQLTFKSDIYSFGVVLLELITGRKAIDNTKTRKDQNLVGWARPL- 325
Query: 290 SRALEEDGNFAELVDPFLEGNYDHQEMIRLAACAASSIRHSAKKRSKMSQIVRAL 344
++ NF ++VDP L+G Y + + + A +A ++ R +S +V AL
Sbjct: 326 ---FKDRRNFPKMVDPLLQGQYPVRGLYQALAISAMCVQEQPTMRPVVSDVVLAL 377
>PSKR_DAUCA (Q8LPB4) Phytosulfokine receptor precursor (EC 2.7.1.37)
(Phytosulfokine LRR receptor kinase)
Length = 1021
Score = 251 bits (642), Expect = 2e-66
Identities = 128/292 (43%), Positives = 186/292 (62%), Gaps = 9/292 (3%)
Query: 62 EELASATKGFANENIIGQGGFGYVHKGILPTGKEIAVKSLKAGSGQGEREFQAEIDIISR 121
+++ +T F NIIG GGFG V+K LP G ++A+K L +GQ +REFQAE++ +SR
Sbjct: 734 DDILKSTSSFNQANIIGCGGFGLVYKATLPDGTKVAIKRLSGDTGQMDREFQAEVETLSR 793
Query: 122 VHHRHLVSLVGYCVSGGQRMLVYEFVPNKTLEYHLHGK--GVPTMDWPTRMRIALGSARG 179
H +LV L+GYC ++L+Y ++ N +L+Y LH K G P++DW TR+RIA G+A G
Sbjct: 794 AQHPNLVHLLGYCNYKNDKLLIYSYMDNGSLDYWLHEKVDGPPSLDWKTRLRIARGAAEG 853
Query: 180 LAYLHEDCSPRIIHRDIKAANVLIDDSFEAKVADFGLAKLTTDTNTHVSTRVMGTFGYMA 239
LAYLH+ C P I+HRDIK++N+L+ D+F A +ADFGLA+L +THV+T ++GT GY+
Sbjct: 854 LAYLHQSCEPHILHRDIKSSNILLSDTFVAHLADFGLARLILPYDTHVTTDLVGTLGYIP 913
Query: 240 PEYASSGKLTEKSDVFSFGVMLLELLTGKRPLDLTNAM-DESLVDWARPLLSRALEEDGN 298
PEY + T K DV+SFGV+LLELLTG+RP+D+ L+ W + + E
Sbjct: 914 PEYGQASVATYKGDVYSFGVVLLELLTGRRPMDVCKPRGSRDLISWVLQMKTEKRE---- 969
Query: 299 FAELVDPFLEGNYDHQEMIRLAACAASSIRHSAKKRSKMSQIVRALEG-DVS 349
+E+ DPF+ +EM+ + A + + K R Q+V LE DVS
Sbjct: 970 -SEIFDPFIYDKDHAEEMLLVLEIACRCLGENPKTRPTTQQLVSWLENIDVS 1020
>NAK_ARATH (P43293) Probable serine/threonine-protein kinase NAK (EC
2.7.1.37)
Length = 389
Score = 248 bits (632), Expect = 3e-65
Identities = 145/356 (40%), Positives = 214/356 (59%), Gaps = 28/356 (7%)
Query: 26 NSGEMSSNYSYGMGPPGSMQSSPGLSLTLKGGTFTYEELASATKGFANENIIGQGGFGYV 85
+ G ++++SY G + + L F+ EL SAT+ F ++++G+GGFG V
Sbjct: 29 SKGSSTASFSYMPRTEGEILQNANLK------NFSLSELKSATRNFRPDSVVGEGGFGCV 82
Query: 86 HKGILP----------TGKEIAVKSLKAGSGQGEREFQAEIDIISRVHHRHLVSLVGYCV 135
KG + TG IAVK L QG RE+ AEI+ + ++ H +LV L+GYC+
Sbjct: 83 FKGWIDESSLAPSKPGTGIVIAVKRLNQEGFQGHREWLAEINYLGQLDHPNLVKLIGYCL 142
Query: 136 SGGQRMLVYEFVPNKTLEYHLHGKGV--PTMDWPTRMRIALGSARGLAYLHEDCSPRIIH 193
R+LVYEF+ +LE HL +G + W TR+R+ALG+ARGLA+LH + P++I+
Sbjct: 143 EEEHRLLVYEFMTRGSLENHLFRRGTFYQPLSWNTRVRMALGAARGLAFLH-NAQPQVIY 201
Query: 194 RDIKAANVLIDDSFEAKVADFGLAKL-TTDTNTHVSTRVMGTFGYMAPEYASSGKLTEKS 252
RD KA+N+L+D ++ AK++DFGLA+ N+HVSTRVMGT GY APEY ++G L+ KS
Sbjct: 202 RDFKASNILLDSNYNAKLSDFGLARDGPMGDNSHVSTRVMGTQGYAAPEYLATGHLSVKS 261
Query: 253 DVFSFGVMLLELLTGKRPLDLTNAMDE-SLVDWARPLLSRALEEDGNFAELVDPFLEGNY 311
DV+SFGV+LLELL+G+R +D + E +LVDWARP L+ ++DP L+G Y
Sbjct: 262 DVYSFGVVLLELLSGRRAIDKNQPVGEHNLVDWARPYLT----NKRRLLRVMDPRLQGQY 317
Query: 312 DHQEMIRLAACAASSIRHSAKKRSKMSQIVRALEGDVSLEDLKESMIKSPAPQTGV 367
+++A A I AK R M++IV+ +E L KE+ + PQ +
