

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148482.4 + phase: 0
(337 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
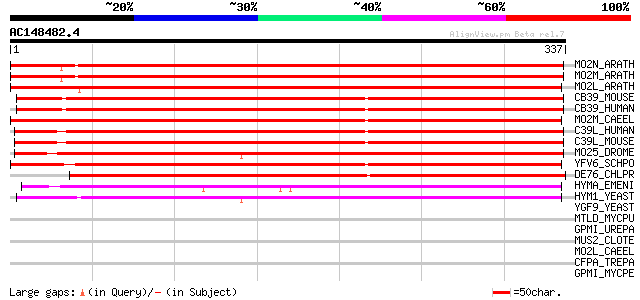
Score E
Sequences producing significant alignments: (bits) Value
MO2N_ARATH (Q9FGK3) Hypothetical MO25-like protein At5g47540 551 e-157
MO2M_ARATH (Q9M0M4) Hypothetical MO25-like protein At4g17270 516 e-146
MO2L_ARATH (Q9ZQ77) Hypothetical MO25-like protein At2g03410 434 e-121
CB39_MOUSE (Q06138) Calcium binding protein 39 (Mo25 protein) 280 3e-75
CB39_HUMAN (Q9Y376) Calcium binding protein 39 (Mo25 protein) (C... 280 3e-75
MO2M_CAEEL (O18211) Hypothetical MO25-like protein Y53C12A.4 in ... 276 4e-74
C39L_HUMAN (Q9H9S4) Calcium binding protein 39-like (Mo25-like p... 271 1e-72
C39L_MOUSE (Q9DB16) Calcium binding protein 39-like (Mo25-like p... 270 3e-72
MO25_DROME (P91891) Mo25 protein (dMo25) 269 6e-72
YFV6_SCHPO (Q9P7Q8) Hypothetical protein C1834.06c in chromosome I 242 1e-63
DE76_CHLPR (Q9XFY6) Degreening related gene dee76 protein 239 7e-63
HYMA_EMENI (O60032) Conidiophore development protein hymA 202 7e-52
HYM1_YEAST (P32464) HYM1 protein 162 1e-39
YGF9_YEAST (P53170) Hypothetical 51.9 kDa protein in PYC1-UBC2 i... 37 0.092
MTLD_MYCPU (Q98PH2) Mannitol-1-phosphate 5-dehydrogenase (EC 1.1... 36 0.12
GPMI_UREPA (Q9PQW1) 2,3-bisphosphoglycerate-independent phosphog... 36 0.16
MUS2_CLOTE (Q891U1) MutS2 protein 34 0.46
MO2L_CAEEL (Q9TZM2) Hypothetical MO25-like protein T27C10.3 in c... 33 0.78
CFPA_TREPA (Q56336) Cytoplasmic filament protein A 33 0.78
GPMI_MYCPE (Q8EW33) 2,3-bisphosphoglycerate-independent phosphog... 33 1.3
>MO2N_ARATH (Q9FGK3) Hypothetical MO25-like protein At5g47540
Length = 343
Score = 551 bits (1421), Expect = e-157
Identities = 282/339 (83%), Positives = 313/339 (92%), Gaps = 4/339 (1%)
Query: 1 MKGLFKPKPRTPTDIVRQTRDLLLFFDRNT---ESRDSKREEKQMTELCKNIRELKSILY 57
MKGLFK KPRTP D+VRQTRDLLLF DR+T + RDSKR+EK M EL +NIR++KSILY
Sbjct: 1 MKGLFKSKPRTPADLVRQTRDLLLFSDRSTSLPDLRDSKRDEK-MAELSRNIRDMKSILY 59
Query: 58 GNSESEPVSEACAQLTQEFFKENTLRLLIKCIPKLNLEARKDATQVVANLQRQPVQSKLI 117
GNSE+EPV+EACAQLTQEFFKE+TLRLLI C+PKLNLE RKDATQVVANLQRQ V S+LI
Sbjct: 60 GNSEAEPVAEACAQLTQEFFKEDTLRLLITCLPKLNLETRKDATQVVANLQRQQVNSRLI 119
Query: 118 ASDYLENNMDLMDILIVGYENTDMALHYGAMLRECIRHQIVAKYVLNSPHMKKFFDYIQL 177
ASDYLE N+DLMD+LI G+ENTDMALHYGAM RECIRHQIVAKYVL S H+KKFFDYIQL
Sbjct: 120 ASDYLEANIDLMDVLIEGFENTDMALHYGAMFRECIRHQIVAKYVLESDHVKKFFDYIQL 179
Query: 178 PNFDIAADAAATFKELMTRHKSTVAEFLSKNYEWFFADYNSKLLESSNYITRRLAVKLLG 237
PNFDIAADAAATFKEL+TRHKSTVAEFL+KN +WFFADYNSKLLESSNYITRR A+KLLG
Sbjct: 180 PNFDIAADAAATFKELLTRHKSTVAEFLTKNEDWFFADYNSKLLESSNYITRRQAIKLLG 239
Query: 238 DMLLDRSNSAVMTQYVSSRENLRILMNLLRESSKSIQIEAFHVFKLFAANQNKPADIVSI 297
D+LLDRSNSAVMT+YVSSR+NLRILMNLLRESSKSIQIEAFHVFKLFAANQNKPADIV+I
Sbjct: 240 DILLDRSNSAVMTKYVSSRDNLRILMNLLRESSKSIQIEAFHVFKLFAANQNKPADIVNI 299
Query: 298 FVANKSKMLRLLDEFKIDKEDEQFEADKAQVMEEIASLE 336
VAN+SK+LRLL + K DKEDE+FEADK+QV+ EIA+LE
Sbjct: 300 LVANRSKLLRLLADLKPDKEDERFEADKSQVLREIAALE 338