Sbjct: 318 SLTRALKIAVLALDCISIDAKSRPTMNEIVKTME---ELHIQKEASKEQQNPQISI 370
>PSKR_ARATH (Q9ZVR7) Putative phytosulfokine receptor precursor (EC
2.7.1.37) (Phytosulfokine LRR receptor kinase)
Length = 1008
Score = 246 bits (629), Expect = 6e-65
Identities = 123/289 (42%), Positives = 182/289 (62%), Gaps = 8/289 (2%)
Query: 60 TYEELASATKGFANENIIGQGGFGYVHKGILPTGKEIAVKSLKAGSGQGEREFQAEIDII 119
+Y++L +T F NIIG GGFG V+K LP GK++A+K L GQ EREF+AE++ +
Sbjct: 723 SYDDLLDSTNSFDQANIIGCGGFGMVYKATLPDGKKVAIKKLSGDCGQIEREFEAEVETL 782
Query: 120 SRVHHRHLVSLVGYCVSGGQRMLVYEFVPNKTLEYHLHGK--GVPTMDWPTRMRIALGSA 177
SR H +LV L G+C R+L+Y ++ N +L+Y LH + G + W TR+RIA G+A
Sbjct: 783 SRAQHPNLVLLRGFCFYKNDRLLIYSYMENGSLDYWLHERNDGPALLKWKTRLRIAQGAA 842
Query: 178 RGLAYLHEDCSPRIIHRDIKAANVLIDDSFEAKVADFGLAKLTTDTNTHVSTRVMGTFGY 237
+GL YLHE C P I+HRDIK++N+L+D++F + +ADFGLA+L + THVST ++GT GY
Sbjct: 843 KGLLYLHEGCDPHILHRDIKSSNILLDENFNSHLADFGLARLMSPYETHVSTDLVGTLGY 902
Query: 238 MAPEYASSGKLTEKSDVFSFGVMLLELLTGKRPLDLTNAMD-ESLVDWARPLLSRALEED 296
+ PEY + T K DV+SFGV+LLELLT KRP+D+ L+ W ++ +
Sbjct: 903 IPPEYGQASVATYKGDVYSFGVVLLELLTDKRPVDMCKPKGCRDLISWV-----VKMKHE 957
Query: 297 GNFAELVDPFLEGNYDHQEMIRLAACAASSIRHSAKKRSKMSQIVRALE 345
+E+ DP + + +EM R+ A + + K+R Q+V L+
Sbjct: 958 SRASEVFDPLIYSKENDKEMFRVLEIACLCLSENPKQRPTTQQLVSWLD 1006
>EXS_ARATH (Q9LYN8) Leucine-rich repeat receptor protein kinase EXS
precursor (EC 2.7.1.37) (Extra sporogenous cells protein)
(EXCESS MICROSPOROCYTES1 protein)
Length = 1192
Score = 238 bits (608), Expect = 2e-62
Identities = 125/287 (43%), Positives = 180/287 (62%), Gaps = 9/287 (3%)
Query: 63 ELASATKGFANENIIGQGGFGYVHKGILPTGKEIAVKSLKAGSGQGEREFQAEIDIISRV 122
++ AT F+ +NIIG GGFG V+K LP K +AVK L QG REF AE++ + +V
Sbjct: 909 DIVEATDHFSKKNIIGDGGFGTVYKACLPGEKTVAVKKLSEAKTQGNREFMAEMETLGKV 968
Query: 123 HHRHLVSLVGYCVSGGQRMLVYEFVPNKTLEYHLHGKG--VPTMDWPTRMRIALGSARGL 180
H +LVSL+GYC +++LVYE++ N +L++ L + + +DW R++IA+G+ARGL
Sbjct: 969 KHPNLVSLLGYCSFSEEKLLVYEYMVNGSLDHWLRNQTGMLEVLDWSKRLKIAVGAARGL 1028
Query: 181 AYLHEDCSPRIIHRDIKAANVLIDDSFEAKVADFGLAKLTTDTNTHVSTRVMGTFGYMAP 240
A+LH P IIHRDIKA+N+L+D FE KVADFGLA+L + +HVST + GTFGY+ P
Sbjct: 1029 AFLHHGFIPHIIHRDIKASNILLDGDFEPKVADFGLARLISACESHVSTVIAGTFGYIPP 1088
Query: 241 EYASSGKLTEKSDVFSFGVMLLELLTGKRPL--DLTNAMDESLVDWARPLLSRALEEDGN 298
EY S + T K DV+SFGV+LLEL+TGK P D + +LV WA +++ G
Sbjct: 1089 EYGQSARATTKGDVYSFGVILLELVTGKEPTGPDFKESEGGNLVGWAIQKINQ-----GK 1143
Query: 299 FAELVDPFLEGNYDHQEMIRLAACAASSIRHSAKKRSKMSQIVRALE 345
+++DP L +RL A + + KR M +++AL+
Sbjct: 1144 AVDVIDPLLVSVALKNSQLRLLQIAMLCLAETPAKRPNMLDVLKALK 1190
>BRI1_ARATH (O22476) BRASSINOSTEROID INSENSITIVE 1 precursor (EC
2.7.1.37) (AtBRI1) (Brassinosteroid LRR receptor kinase)
Length = 1196
Score = 238 bits (608), Expect = 2e-62
Identities = 118/230 (51%), Positives = 166/230 (71%), Gaps = 3/230 (1%)