>MO2M_ARATH (Q9M0M4) Hypothetical MO25-like protein At4g17270
Length = 343
Score = 516 bits (1329), Expect = e-146
Identities = 264/339 (77%), Positives = 303/339 (88%), Gaps = 4/339 (1%)
Query: 1 MKGLFKPKPRTPTDIVRQTRDLLLFFDRNT---ESRDSKREEKQMTELCKNIRELKSILY 57
M+GLFK KPRTP DIVRQTRDLLL+ DR+ + R+SKREEK M EL K+IR+LK ILY
Sbjct: 1 MRGLFKSKPRTPADIVRQTRDLLLYADRSNSFPDLRESKREEK-MVELSKSIRDLKLILY 59
Query: 58 GNSESEPVSEACAQLTQEFFKENTLRLLIKCIPKLNLEARKDATQVVANLQRQPVQSKLI 117
GNSE+EPV+EACAQLTQEFFK +TLR L+ +P LNLEARKDATQVVANLQRQ V S+LI
Sbjct: 60 GNSEAEPVAEACAQLTQEFFKADTLRRLLTSLPNLNLEARKDATQVVANLQRQQVNSRLI 119
Query: 118 ASDYLENNMDLMDILIVGYENTDMALHYGAMLRECIRHQIVAKYVLNSPHMKKFFDYIQL 177
A+DYLE+N+DLMD L+ G+ENTDMALHYG M RECIRHQIVAKYVL+S H+KKFF YIQL
Sbjct: 120 AADYLESNIDLMDFLVDGFENTDMALHYGTMFRECIRHQIVAKYVLDSEHVKKFFYYIQL 179
Query: 178 PNFDIAADAAATFKELMTRHKSTVAEFLSKNYEWFFADYNSKLLESSNYITRRLAVKLLG 237
PNFDIAADAAATFKEL+TRHKSTVAEFL KN +WFFADYNSKLLES+NYITRR A+KLLG
Sbjct: 180 PNFDIAADAAATFKELLTRHKSTVAEFLIKNEDWFFADYNSKLLESTNYITRRQAIKLLG 239
Query: 238 DMLLDRSNSAVMTQYVSSRENLRILMNLLRESSKSIQIEAFHVFKLFAANQNKPADIVSI 297
D+LLDRSNSAVMT+YVSS +NLRILMNLLRESSK+IQIEAFHVFKLF ANQNKP+DI +I
Sbjct: 240 DILLDRSNSAVMTKYVSSMDNLRILMNLLRESSKTIQIEAFHVFKLFVANQNKPSDIANI 299
Query: 298 FVANKSKMLRLLDEFKIDKEDEQFEADKAQVMEEIASLE 336
VAN++K+LRLL + K DKEDE+F+ADKAQV+ EIA+L+
Sbjct: 300 LVANRNKLLRLLADIKPDKEDERFDADKAQVVREIANLK 338
>MO2L_ARATH (Q9ZQ77) Hypothetical MO25-like protein At2g03410
Length = 348
Score = 434 bits (1116), Expect = e-121
Identities = 213/338 (63%), Positives = 276/338 (81%), Gaps = 3/338 (0%)
Query: 1 MKGLFKPKPRTPTDIVRQTRDLLLFFDRNTESRDSKREEKQ--MTELCKNIRELKSILYG 58
MKGLFK K R P +IVRQTRDL+ + E D++ ++ ELC+NIR+LKSILYG
Sbjct: 1 MKGLFKNKSRLPGEIVRQTRDLIALAESEEEETDARNSKRLGICAELCRNIRDLKSILYG 60
Query: 59 NSESEPVSEACAQLTQEFFKENTLRLLIKCIPKLNLEARKDATQVVANLQRQPVQSKLIA 118
N E+EPV EAC LTQEFF+ +TLR LIK IPKL+LEARKDATQ+VANLQ+Q V+ +L+A
Sbjct: 61 NGEAEPVPEACLLLTQEFFRADTLRPLIKSIPKLDLEARKDATQIVANLQKQQVEFRLVA 120
Query: 119 SDYLENNMDLMDILIVGYENT-DMALHYGAMLRECIRHQIVAKYVLNSPHMKKFFDYIQL 177
S+YLE+N+D++D L+ G ++ ++ALHY ML+EC+RHQ+VAKY+L S +++KFFDY+QL
Sbjct: 121 SEYLESNLDVIDSLVEGIDHDHELALHYTGMLKECVRHQVVAKYILESKNLEKFFDYVQL 180
Query: 178 PNFDIAADAAATFKELMTRHKSTVAEFLSKNYEWFFADYNSKLLESSNYITRRLAVKLLG 237
P FD+A DA+ F+EL+TRHKSTVAE+L+KNYEWFFA+YN+KLLE +Y T+R A KLLG
Sbjct: 181 PYFDVATDASKIFRELLTRHKSTVAEYLAKNYEWFFAEYNTKLLEKGSYFTKRQASKLLG 240
Query: 238 DMLLDRSNSAVMTQYVSSRENLRILMNLLRESSKSIQIEAFHVFKLFAANQNKPADIVSI 297
D+L+DRSNS VM +YVSS +NLRI+MNLLRE +K+IQ+EAFH+FKLF AN+NKP DIV+I
Sbjct: 241 DVLMDRSNSGVMVKYVSSLDNLRIMMNLLREPTKNIQLEAFHIFKLFVANENKPEDIVAI 300
Query: 298 FVANKSKMLRLLDEFKIDKEDEQFEADKAQVMEEIASL 335
VAN++K+LRL + K +KED FE DKA VM EIA+L
Sbjct: 301 LVANRTKILRLFADLKPEKEDVGFETDKALVMNEIATL 338
>CB39_MOUSE (Q06138) Calcium binding protein 39 (Mo25 protein)
Length = 341
Score = 280 bits (717), Expect = 3e-75
Identities = 140/333 (42%), Positives = 223/333 (66%), Gaps = 4/333 (1%)
Query: 5 FKPKPRTPTDIVRQTRDLLLFFDRNTESRDSKREEKQMTELCKNIRELKSILYGNSESEP 64
F ++P DIV+ ++ + ++ S K+ EK E+ KN+ +K ILYG +E EP