Query: 60 TYEELASATKGFANENIIGQGGFGYVHKGILPTGKEIAVKSLKAGSGQGEREFQAEIDII 119
T+ +L AT GF N+++IG GGFG V+K IL G +A+K L SGQG+REF AE++ I
Sbjct: 872 TFADLLQATNGFHNDSLIGSGGFGDVYKAILKDGSAVAIKKLIHVSGQGDREFMAEMETI 931
Query: 120 SRVHHRHLVSLVGYCVSGGQRMLVYEFVPNKTLEYHLHG--KGVPTMDWPTRMRIALGSA 177
++ HR+LV L+GYC G +R+LVYEF+ +LE LH K ++W TR +IA+GSA
Sbjct: 932 GKIKHRNLVPLLGYCKVGDERLLVYEFMKYGSLEDVLHDPKKAGVKLNWSTRRKIAIGSA 991
Query: 178 RGLAYLHEDCSPRIIHRDIKAANVLIDDSFEAKVADFGLAKLTTDTNTHVSTRVM-GTFG 236
RGLA+LH +CSP IIHRD+K++NVL+D++ EA+V+DFG+A+L + +TH+S + GT G
Sbjct: 992 RGLAFLHHNCSPHIIHRDMKSSNVLLDENLEARVSDFGMARLMSAMDTHLSVSTLAGTPG 1051
Query: 237 YMAPEYASSGKLTEKSDVFSFGVMLLELLTGKRPLDLTNAMDESLVDWAR 286
Y+ PEY S + + K DV+S+GV+LLELLTGKRP D + D +LV W +
Sbjct: 1052 YVPPEYYQSFRCSTKGDVYSYGVVLLELLTGKRPTDSPDFGDNNLVGWVK 1101
>APKA_ARATH (Q06548) Protein kinase APK1A, chloroplast precursor (EC
2.7.1.-)
Length = 410
Score = 238 bits (608), Expect = 2e-62
Identities = 143/361 (39%), Positives = 212/361 (58%), Gaps = 28/361 (7%)
Query: 6 RVQQNPNGASGVWGAPHPLMNSGEMSSNYSYGMGP--PGSMQSSPGLSLTLKGGTFTYEE 63
+V+ +GAS + A + + G +S+ S P G + SP L +F++ E
Sbjct: 8 QVKAESSGASTKYDAKD-IGSLGSKASSVSVRPSPRTEGEILQSPNLK------SFSFAE 60
Query: 64 LASATKGFANENIIGQGGFGYVHKGILP----------TGKEIAVKSLKAGSGQGEREFQ 113
L SAT+ F ++++G+GGFG V KG + TG IAVK L QG +E+
Sbjct: 61 LKSATRNFRPDSVLGEGGFGCVFKGWIDEKSLTASRPGTGLVIAVKKLNQDGWQGHQEWL 120
Query: 114 AEIDIISRVHHRHLVSLVGYCVSGGQRMLVYEFVPNKTLEYHLHGKGV--PTMDWPTRMR 171
AE++ + + HRHLV L+GYC+ R+LVYEF+P +LE HL +G+ + W R++
Sbjct: 121 AEVNYLGQFSHRHLVKLIGYCLEDEHRLLVYEFMPRGSLENHLFRRGLYFQPLSWKLRLK 180
Query: 172 IALGSARGLAYLHEDCSPRIIHRDIKAANVLIDDSFEAKVADFGLAKL-TTDTNTHVSTR 230
+ALG+A+GLA+LH R+I+RD K +N+L+D + AK++DFGLAK +HVSTR
Sbjct: 181 VALGAAKGLAFLHSS-ETRVIYRDFKTSNILLDSEYNAKLSDFGLAKDGPIGDKSHVSTR 239
Query: 231 VMGTFGYMAPEYASSGKLTEKSDVFSFGVMLLELLTGKRPLDLTNAMDE-SLVDWARPLL 289
VMGT GY APEY ++G LT KSDV+SFGV+LLELL+G+R +D E +LV+WA+P L
Sbjct: 240 VMGTHGYAAPEYLATGHLTTKSDVYSFGVVLLELLSGRRAVDKNRPSGERNLVEWAKPYL 299
Query: 290 SRALEEDGNFAELVDPFLEGNYDHQEMIRLAACAASSIRHSAKKRSKMSQIVRALEGDVS 349
+ ++D L+ Y +E ++A + + K R MS++V LE S
Sbjct: 300 VNKRK----IFRVIDNRLQDQYSMEEACKVATLSLRCLTTEIKLRPNMSEVVSHLEHIQS 355
Query: 350 L 350
L
Sbjct: 356 L 356
>APKB_ARATH (P46573) Protein kinase APK1B, chloroplast precursor (EC
2.7.1.-)
Length = 412
Score = 236 bits (601), Expect = 1e-61
Identities = 136/349 (38%), Positives = 207/349 (58%), Gaps = 27/349 (7%)
Query: 13 GASGVWGAPHPLMNSGEMSSNYSYGMGP--PGSMQSSPGLSLTLKGGTFTYEELASATKG 70
GAS + + + G SS+ S P G + SP L +FT+ EL +AT+
Sbjct: 15 GASPKYMSSEANDSLGSKSSSVSIRTNPRTEGEILQSPNLK------SFTFAELKAATRN 68
Query: 71 FANENIIGQGGFGYVHKGILP----------TGKEIAVKSLKAGSGQGEREFQAEIDIIS 120
F ++++G+GGFG V KG + TG IAVK L QG +E+ AE++ +
Sbjct: 69 FRPDSVLGEGGFGSVFKGWIDEQTLTASKPGTGVVIAVKKLNQDGWQGHQEWLAEVNYLG 128