Sbjct: 5 FGKSHKSPADIVKNLKESMAVLEKQDIS--DKKAEKATEEVSKNLVAMKEILYGTNEKEP 62
Query: 65 VSEACAQLTQEFFKENTLRLLIKCIPKLNLEARKDATQVVANLQRQPVQSKLIASDYLEN 124
+EA AQL QE + L L+ + ++ E +KD Q+ N+ R+ + ++ +Y+
Sbjct: 63 QTEAVAQLAQELYNSGLLGTLVADLQLIDFEGKKDVAQIFNNILRRQIGTRTPTVEYICT 122
Query: 125 NMDLMDILIVGYENTDMALHYGAMLRECIRHQIVAKYVLNSPHMKKFFDYIQLPNFDIAA 184
+++ +L+ GYE+ ++AL+ G MLRECIRH+ +AK +L S FF Y+++ FDIA+
Sbjct: 123 QQNILFMLLKGYESPEIALNCGIMLRECIRHEPLAKIILWSEQFYDFFRYVEMSTFDIAS 182
Query: 185 DAAATFKELMTRHKSTVAEFLSKNYEWFFADYNSKLLESSNYITRRLAVKLLGDMLLDRS 244
DA ATFK+L+TRHK AEFL ++Y+ FF++Y KLL S NY+T+R ++KLLG++LLDR
Sbjct: 183 DAFATFKDLLTRHKLLSAEFLEQHYDRFFSEY-EKLLHSENYVTKRQSLKLLGELLLDRH 241
Query: 245 NSAVMTQYVSSRENLRILMNLLRESSKSIQIEAFHVFKLFAANQNKPADIVSIFVANKSK 304
N +MT+Y+S ENL+++MNLLR+ S++IQ EAFHVFK+F AN NK I+ I + N++K
Sbjct: 242 NFTIMTKYISKPENLKLMMNLLRDKSRNIQFEAFHVFKVFVANPNKTQPILDILLKNQTK 301
Query: 305 MLRLLDEFKIDK-EDEQFEADKAQVMEEIASLE 336
++ L +F+ D+ EDEQF +K ++++I +L+
Sbjct: 302 LIEFLSKFQNDRTEDEQFNDEKTYLVKQIRNLK 334
>CB39_HUMAN (Q9Y376) Calcium binding protein 39 (Mo25 protein)
(CGI-66)
Length = 341
Score = 280 bits (717), Expect = 3e-75
Identities = 140/333 (42%), Positives = 222/333 (66%), Gaps = 4/333 (1%)
Query: 5 FKPKPRTPTDIVRQTRDLLLFFDRNTESRDSKREEKQMTELCKNIRELKSILYGNSESEP 64
F ++P DIV+ ++ + ++ S K+ EK E+ KN+ +K ILYG +E EP
Sbjct: 5 FGKSHKSPADIVKNLKESMAVLEKQDIS--DKKAEKATEEVSKNLVAMKEILYGTNEKEP 62
Query: 65 VSEACAQLTQEFFKENTLRLLIKCIPKLNLEARKDATQVVANLQRQPVQSKLIASDYLEN 124
+EA AQL QE + L L+ + ++ E +KD Q+ N+ R+ + ++ +Y+
Sbjct: 63 QTEAVAQLAQELYNSGLLSTLVADLQLIDFEGKKDVAQIFNNILRRQIGTRTPTVEYICT 122
Query: 125 NMDLMDILIVGYENTDMALHYGAMLRECIRHQIVAKYVLNSPHMKKFFDYIQLPNFDIAA 184
+++ +L+ GYE+ ++AL+ G MLRECIRH+ +AK +L S FF Y+++ FDIA+
Sbjct: 123 QQNILFMLLKGYESPEIALNCGIMLRECIRHEPLAKIILWSEQFYDFFRYVEMSTFDIAS 182
Query: 185 DAAATFKELMTRHKSTVAEFLSKNYEWFFADYNSKLLESSNYITRRLAVKLLGDMLLDRS 244
DA ATFK+L+TRHK AEFL ++Y+ FF++Y KLL S NY+T+R ++KLLG++LLDR
Sbjct: 183 DAFATFKDLLTRHKLLSAEFLEQHYDRFFSEY-EKLLHSENYVTKRQSLKLLGELLLDRH 241
Query: 245 NSAVMTQYVSSRENLRILMNLLRESSKSIQIEAFHVFKLFAANQNKPADIVSIFVANKSK 304
N +MT+Y+S ENL+++MNLLR+ S++IQ EAFHVFK+F AN NK I+ I + N++K
Sbjct: 242 NFTIMTKYISKPENLKLMMNLLRDKSRNIQFEAFHVFKVFVANPNKTQPILDILLKNQAK 301
Query: 305 MLRLLDEFKIDK-EDEQFEADKAQVMEEIASLE 336
++ L +F+ D+ EDEQF +K ++++I L+
Sbjct: 302 LIEFLSKFQNDRTEDEQFNDEKTYLVKQIRDLK 334
>MO2M_CAEEL (O18211) Hypothetical MO25-like protein Y53C12A.4 in
chromosome II
Length = 338
Score = 276 bits (707), Expect = 4e-74
Identities = 142/336 (42%), Positives = 212/336 (62%), Gaps = 2/336 (0%)
Query: 1 MKGLFKPKPRTPTDIVRQTRDLLLFFDRNTESRDSKREEKQMTELCKNIRELKSILYGNS 60
+K LF +TP D+V+ RD LL DR+ + ++ EK + E K + K+ +YG+
Sbjct: 2 LKPLFGKADKTPADVVKNLRDALLVIDRHGTNTSERKVEKAIEETAKMLALAKTFIYGSD 61
Query: 61 ESEPVSEACAQLTQEFFKENTLRLLIKCIPKLNLEARKDATQVVANLQRQPVQSKLIASD 120
+EP +E QL QE + N L +LIK + K E +KD V NL R+ + ++ +
Sbjct: 62 ANEPNNEQVTQLAQEVYNANVLPMLIKHLHKFEFECKKDVASVFNNLLRRQIGTRSPTVE 121
Query: 121 YLENNMDLMDILIVGYENTDMALHYGAMLRECIRHQIVAKYVLNSPHMKKFFDYIQLPNF 180