Query: 121 RVHHRHLVSLVGYCVSGGQRMLVYEFVPNKTLEYHLHGKG--VPTMDWPTRMRIALGSAR 178
+ H +LV L+GYC+ R+LVYEF+P +LE HL +G + W R+++ALG+A+
Sbjct: 129 QFSHPNLVKLIGYCLEDEHRLLVYEFMPRGSLENHLFRRGSYFQPLSWTLRLKVALGAAK 188
Query: 179 GLAYLHEDCSPRIIHRDIKAANVLIDDSFEAKVADFGLAKL-TTDTNTHVSTRVMGTFGY 237
GLA+LH + +I+RD K +N+L+D + AK++DFGLAK T +HVSTR+MGT+GY
Sbjct: 189 GLAFLH-NAETSVIYRDFKTSNILLDSEYNAKLSDFGLAKDGPTGDKSHVSTRIMGTYGY 247
Query: 238 MAPEYASSGKLTEKSDVFSFGVMLLELLTGKRPLDLTNAM-DESLVDWARPLLSRALEED 296
APEY ++G LT KSDV+S+GV+LLE+L+G+R +D ++ LV+WARPLL+ +
Sbjct: 248 AAPEYLATGHLTTKSDVYSYGVVLLEVLSGRRAVDKNRPPGEQKLVEWARPLLANKRK-- 305
Query: 297 GNFAELVDPFLEGNYDHQEMIRLAACAASSIRHSAKKRSKMSQIVRALE 345
++D L+ Y +E ++A A + K R M+++V LE
Sbjct: 306 --LFRVIDNRLQDQYSMEEACKVATLALRCLTFEIKLRPNMNEVVSHLE 352
>BRI1_LYCPE (Q8L899) Systemin receptor SR160 precursor (EC 2.7.1.37)
(Brassinosteroid LRR receptor kinase)
Length = 1207
Score = 230 bits (587), Expect = 4e-60
Identities = 126/290 (43%), Positives = 189/290 (64%), Gaps = 17/290 (5%)
Query: 60 TYEELASATKGFANENIIGQGGFGYVHKGILPTGKEIAVKSLKAGSGQGEREFQAEIDII 119
T+ +L AT GF N++++G GGFG V+K L G +A+K L SGQG+REF AE++ I
Sbjct: 877 TFADLLEATNGFHNDSLVGSGGFGDVYKAQLKDGSVVAIKKLIHVSGQGDREFTAEMETI 936
Query: 120 SRVHHRHLVSLVGYCVSGGQRMLVYEFVPNKTLEYHLHGK---GVPTMDWPTRMRIALGS 176
++ HR+LV L+GYC G +R+LVYE++ +LE LH + G+ ++WP R +IA+G+
Sbjct: 937 GKIKHRNLVPLLGYCKVGEERLLVYEYMKYGSLEDVLHDRKKTGI-KLNWPARRKIAIGA 995
Query: 177 ARGLAYLHEDCSPRIIHRDIKAANVLIDDSFEAKVADFGLAKLTTDTNTHVSTRVM-GTF 235
ARGLA+LH +C P IIHRD+K++NVL+D++ EA+V+DFG+A+L + +TH+S + GT
Sbjct: 996 ARGLAFLHHNCIPHIIHRDMKSSNVLLDENLEARVSDFGMARLMSAMDTHLSVSTLAGTP 1055
Query: 236 GYMAPEYASSGKLTEKSDVFSFGVMLLELLTGKRPLDLTNAMDESLVDWARPLLSRALEE 295
GY+ PEY S + + K DV+S+GV+LLELLTGK+P D + D +LV W + L
Sbjct: 1056 GYVPPEYYQSFRCSTKGDVYSYGVVLLELLTGKQPTDSADFGDNNLVGWVK------LHA 1109
Query: 296 DGNFAELVDPFL---EGNYDHQEMIRL-AACAASSIRHSAKKRSKMSQIV 341
G ++ D L + + + + + L ACA RH KR M Q++
Sbjct: 1110 KGKITDVFDRELLKEDASIEIELLQHLKVACACLDDRH--WKRPTMIQVM 1157
>BRI1_LYCES (Q8GUQ5) Brassinosteroid LRR receptor kinase precursor (EC
2.7.1.37) (tBRI1) (Altered brassinolide sensitivity 1)
(Systemin receptor SR160)
Length = 1207
Score = 230 bits (587), Expect = 4e-60
Identities = 126/290 (43%), Positives = 189/290 (64%), Gaps = 17/290 (5%)
Query: 60 TYEELASATKGFANENIIGQGGFGYVHKGILPTGKEIAVKSLKAGSGQGEREFQAEIDII 119
T+ +L AT GF N++++G GGFG V+K L G +A+K L SGQG+REF AE++ I
Sbjct: 877 TFADLLEATNGFHNDSLVGSGGFGDVYKAQLKDGSVVAIKKLIHVSGQGDREFTAEMETI 936
Query: 120 SRVHHRHLVSLVGYCVSGGQRMLVYEFVPNKTLEYHLHGK---GVPTMDWPTRMRIALGS 176
++ HR+LV L+GYC G +R+LVYE++ +LE LH + G+ ++WP R +IA+G+
Sbjct: 937 GKIKHRNLVPLLGYCKVGEERLLVYEYMKYGSLEDVLHDRKKIGI-KLNWPARRKIAIGA 995
Query: 177 ARGLAYLHEDCSPRIIHRDIKAANVLIDDSFEAKVADFGLAKLTTDTNTHVSTRVM-GTF 235
ARGLA+LH +C P IIHRD+K++NVL+D++ EA+V+DFG+A+L + +TH+S + GT
Sbjct: 996 ARGLAFLHHNCIPHIIHRDMKSSNVLLDENLEARVSDFGMARLMSAMDTHLSVSTLAGTP 1055
Query: 236 GYMAPEYASSGKLTEKSDVFSFGVMLLELLTGKRPLDLTNAMDESLVDWARPLLSRALEE 295
GY+ PEY S + + K DV+S+GV+LLELLTGK+P D + D +LV W + L
Sbjct: 1056 GYVPPEYYQSFRCSTKGDVYSYGVVLLELLTGKQPTDSADFGDNNLVGWVK------LHA 1109
Query: 296 DGNFAELVDPFL---EGNYDHQEMIRL-AACAASSIRHSAKKRSKMSQIV 341
G ++ D L + + + + + L ACA RH KR M Q++
Sbjct: 1110 KGKITDVFDRELLKEDASIEIELLQHLKVACACLDDRH--WKRPTMIQVM 1157
>CRI4_MAIZE (O24585) Putative receptor protein kinase CRINKLY4
precursor (EC 2.7.1.-)
Length = 901
Score = 226 bits (577), Expect = 6e-59
Identities = 124/298 (41%), Positives = 191/298 (63%), Gaps = 12/298 (4%)
Query: 59 FTYEELASATKGFANENIIGQGGFGYVHKGILPTGKEIAVK-SLKAGS-GQGEREFQAEI 116
F+YEEL AT GF+ ++ +G+G F V KGIL G +AVK ++KA + +EF E+
Sbjct: 493 FSYEELEQATGGFSEDSQVGKGSFSCVFKGILRDGTVVAVKRAIKASDVKKSSKEFHNEL 552
Query: 117 DIISRVHHRHLVSLVGYCVSGGQRMLVYEFVPNKTLEYHLHGKG---VPTMDWPTRMRIA 173
D++SR++H HL++L+GYC G +R+LVYEF+ + +L HLHGK ++W R+ IA
Sbjct: 553 DLLSRLNHAHLLNLLGYCEDGSERLLVYEFMAHGSLYQHLHGKDPNLKKRLNWARRVTIA 612
Query: 174 LGSARGLAYLHEDCSPRIIHRDIKAANVLIDDSFEAKVADFGLAKL-TTDTNTHVSTRVM 232
+ +ARG+ YLH P +IHRDIK++N+LID+ A+VADFGL+ L D+ T +S
Sbjct: 613 VQAARGIEYLHGYACPPVIHRDIKSSNILIDEDHNARVADFGLSILGPADSGTPLSELPA 672
Query: 233 GTFGYMAPEYASSGKLTEKSDVFSFGVMLLELLTGKRPLDLTNAMDESLVDWARPLLSRA 292
GT GY+ PEY LT KSDV+SFGV+LLE+L+G++ +D+ + ++V+WA PL+
Sbjct: 673 GTLGYLDPEYYRLHYLTTKSDVYSFGVVLLEILSGRKAIDM-QFEEGNIVEWAVPLI--- 728
Query: 293 LEEDGNFAELVDPFLEGNYDHQEMIRLAACAASSIRHSAKKRSKMSQIVRALEGDVSL 350
+ G+ ++DP L D + + ++A+ A +R K R M ++ ALE ++L
Sbjct: 729 --KAGDIFAILDPVLSPPSDLEALKKIASVACKCVRMRGKDRPSMDKVTTALEHALAL 784
>CX32_ARATH (P27450) Probable serine/threonine-protein kinase Cx32,
chloroplast precursor (EC 2.7.1.37)
Length = 419
Score = 222 bits (565), Expect = 1e-57
Identities = 127/321 (39%), Positives = 192/321 (59%), Gaps = 34/321 (10%)
Query: 42 GSMQSSPGLSLTLKGGTFTYEELASATKGFANENIIGQGGFGYVHKGILP---------- 91
G + SP L + + + +L +ATK F ++++GQGGFG V++G +
Sbjct: 63 GKLLESPNLKV------YNFLDLKTATKNFKPDSMLGQGGFGKVYRGWVDATTLAPSRVG 116
Query: 92 TGKEIAVKSLKAGSGQGEREFQAEIDIISRVHHRHLVSLVGYCVSGGQRMLVYEFVPNKT 151
+G +A+K L + S QG E+++E++ + + HR+LV L+GYC + +LVYEF+P +
Sbjct: 117 SGMIVAIKRLNSESVQGFAEWRSEVNFLGMLSHRNLVKLLGYCREDKELLLVYEFMPKGS 176
Query: 152 LEYHLHGKGVPTMDWPTRMRIALGSARGLAYLHEDCSPRIIHRDIKAANVLIDDSFEAKV 211
LE HL + P W R++I +G+ARGLA+LH +I+RD KA+N+L+D +++AK+
Sbjct: 177 LESHLFRRNDP-FPWDLRIKIVIGAARGLAFLHS-LQREVIYRDFKASNILLDSNYDAKL 234
Query: 212 ADFGLAKL-TTDTNTHVSTRVMGTFGYMAPEYASSGKLTEKSDVFSFGVMLLELLTG--- 267
+DFGLAKL D +HV+TR+MGT+GY APEY ++G L KSDVF+FGV+LLE++TG
Sbjct: 235 SDFGLAKLGPADEKSHVTTRIMGTYGYAAPEYMATGHLYVKSDVFAFGVVLLEIMTGLTA 294