YL +++ L++GYE D+AL G+MLRE +RH+ +A+ VL S + ++FF ++Q F
Sbjct: 122 YLAARPEILITLLLGYEQPDIALTCGSMLREAVRHEHLARIVLYSEYFQRFFVFVQSDVF 181
Query: 181 DIAADAAATFKELMTRHKSTVAEFLSKNYEWFFADYNSKLLESSNYITRRLAVKLLGDML 240
DIA DA +TFK+LMT+HK+ AE+L NY+ FF Y S L S NY+TRR ++KLLG++L
Sbjct: 182 DIATDAFSTFKDLMTKHKNMCAEYLDNNYDRFFGQY-SALTNSENYVTRRQSLKLLGELL 240
Query: 241 LDRSNSAVMTQYVSSRENLRILMNLLRESSKSIQIEAFHVFKLFAANQNKPADIVSIFVA 300
LDR N + M +Y++S ENL+ +M LLR+ ++IQ EAFHVFK+F AN NKP I I
Sbjct: 241 LDRHNFSTMNKYITSPENLKTVMELLRDKRRNIQYEAFHVFKIFVANPNKPRPITDILTR 300
Query: 301 NKSKMLRLLDEFKIDK-EDEQFEADKAQVMEEIASL 335
N+ K++ L F D+ DEQF +KA ++++I L
Sbjct: 301 NRDKLVEFLTAFHNDRTNDEQFNDEKAYLIKQIQEL 336
>C39L_HUMAN (Q9H9S4) Calcium binding protein 39-like (Mo25-like
protein) (Antigen MLAA-34)
Length = 337
Score = 271 bits (694), Expect = 1e-72
Identities = 139/334 (41%), Positives = 218/334 (64%), Gaps = 7/334 (2%)
Query: 4 LFKPKPRTPTDIVRQTRDLLLFFDRNTESRDSKREEKQMTELCKNIRELKSILYGNSESE 63
LF + P +IV+ +D L ++ K+ +K E+ K+++ +K IL G +E E
Sbjct: 6 LFSKSHKNPAEIVKILKDNLAILEKQ-----DKKTDKASEEVSKSLQAMKEILCGTNEKE 60
Query: 64 PVSEACAQLTQEFFKENTLRLLIKCIPKLNLEARKDATQVVANLQRQPVQSKLIASDYLE 123
P +EA AQL QE + L LI + ++ E +KD TQ+ N+ R+ + ++ +Y+
Sbjct: 61 PPTEAVAQLAQELYSSGLLVTLIADLQLIDFEGKKDVTQIFNNILRRQIGTRSPTVEYIS 120
Query: 124 NNMDLMDILIVGYENTDMALHYGAMLRECIRHQIVAKYVLNSPHMKKFFDYIQLPNFDIA 183
+ ++ +L+ GYE +AL G MLRECIRH+ +AK +L S + FF Y++L FDIA
Sbjct: 121 AHPHILFMLLKGYEAPQIALRCGIMLRECIRHEPLAKIILFSNQFRDFFKYVELSTFDIA 180
Query: 184 ADAAATFKELMTRHKSTVAEFLSKNYEWFFADYNSKLLESSNYITRRLAVKLLGDMLLDR 243
+DA ATFK+L+TRHK VA+FL +NY+ F DY KLL+S NY+T+R ++KLLG+++LDR
Sbjct: 181 SDAFATFKDLLTRHKVLVADFLEQNYDTIFEDY-EKLLQSENYVTKRQSLKLLGELILDR 239
Query: 244 SNSAVMTQYVSSRENLRILMNLLRESSKSIQIEAFHVFKLFAANQNKPADIVSIFVANKS 303
N A+MT+Y+S ENL+++MNLLR+ S +IQ EAFHVFK+F A+ +K IV I + N+
Sbjct: 240 HNFAIMTKYISKPENLKLMMNLLRDKSPNIQFEAFHVFKVFVASPHKTQPIVEILLKNQP 299
Query: 304 KMLRLLDEFKIDK-EDEQFEADKAQVMEEIASLE 336
K++ L F+ ++ +DEQF +K ++++I L+
Sbjct: 300 KLIEFLSSFQKERTDDEQFADEKNYLIKQIRDLK 333
>C39L_MOUSE (Q9DB16) Calcium binding protein 39-like (Mo25-like
protein)
Length = 337
Score = 270 bits (691), Expect = 3e-72
Identities = 137/334 (41%), Positives = 218/334 (65%), Gaps = 7/334 (2%)
Query: 4 LFKPKPRTPTDIVRQTRDLLLFFDRNTESRDSKREEKQMTELCKNIRELKSILYGNSESE 63
LF + P +IV+ +D L ++ K+ +K E+ K+++ +K IL G ++ E
Sbjct: 6 LFSKSHKNPAEIVKILKDNLAILEKQ-----DKKTDKASEEVSKSLQAMKEILCGTNDKE 60
Query: 64 PVSEACAQLTQEFFKENTLRLLIKCIPKLNLEARKDATQVVANLQRQPVQSKLIASDYLE 123
P +EA AQL QE + L LI + ++ E +KD TQ+ N+ R+ + ++ +Y+
Sbjct: 61 PPTEAVAQLAQELYSSGLLVTLIADLQLIDFEGKKDVTQIFNNILRRQIGTRCPTVEYIS 120
Query: 124 NNMDLMDILIVGYENTDMALHYGAMLRECIRHQIVAKYVLNSPHMKKFFDYIQLPNFDIA 183
++ ++ +L+ GYE +AL G MLRECIRH+ +AK +L S + FF Y++L FDIA
Sbjct: 121 SHPHILFMLLKGYEAPQIALRCGIMLRECIRHEPLAKIILFSNQFRDFFKYVELSTFDIA 180
Query: 184 ADAAATFKELMTRHKSTVAEFLSKNYEWFFADYNSKLLESSNYITRRLAVKLLGDMLLDR 243
+DA ATFK+L+TRHK VA+FL +NY+ F DY KLL+S NY+T+R ++KLLG+++LDR
Sbjct: 181 SDAFATFKDLLTRHKVLVADFLEQNYDTIFEDY-EKLLQSENYVTKRQSLKLLGELILDR 239