Query: 268 ---KRPLDLTNAMDESLVDWARPLLSRALEEDGNFAELVDPFLEGNYDHQEMIRLAACAA 324
KRP ESLVDW RP LS +++D ++G Y + +A
Sbjct: 295 HNTKRPRG-----QESLVDWLRPELS----NKHRVKQIMDKGIKGQYTTKVATEMARITL 345
Query: 325 SSIRHSAKKRSKMSQIVRALE 345
S I K R M ++V LE
Sbjct: 346 SCIEPDPKNRPHMKEVVEVLE 366
>TMK1_ARATH (P43298) Putative receptor protein kinase TMK1 precursor
(EC 2.7.1.-)
Length = 942
Score = 219 bits (559), Expect = 7e-57
Identities = 122/331 (36%), Positives = 195/331 (58%), Gaps = 15/331 (4%)
Query: 28 GEMSSNYSYGMGPPGSMQSSPGLSLTLKGGTF-TYEELASATKGFANENIIGQGGFGYVH 86
G +S Y+ PG+ + + + G + + L S T F+++NI+G GGFG V+
Sbjct: 548 GGISDTYTL----PGTSEVGDNIQMVEAGNMLISIQVLRSVTNNFSSDNILGSGGFGVVY 603
Query: 87 KGILPTGKEIAVKSLKAG--SGQGEREFQAEIDIISRVHHRHLVSLVGYCVSGGQRMLVY 144
KG L G +IAVK ++ G +G+G EF++EI ++++V HRHLV+L+GYC+ G +++LVY
Sbjct: 604 KGELHDGTKIAVKRMENGVIAGKGFAEFKSEIAVLTKVRHRHLVTLLGYCLDGNEKLLVY 663
Query: 145 EFVPNKTLEYHLHG---KGVPTMDWPTRMRIALGSARGLAYLHEDCSPRIIHRDIKAANV 201
E++P TL HL +G+ + W R+ +AL ARG+ YLH IHRD+K +N+
Sbjct: 664 EYMPQGTLSRHLFEWSEEGLKPLLWKQRLTLALDVARGVEYLHGLAHQSFIHRDLKPSNI 723
Query: 202 LIDDSFEAKVADFGLAKLTTDTNTHVSTRVMGTFGYMAPEYASSGKLTEKSDVFSFGVML 261
L+ D AKVADFGL +L + + TR+ GTFGY+APEYA +G++T K DV+SFGV+L
Sbjct: 724 LLGDDMRAKVADFGLVRLAPEGKGSIETRIAGTFGYLAPEYAVTGRVTTKVDVYSFGVIL 783
Query: 262 LELLTGKRPLDLTNAMDE-SLVDWARPLLSRALEEDGNFAELVDPFLEGNYDHQEMIR-L 319
+EL+TG++ LD + + LV W + + + ++ +F + +D ++ + + + +
Sbjct: 784 MELITGRKSLDESQPEESIHLVSWFKRMY---INKEASFKKAIDTTIDLDEETLASVHTV 840
Query: 320 AACAASSIRHSAKKRSKMSQIVRALEGDVSL 350
A A +R M V L V L
Sbjct: 841 AELAGHCCAREPYQRPDMGHAVNILSSLVEL 871
>SIRK_ARATH (O64483) Senescence-induced receptor-like
serine/threonine kinase precursor (FLG22-induced
receptor-like kinase 1)
Length = 876
Score = 217 bits (553), Expect = 4e-56
Identities = 112/288 (38%), Positives = 181/288 (61%), Gaps = 9/288 (3%)
Query: 59 FTYEELASATKGFANENIIGQGGFGYVHKGILPTGKEIAVKSLKAGSGQGEREFQAEIDI 118
F Y E+ + T F E +IG+GGFG V+ G++ G+++AVK L S QG +EF+AE+D+
Sbjct: 564 FKYSEVVNITNNF--ERVIGKGGFGKVYHGVI-NGEQVAVKVLSEESAQGYKEFRAEVDL 620
Query: 119 ISRVHHRHLVSLVGYCVSGGQRMLVYEFVPNKTLEYHLHGKGVPTMDWPTRMRIALGSAR 178
+ RVHH +L SLVGYC +L+YE++ N+ L +L GK + W R++I+L +A+
Sbjct: 621 LMRVHHTNLTSLVGYCNEINHMVLIYEYMANENLGDYLAGKRSFILSWEERLKISLDAAQ 680
Query: 179 GLAYLHEDCSPRIIHRDIKAANVLIDDSFEAKVADFGLAK-LTTDTNTHVSTRVMGTFGY 237
GL YLH C P I+HRD+K N+L+++ +AK+ADFGL++ + + + +ST V G+ GY
Sbjct: 681 GLEYLHNGCKPPIVHRDVKPTNILLNEKLQAKMADFGLSRSFSVEGSGQISTVVAGSIGY 740
Query: 238 MAPEYASSGKLTEKSDVFSFGVMLLELLTGKRPLDLTNAMDESLVDWARPLLSRALEEDG 297
+ PEY S+ ++ EKSDV+S GV+LLE++TG+ + + + D R +L+ +G
Sbjct: 741 LDPEYYSTRQMNEKSDVYSLGVVLLEVITGQPAIASSKTEKVHISDHVRSILA-----NG 795
Query: 298 NFAELVDPFLEGNYDHQEMIRLAACAASSIRHSAKKRSKMSQIVRALE 345
+ +VD L YD +++ A + H++ +R MSQ+V L+
Sbjct: 796 DIRGIVDQRLRERYDVGSAWKMSEIALACTEHTSAQRPTMSQVVMELK 843
>CLV1_ARATH (Q9SYQ8) Receptor protein kinase CLAVATA1 precursor (EC
2.7.1.-)
Length = 980
Score = 211 bits (536), Expect = 3e-54
Identities = 114/276 (41%), Positives = 168/276 (60%), Gaps = 8/276 (2%)
Query: 74 ENIIGQGGFGYVHKGILPTGKEIAVKSLKA-GSGQGEREFQAEIDIISRVHHRHLVSLVG 132
ENIIG+GG G V++G +P ++A+K L G+G+ + F AEI + R+ HRH+V L+G
Sbjct: 695 ENIIGKGGAGIVYRGSMPNNVDVAIKRLVGRGTGRSDHGFTAEIQTLGRIRHRHIVRLLG 754
Query: 133 YCVSGGQRMLVYEFVPNKTLEYHLHGKGVPTMDWPTRMRIALGSARGLAYLHEDCSPRII 192
Y + +L+YE++PN +L LHG + W TR R+A+ +A+GL YLH DCSP I+
Sbjct: 755 YVANKDTNLLLYEYMPNGSLGELLHGSKGGHLQWETRHRVAVEAAKGLCYLHHDCSPLIL 814
Query: 193 HRDIKAANVLIDDSFEAKVADFGLAKLTTD-TNTHVSTRVMGTFGYMAPEYASSGKLTEK 251
HRD+K+ N+L+D FEA VADFGLAK D + + + G++GY+APEYA + K+ EK
Sbjct: 815 HRDVKSNNILLDSDFEAHVADFGLAKFLVDGAASECMSSIAGSYGYIAPEYAYTLKVDEK 874
Query: 252 SDVFSFGVMLLELLTGKRPL-DLTNAMDESLVDWARPLLSRALE--EDGNFAELVDPFLE 308
SDV+SFGV+LLEL+ GK+P+ + +D +V W R + + +VDP L
Sbjct: 875 SDVYSFGVVLLELIAGKKPVGEFGEGVD--IVRWVRNTEEEITQPSDAAIVVAIVDPRLT 932
Query: 309 GNYDHQEMIRLAACAASSIRHSAKKRSKMSQIVRAL 344
G Y +I + A + A R M ++V L
Sbjct: 933 G-YPLTSVIHVFKIAMMCVEEEAAARPTMREVVHML 967
>SRK6_BRAOL (Q09092) Putative serine/threonine-protein kinase
receptor precursor (EC 2.7.1.37) (S-receptor kinase)
(SRK)
Length = 849
Score = 209 bits (533), Expect = 8e-54
Identities = 117/291 (40%), Positives = 180/291 (61%), Gaps = 17/291 (5%)
Query: 62 EELASATKGFANENIIGQGGFGYVHKGILPTGKEIAVKSLKAGSGQGEREFQAEIDIISR 121
E + AT+ F++ N +GQGGFG V+KG L GKEIAVK L S QG EF E+ +I+R
Sbjct: 519 ETVVKATENFSSCNKLGQGGFGIVYKGRLLDGKEIAVKRLSKTSVQGTDEFMNEVTLIAR 578
Query: 122 VHHRHLVSLVGYCVSGGQRMLVYEFVPNKTLEYHLHGKGVPT-MDWPTRMRIALGSARGL 180
+ H +LV ++G C+ G ++ML+YE++ N +L+ +L GK + ++W R I G ARGL
Sbjct: 579 LQHINLVQVLGCCIEGDEKMLIYEYLENLSLDSYLFGKTRRSKLNWNERFDITNGVARGL 638
Query: 181 AYLHEDCSPRIIHRDIKAANVLIDDSFEAKVADFGLAKLTTDTNTHVST-RVMGTFGYMA 239
YLH+D RIIHRD+K +N+L+D + K++DFG+A++ T +T +V+GT+GYM+
Sbjct: 639 LYLHQDSRFRIIHRDLKVSNILLDKNMIPKISDFGMARIFERDETEANTMKVVGTYGYMS 698
Query: 240 PEYASSGKLTEKSDVFSFGVMLLELLTGKRPLDLTNAMDESLVDWARPLLSRALE--EDG 297
PEYA G +EKSDVFSFGV++LE+++GK+ N +D+ LLS ++G
Sbjct: 699 PEYAMYGIFSEKSDVFSFGVIVLEIVSGKKNRGFYN------LDYENDLLSYVWSRWKEG 752
Query: 298 NFAELVDPFLEGN-------YDHQEMIRLAACAASSIRHSAKKRSKMSQIV 341
E+VDP + + + QE+++ ++ A+ R MS +V
Sbjct: 753 RALEIVDPVIVDSLSSQPSIFQPQEVLKCIQIGLLCVQELAEHRPAMSSVV 803
>KPRO_MAIZE (P17801) Putative receptor protein kinase ZmPK1
precursor (EC 2.7.1.37)
Length = 817
Score = 190 bits (483), Expect = 5e-48
Identities = 117/294 (39%), Positives = 168/294 (56%), Gaps = 13/294 (4%)
Query: 59 FTYEELASATKGFANENIIGQGGFGYVHKGILPTGKEIAVKSLKAGSGQGEREFQAEIDI 118
++Y EL AT+ F E +G+G G V+KG+L + +AVK L+ QG+ FQAE+ +
Sbjct: 524 YSYRELVKATRKFKVE--LGRGESGTVYKGVLEDDRHVAVKKLE-NVRQGKEVFQAELSV 580