Query: 244 SNSAVMTQYVSSRENLRILMNLLRESSKSIQIEAFHVFKLFAANQNKPADIVSIFVANKS 303
N +MT+Y+S ENL+++MNLLR+ S +IQ EAFHVFK+F A+ +K IV I + N+
Sbjct: 240 HNFTIMTKYISKPENLKLMMNLLRDKSPNIQFEAFHVFKVFVASPHKTQPIVEILLKNQP 299
Query: 304 KMLRLLDEFKIDK-EDEQFEADKAQVMEEIASLE 336
K++ L F+ ++ +DEQF +K ++++I L+
Sbjct: 300 KLIEFLSSFQKERTDDEQFADEKNYLIKQIRDLK 333
>MO25_DROME (P91891) Mo25 protein (dMo25)
Length = 339
Score = 269 bits (688), Expect = 6e-72
Identities = 136/337 (40%), Positives = 224/337 (66%), Gaps = 9/337 (2%)
Query: 4 LFKPKPRTPTDIVRQTRDLLLFFDRNTESRDSKREEKQMTELCKNIRELKSILYGNSESE 63
LF ++P ++V+ ++ + N ++ EK ++ KN+ +K++LYG+S++E
Sbjct: 3 LFGKSQKSPVELVKSLKEAI-----NALEAGDRKVEKAQEDVSKNLVSIKNMLYGSSDAE 57
Query: 64 PVSE-ACAQLTQEFFKENTLRLLIKCIPKLNLEARKDATQVVANLQRQPVQSKLIASDYL 122
P ++ AQL+QE + N L LLI+ + +++ E +K + N+ R+ + ++ +Y+
Sbjct: 58 PPADYVVAQLSQELYNSNLLLLLIQNLHRIDFEGKKHVALIFNNVLRRQIGTRSPTVEYI 117
Query: 123 ENNMDLMDILIVGYENT--DMALHYGAMLRECIRHQIVAKYVLNSPHMKKFFDYIQLPNF 180
+++ L+ GYE+ ++AL+ G MLREC R++ +AK +L+S KFF Y+++ F
Sbjct: 118 CTKPEILFTLMAGYEDAHPEIALNSGTMLRECARYEALAKIMLHSDEFFKFFRYVEVSTF 177
Query: 181 DIAADAAATFKELMTRHKSTVAEFLSKNYEWFFADYNSKLLESSNYITRRLAVKLLGDML 240
DIA+DA +TFKEL+TRHK AEFL NY+ FF+ + +LL S NY+TRR ++KLLG++L
Sbjct: 178 DIASDAFSTFKELLTRHKLLCAEFLDANYDKFFSQHYQRLLNSENYVTRRQSLKLLGELL 237
Query: 241 LDRSNSAVMTQYVSSRENLRILMNLLRESSKSIQIEAFHVFKLFAANQNKPADIVSIFVA 300
LDR N VMT+Y+S ENL+++MN+L+E S++IQ EAFHVFK+F AN NKP I+ I +
Sbjct: 238 LDRHNFTVMTRYISEPENLKLMMNMLKEKSRNIQFEAFHVFKVFVANPNKPKPILDILLR 297
Query: 301 NKSKMLRLLDEFKIDK-EDEQFEADKAQVMEEIASLE 336
N++K++ L F D+ EDEQF +KA ++++I L+
Sbjct: 298 NQTKLVDFLTNFHTDRSEDEQFNDEKAYLIKQIKELK 334
>YFV6_SCHPO (Q9P7Q8) Hypothetical protein C1834.06c in chromosome I
Length = 329
Score = 242 bits (617), Expect = 1e-63
Identities = 132/336 (39%), Positives = 211/336 (62%), Gaps = 8/336 (2%)
Query: 1 MKGLFKPKPRTPTDIVRQTRDLLLFFDRNTESRDSKREEKQMTELCKNIRELKSILYGNS 60
M LF +P++ D+VR D L + N + + K E+ K ++ L+ L G +
Sbjct: 1 MSFLFNKRPKSTQDVVRCLCDNLPKLEINNDKK------KSFEEVSKCLQNLRVSLCGTA 54
Query: 61 ESEPVSEACAQLTQEFFKENTLRLLIKCIPKLNLEARKDATQVVANLQRQPVQSKLIASD 120
E EP ++ + L+ + ++ N LL++ +PKL E++KD + + L R+ V S+ D
Sbjct: 55 EVEPDADLVSDLSFQIYQSNLPFLLVRYLPKLEFESKKDTGLIFSALLRRHVASRYPTVD 114
Query: 121 YLENNMDLMDILIVGYENTDMALHYGAMLRECIRHQIVAKYVLNSPHMKKFFDYIQLPNF 180
Y+ + + +L+ Y ++A G++LREC RH+ + + +LNS FF IQ +F
Sbjct: 115 YMLAHPQIFPVLVSYYRYQEVAFTAGSILRECSRHEALNEVLLNSRDFWTFFSLIQASSF 174
Query: 181 DIAADAAATFKELMTRHKSTVAEFLSKNYEWFFADYNSKLLESSNYITRRLAVKLLGDML 240
D+A+DA +TFK ++ HKS VAEF+S +++ FF Y + LL+S NY+T+R ++KLLG++L
Sbjct: 175 DMASDAFSTFKSILLNHKSQVAEFISYHFDEFFKQY-TVLLKSENYVTKRQSLKLLGEIL 233
Query: 241 LDRSNSAVMTQYVSSRENLRILMNLLRESSKSIQIEAFHVFKLFAANQNKPADIVSIFVA 300
L+R+N +VMT+Y+SS ENL+++M LLR+ SK+IQ EAFHVFKLF AN K +++ I
Sbjct: 234 LNRANRSVMTRYISSAENLKLMMILLRDKSKNIQFEAFHVFKLFVANPEKSEEVIEILRR 293
Query: 301 NKSKMLRLLDEFKID-KEDEQFEADKAQVMEEIASL 335
NKSK++ L F D K DEQF ++A V+++I L
Sbjct: 294 NKSKLISYLSAFHTDRKNDEQFNDERAFVIKQIERL 329
>DE76_CHLPR (Q9XFY6) Degreening related gene dee76 protein