Query: 119 ISRVHHRHLVSLVGYCVSGGQRMLVYEFVPNKTLEYHLHGKGVPTM-DWPTRMRIALGSA 177
I R++H +LV + G+C G R+LV E+V N +L L +G + DW R IALG A
Sbjct: 581 IGRINHMNLVRIWGFCSEGSHRLLVSEYVENGSLANILFSEGGNILLDWEGRFNIALGVA 640
Query: 178 RGLAYLHEDCSPRIIHRDIKAANVLIDDSFEAKVADFGLAK-LTTDTNTHVSTRVMGTFG 236
+GLAYLH +C +IH D+K N+L+D +FE K+ DFGL K L +T + V GT G
Sbjct: 641 KGLAYLHHECLEWVIHCDVKPENILLDQAFEPKITDFGLVKLLNRGGSTQNVSHVRGTLG 700
Query: 237 YMAPEYASSGKLTEKSDVFSFGVMLLELLTGKRPLDLTNAMDE--SLVDWARPLLSRALE 294
Y+APE+ SS +T K DV+S+GV+LLELLTG R +L DE S++ +LS LE
Sbjct: 701 YIAPEWVSSLPITAKVDVYSYGVVLLELLTGTRVSELVGGTDEVHSMLRKLVRMLSAKLE 760
Query: 295 EDGNFAELVDPFLEGN----YDHQEMIRLAACAASSIRHSAKKRSKMSQIVRAL 344
G +D +L+ ++ + L A S + KR M V+ L
Sbjct: 761 --GEEQSWIDGYLDSKLNRPVNYVQARTLIKLAVSCLEEDRSKRPTMEHAVQTL 812
>RLK5_ARATH (P47735) Receptor-like protein kinase 5 precursor (EC
2.7.1.37)
Length = 999
Score = 181 bits (458), Expect = 4e-45
Identities = 106/292 (36%), Positives = 170/292 (57%), Gaps = 23/292 (7%)
Query: 74 ENIIGQGGFGYVHKGILPTGKEIAVKSLKAGSGQGERE----------FQAEIDIISRVH 123
+N+IG G G V+K L G+ +AVK L G+ E F AE++ + +
Sbjct: 686 KNVIGFGSSGKVYKVELRGGEVVAVKKLNKSVKGGDDEYSSDSLNRDVFAAEVETLGTIR 745
Query: 124 HRHLVSLVGYCVSGGQRMLVYEFVPNKTLEYHLHG--KGVPTMDWPTRMRIALGSARGLA 181
H+ +V L C SG ++LVYE++PN +L LHG KG + WP R+RIAL +A GL+
Sbjct: 746 HKSIVRLWCCCSSGDCKLLVYEYMPNGSLADVLHGDRKGGVVLGWPERLRIALDAAEGLS 805
Query: 182 YLHEDCSPRIIHRDIKAANVLIDDSFEAKVADFGLAK---LTTDTNTHVSTRVMGTFGYM 238
YLH DC P I+HRD+K++N+L+D + AKVADFG+AK ++ + + G+ GY+
Sbjct: 806 YLHHDCVPPIVHRDVKSSNILLDSDYGAKVADFGIAKVGQMSGSKTPEAMSGIAGSCGYI 865
Query: 239 APEYASSGKLTEKSDVFSFGVMLLELLTGKRPLDLTNAMDESLVDWARPLLSRALEEDGN 298
APEY + ++ EKSD++SFGV+LLEL+TGK+P D + D+ + W + AL++ G
Sbjct: 866 APEYVYTLRVNEKSDIYSFGVVLLELVTGKQPTD-SELGDKDMAKW----VCTALDKCG- 919
Query: 299 FAELVDPFLEGNYDHQ--EMIRLAACAASSIRHSAKKRSKMSQIVRALEGDV 348
++DP L+ + + ++I + S + + K+ +++ + G V
Sbjct: 920 LEPVIDPKLDLKFKEEISKVIHIGLLCTSPLPLNRPSMRKVVIMLQEVSGAV 971
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.316 0.133 0.385
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 47,734,131
Number of Sequences: 164201
Number of extensions: 2125360
Number of successful extensions: 8779
Number of sequences better than 10.0: 1637
Number of HSP's better than 10.0 without gapping: 1208
Number of HSP's successfully gapped in prelim test: 429
Number of HSP's that attempted gapping in prelim test: 5572
Number of HSP's gapped (non-prelim): 1821
length of query: 398
length of database: 59,974,054
effective HSP length: 112
effective length of query: 286
effective length of database: 41,583,542
effective search space: 11892893012
effective search space used: 11892893012
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 67 (30.4 bits)
Medicago: description of AC148484.8