Length = 321
Score = 239 bits (610), Expect = 7e-63
Identities = 123/302 (40%), Positives = 194/302 (63%), Gaps = 2/302 (0%)
Query: 37 REEKQMTELCKNIRELKSILYGNSESEPVSEACAQLTQEFFKENTLRLLIKCIPKLNLEA 96
++++ + ++ K I +K ++G E E + E + + L+ + L+ E
Sbjct: 21 KQDRVVEDISKAIMSIKEAIFGEDEQSSSKEHAQGIASEACRVGLVSDLVTYLTVLDFET 80
Query: 97 RKDATQVVANLQRQPVQSK-LIASDYLENNMDLMDILIVGYENTDMALHYGAMLRECIRH 155
RKD Q+ + R ++ DY+ + D++ L GYE+ ++AL+ G M RECIRH
Sbjct: 81 RKDVVQIFCAIIRITLEDGGRPGRDYVLAHPDVLSTLFYGYEDPEIALNCGQMFRECIRH 140
Query: 156 QIVAKYVLNSPHMKKFFDYIQLPNFDIAADAAATFKELMTRHKSTVAEFLSKNYEWFFAD 215
+ +AK+VL ++ F+ + + +F++A+DA ATFK+L+TRHK VA FL +NYE FF+
Sbjct: 141 EDIAKFVLECNLFEELFEKLNVQSFEVASDAFATFKDLLTRHKQLVAAFLQENYEDFFSQ 200
Query: 216 YNSKLLESSNYITRRLAVKLLGDMLLDRSNSAVMTQYVSSRENLRILMNLLRESSKSIQI 275
+ KLL S NY+TRR ++KLLG++LLDR N +M QYVS NL ++MNLL++SS+SIQ
Sbjct: 201 LD-KLLTSDNYVTRRQSLKLLGELLLDRVNVKIMMQYVSDVNNLILMMNLLKDSSRSIQF 259
Query: 276 EAFHVFKLFAANQNKPADIVSIFVANKSKMLRLLDEFKIDKEDEQFEADKAQVMEEIASL 335
EAFHVFK+F AN NK + I V NK+K+L L++F D++DEQF+ +KA +++EI+ +
Sbjct: 260 EAFHVFKVFVANPNKTKPVADILVNNKNKLLTYLEDFHNDRDDEQFKEEKAVIIKEISMM 319
Query: 336 EA 337
A
Sbjct: 320 HA 321
>HYMA_EMENI (O60032) Conidiophore development protein hymA
Length = 384
Score = 202 bits (515), Expect = 7e-52
Identities = 119/357 (33%), Positives = 199/357 (55%), Gaps = 35/357 (9%)
Query: 8 KPRTPTDIVRQTRDLLLFFDRNTESRDSKREEKQMTELCKNIRELKSILYGNSESEPVSE 67
+ R P+D+VR +DLLL R+ K EL K + ++K ++ G E E ++
Sbjct: 9 RSRQPSDVVRSIKDLLL------RLREPSTASKVEDELAKQLSQMKLMVQGTQELEASTD 62
Query: 68 ACAQLTQEFFKENTLRLLIKCIPKLNLEARKDATQVVANLQR-QPVQSKL----IASDYL 122
L Q E+ L L + L EARKD + +++ R +P + S +
Sbjct: 63 QVHALVQAMLHEDLLYELAVALHNLPFEARKDTQTIFSHILRFKPPHGNSPDPPVISYIV 122
Query: 123 ENNMDLMDILIVGYENTDMALHYGAMLRECIRHQIVAKYVL------------------N 164
N +++ L GYE++ A+ G +LRE ++ ++A +L N
Sbjct: 123 HNRPEIIIELCRGYEHSQSAMPCGTILREALKFDVIAAIILYDQSKEGEPAIRLTEVQPN 182
Query: 165 SPHMK-----KFFDYIQLPNFDIAADAAATFKELMTRHKSTVAEFLSKNYEWFFADYNSK 219
P +FF +I F+++ADA TF+E++TRHKS V +L+ N+++FFA +N+
Sbjct: 183 VPQRGTGVFWRFFHWIDRGTFELSADAFTTFREILTRHKSLVTGYLATNFDYFFAQFNTF 242
Query: 220 LLESSNYITRRLAVKLLGDMLLDRSNSAVMTQYVSSRENLRILMNLLRESSKSIQIEAFH 279
L++S +Y+T+R ++KLLG++LLDR+N +VM +YV S ENL++ M LLR+ K +Q E FH
Sbjct: 243 LVQSESYVTKRQSIKLLGEILLDRANYSVMMRYVESGENLKLCMKLLRDDRKMVQYEGFH 302
Query: 280 VFKLFAANQNKPADIVSIFVANKSKMLRLLDEFKIDK-EDEQFEADKAQVMEEIASL 335
VFK+F AN +K + I + N+ ++LR L +F D+ +D+QF +K+ ++ +I L
Sbjct: 303 VFKVFVANPDKSVAVQRILINNRDRLLRFLPKFLEDRTDDDQFTDEKSFLVRQIELL 359
>HYM1_YEAST (P32464) HYM1 protein
Length = 399
Score = 162 bits (410), Expect = 1e-39
Identities = 97/340 (28%), Positives = 178/340 (51%), Gaps = 11/340 (3%)
Query: 5 FKPKPRTPTDIVRQTRDLLLFFDRNTESRDSKREEKQMTELCKNIRELKSILYGNSESEP 64
+K P+TP+D R + L F + ++D+KR+ ++ E K + K + G+++ P
Sbjct: 16 WKKNPKTPSDYARLIIEQLNKFSSPSLTQDNKRKVQE--ECTKYLIGTKHFIVGDTDPHP 73
Query: 65 VSEACAQLTQEFFKENTLRLLIKCIPKLNLEARKDATQVVANLQRQPVQSKLIASDYLEN 124
EA +L + + L+ L EAR++ + + +K + DYL +
Sbjct: 74 TPEAIDELYTAMHRADVFYELLLHFVDLEFEARRECMLIFSICLGYSKDNKFVTVDYLVS 133
Query: 125 NMDLMDILIVGYENT-------DMALHYGAMLRECIRHQIVAKYVLNSPHMKKFFDYIQL 177
+ +++ E D+ L G M+ ECI+++ + + +L P + KFF++ +L
Sbjct: 134 QPKTISLMLRTAEVALQQKGCQDIFLTVGNMIIECIKYEQLCRIILKDPQLWKFFEFAKL 193
Query: 178 PNFDIAADAAATFKELMTRHKSTVA-EFLSKNYEWF-FADYNSKLLESSNYITRRLAVKL 235
NF+I+ ++ T H V+ EF S F +KL+ +Y+T+R + KL
Sbjct: 194 GNFEISTESLQILSAAFTAHPKLVSKEFFSNEINIIRFIKCINKLMAHGSYVTKRQSTKL 253
Query: 236 LGDMLLDRSNSAVMTQYVSSRENLRILMNLLRESSKSIQIEAFHVFKLFAANQNKPADIV 295
L +++ RSN+A+M Y++S ENL+++M L+ + SK++Q+EAF+VFK+ AN K +
Sbjct: 254 LASLIVIRSNNALMNIYINSPENLKLIMTLMTDKSKNLQLEAFNVFKVMVANPRKSKPVF 313
Query: 296 SIFVANKSKMLRLLDEFKIDKEDEQFEADKAQVMEEIASL 335
I V N+ K+L F +D +D F ++ +++EI SL
Sbjct: 314 DILVKNRDKLLTYFKTFGLDSQDSTFLDEREFIVQEIDSL 353
>YGF9_YEAST (P53170) Hypothetical 51.9 kDa protein in PYC1-UBC2
intergenic region
Length = 445
Score = 36.6 bits (83), Expect = 0.092
Identities = 38/137 (27%), Positives = 58/137 (41%), Gaps = 16/137 (11%)
Query: 92 LNLEARKDATQVVANLQRQPVQSKLIASDYLENNMDLMDILIVGYENTDMALHYGAMLRE 151
L+L R+ Q++++L R + +LI +++ + G E + L G + +E
Sbjct: 193 LDLYPREKMDQLLSDLLRARISRRLIVEEHVVYTANYTS----GKEENTLVL--GDIFQE 246
Query: 152 CIRHQIVAKYVLN-SPHMKKF-----FDYIQLPNFDIAADAAATFKELMTRHKSTVAEFL 205
C KY+L S +KF F I +P F I D +F L T K + E L
Sbjct: 247 CS----AKKYLLEASEESQKFIQDMYFKDIPMPEFIIEGDTQLSFYFLPTHLKYLLGEIL 302
Query: 206 SKNYEWFFADYNSKLLE 222
YE Y K LE
Sbjct: 303 RNTYEATMKHYIRKGLE 319
>MTLD_MYCPU (Q98PH2) Mannitol-1-phosphate 5-dehydrogenase (EC
1.1.1.17)
Length = 360
Score = 36.2 bits (82), Expect = 0.12
Identities = 29/122 (23%), Positives = 57/122 (45%), Gaps = 7/122 (5%)
Query: 201 VAEFLSKNYEWFFADYNSKLLESSNYITRRLAVKLLGDMLLDRSNSAVMTQYVSSRENLR 260
+ E LSK Y F DY + LE + +R ++K D++ + + + Q +S E
Sbjct: 235 INEILSKEYLLFKVDYLNDYLEKN---LKRFSIKENQDLISRVARNPI--QKLSKNERYF 289
Query: 261 ILMNLLRESSKSIQI--EAFHVFKLFAANQNKPADIVSIFVANKSKMLRLLDEFKIDKED 318
++ NL+++ + I I E + + +K + + + NKS L +D+ED
Sbjct: 290 LIYNLVKKHNLEIDILLEIYKSIFYYDNKMDKESSKIQSTIENKSLAYALKKFSNLDQED 349
Query: 319 EQ 320
++
Sbjct: 350 QE 351
>GPMI_UREPA (Q9PQW1) 2,3-bisphosphoglycerate-independent
phosphoglycerate mutase (EC 5.4.2.1)
(Phosphoglyceromutase) (BPG-independent PGAM) (iPGM)
Length = 502
Score = 35.8 bits (81), Expect = 0.16
Identities = 39/157 (24%), Positives = 65/157 (40%), Gaps = 14/157 (8%)
Query: 101 TQVVANLQRQPVQSK-LIASDYLENNMDLMDILIVGYENTDMALHYGAMLRECIRHQIVA 159
+Q VA P S LI +++ D D+ IV Y N DM H G M Q
Sbjct: 350 SQKVATYDLCPQMSAHLITKTIIDHYFD-HDVFIVNYANPDMVGHSGNM-------QQTI 401
Query: 160 KYVLNSPH-MKKFFDYIQLPNFDIAADAAATFKELMTRHKSTVAEFLSKNYEWFFADYNS 218
+ +L+ H ++K +D+ + N + E+M + V S N WF N
Sbjct: 402 EAILSVDHEIRKLYDFFEKNNGVLMITGDHGNAEMMLDDQKQVITSHSVNDVWFIITDNK 461
Query: 219 KLLESSNYITRRLAVKLLGDMLLDRSNSAVMTQYVSS 255
+L S T++ ++ + +L+ N A Q ++
Sbjct: 462 VILNS----TQKFSLANIAPTILEYLNIAQPIQMTAT 494
>MUS2_CLOTE (Q891U1) MutS2 protein
Length = 786
Score = 34.3 bits (77), Expect = 0.46
Identities = 26/94 (27%), Positives = 45/94 (47%), Gaps = 5/94 (5%)
Query: 6 KPKPRTPTDIVRQTRDLLLFFDRNTESRDSKREEKQMTELCKNIRELKSILYGNSESEPV 65
K K IV +TR+ LL + + K +++ E+ KNIREL+ + Y ++ + +
Sbjct: 548 KDKYEDKLKIVEETREKLLKGAQEEAKKLIKEAKEEADEILKNIRELEKMGYSSTARQKL 607
Query: 66 SEACAQLTQEFFK-----ENTLRLLIKCIPKLNL 94
E +L + K EN + K I ++NL
Sbjct: 608 EEERKKLNSKIHKLEEKEENLNKEKGKKIKEINL 641
>MO2L_CAEEL (Q9TZM2) Hypothetical MO25-like protein T27C10.3 in
chromosome I
Length = 339
Score = 33.5 bits (75), Expect = 0.78
Identities = 17/128 (13%), Positives = 57/128 (44%), Gaps = 1/128 (0%)
Query: 179 NFDIAADAAATFKELMTRHKSTVAEFLSKNYEWFFADYNSKLLESSNYITRRLAVKLLGD 238
+FD+ T + + + + F+ N F + KL+ SN+ + + K L +
Sbjct: 108 DFDVIQGTFDTLQIIFFTNHESANNFIKNNLPRFMQTLH-KLIACSNFFIQAKSFKFLNE 166
Query: 239 MLLDRSNSAVMTQYVSSRENLRILMNLLRESSKSIQIEAFHVFKLFAANQNKPADIVSIF 298
+ ++N + +++ +++++ ++ + +++ A + ++F N ++
Sbjct: 167 LFTAQTNYETRSLWMAEPAFIKLVVLAIQSNKHAVRSRAVSILEIFIRNPRNSPEVHEFI 226
Query: 299 VANKSKML 306
N++ ++
Sbjct: 227 GRNRNVLI 234
>CFPA_TREPA (Q56336) Cytoplasmic filament protein A
Length = 677
Score = 33.5 bits (75), Expect = 0.78
Identities = 43/185 (23%), Positives = 80/185 (43%), Gaps = 27/185 (14%)
Query: 25 FFDRNTESRDSKREEKQMTELCKNIRELKSILYGNSESEPVSEACAQLTQEFFKENTLRL 84
F D + E+ DSKR E EL + I L+ A++ E F + +R
Sbjct: 278 FTDDDCENPDSKRYELISRELMERISNLR----------------AEIDPETFDQLNVRE 321
Query: 85 LIKCIPKLNLEARKDATQVVANLQRQPVQSKLIASDYLENNMDLMDILIVGYENTDMALH 144
IK I L R N + + + Y+EN + ++++ Y++TD++
Sbjct: 322 NIKKIVDLE-NIRNRGFNTAINSITSILDTSRMGYQYIENFKNARELILREYDDTDISNL 380
Query: 145 YGAMLRECIRHQIVAKYVLNSPHMKKFFDY-IQLPNFDIAADAAATFKELMTRH-KSTVA 202
R+Q+ KY+ N+ +++ Y + L +F+ D + L T++ KS +
Sbjct: 381 PDE------RYQLRLKYLDNAQLIEERKGYEVMLRSFETEVD--HLWDVLRTKYDKSKAS 432
Query: 203 EFLSK 207
F++K
Sbjct: 433 RFMAK 437
>GPMI_MYCPE (Q8EW33) 2,3-bisphosphoglycerate-independent
phosphoglycerate mutase (EC 5.4.2.1)
(Phosphoglyceromutase) (BPG-independent PGAM) (iPGM)
Length = 502
Score = 32.7 bits (73), Expect = 1.3
Identities = 18/71 (25%), Positives = 35/71 (48%), Gaps = 5/71 (7%)
Query: 76 FFKENTLRLLIKCIPKLNLEARKDATQVVANLQRQPVQSKLIASDYLENNMDLMDILIVG 135
FF + + + +K K+ ++++K V P S + +D L N+D D++++
Sbjct: 335 FFFDGGVEVDLKNESKILVDSKK-----VKTYDEVPAMSAVEITDKLIENLDKFDVIVLN 389
Query: 136 YENTDMALHYG 146
+ N DM H G
Sbjct: 390 FANADMVGHTG 400
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.320 0.133 0.364
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 34,067,074
Number of Sequences: 164201
Number of extensions: 1285750
Number of successful extensions: 4353
Number of sequences better than 10.0: 36
Number of HSP's better than 10.0 without gapping: 16
Number of HSP's successfully gapped in prelim test: 20
Number of HSP's that attempted gapping in prelim test: 4309
Number of HSP's gapped (non-prelim): 37
length of query: 337
length of database: 59,974,054
effective HSP length: 111
effective length of query: 226
effective length of database: 41,747,743
effective search space: 9434989918
effective search space used: 9434989918
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 66 (30.0 bits)
Medicago: description of AC148482.4


