

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148481.8 + phase: 0 /pseudo
(339 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
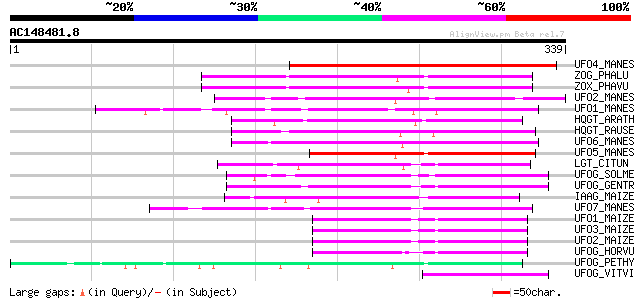
Score E
Sequences producing significant alignments: (bits) Value
UFO4_MANES (Q40286) Flavonol 3-O-glucosyltransferase 4 (EC 2.4.1... 138 2e-32
ZOG_PHALU (Q9ZSK5) Zeatin O-glucosyltransferase (EC 2.4.1.203) (... 135 2e-31
ZOX_PHAVU (P56725) Zeatin O-xylosyltransferase (EC 2.4.2.40) (Ze... 132 2e-30
UFO2_MANES (Q40285) Flavonol 3-O-glucosyltransferase 2 (EC 2.4.1... 129 1e-29
UFO1_MANES (Q40284) Flavonol 3-O-glucosyltransferase 1 (EC 2.4.1... 127 3e-29
HQGT_ARATH (Q9M156) Probable hydroquinone glucosyltransferase (E... 125 2e-28
HQGT_RAUSE (Q9AR73) Hydroquinone glucosyltransferase (EC 2.4.1.2... 124 4e-28
UFO6_MANES (Q40288) Flavonol 3-O-glucosyltransferase 6 (EC 2.4.1... 123 8e-28
UFO5_MANES (Q40287) Flavonol 3-O-glucosyltransferase 5 (EC 2.4.1... 122 1e-27
LGT_CITUN (Q9MB73) Limonoid UDP-glucosyltransferase (EC 2.4.1.21... 120 6e-27
UFOG_SOLME (Q43641) Flavonol 3-O-glucosyltransferase (EC 2.4.1.9... 109 1e-23
UFOG_GENTR (Q96493) Flavonol 3-O-glucosyltransferase (EC 2.4.1.9... 102 1e-21
IAAG_MAIZE (Q41819) Indole-3-acetate beta-glucosyltransferase (E... 100 4e-21
UFO7_MANES (Q40289) Flavonol 3-O-glucosyltransferase 7 (EC 2.4.1... 92 2e-18
UFO1_MAIZE (P16166) Flavonol 3-O-glucosyltransferase (EC 2.4.1.9... 89 1e-17
UFO3_MAIZE (P16167) Flavonol 3-O-glucosyltransferase (EC 2.4.1.9... 86 1e-16
UFO2_MAIZE (P16165) Flavonol 3-O-glucosyltransferase (EC 2.4.1.9... 84 5e-16
UFOG_HORVU (P14726) Flavonol 3-O-glucosyltransferase (EC 2.4.1.9... 80 1e-14
UFOG_PETHY (Q43716) Flavonol 3-O-glucosyltransferase (EC 2.4.1.9... 74 7e-13
UFOG_VITVI (P51094) Flavonol 3-O-glucosyltransferase (EC 2.4.1.9... 57 7e-08
>UFO4_MANES (Q40286) Flavonol 3-O-glucosyltransferase 4 (EC
2.4.1.91) (UDP-glucose flavonoid 3-O-glucosyltransferase
4) (Fragment)
Length = 241
Score = 138 bits (347), Expect = 2e-32
Identities = 72/165 (43%), Positives = 102/165 (61%), Gaps = 2/165 (1%)
Query: 172 KKEQNEDVIVES-ELLNWLNSKPNDSVLYVSFGSLTRLSHSQIVEIAHGLENSGHNFIWV 230
K E+ + V++ ELL WL+ SV+Y GS++ L+ Q+ E+ GLE++ FIWV
Sbjct: 8 KAERGDKASVDNTELLKWLDLWEPGSVIYACLGSISGLTSWQLAELGLGLESTNQPFIWV 67
Query: 231 VRKKD-GEGDEDGFLDDFKQRMKENKKGYIIWNWAPQLLILGHPATAGVVTHCGWNSILE 289
+R+ + EG E L++ + K ++ + I W+PQ+LIL HPA THCGWNS LE
Sbjct: 68 IREGEKSEGLEKWILEEGYEERKRKREDFWIRGWSPQVLILSHPAIGAFFTHCGWNSTLE 127
Query: 290 SLSVSLPIITWPMFAEQFYNEKLLVFVLKIVVSVGSKVNTFWSNE 334
+S +PI+ P+FAEQFYNEKL+V VL I VSVG + W E
Sbjct: 128 GISAGVPIVACPLFAEQFYNEKLVVEVLGIGVSVGVEAAVTWGLE 172
>ZOG_PHALU (Q9ZSK5) Zeatin O-glucosyltransferase (EC 2.4.1.203)
(Trans-zeatin O-beta-D-glucosyltransferase)
Length = 459
Score = 135 bits (339), Expect = 2e-31
Identities = 76/210 (36%), Positives = 117/210 (55%), Gaps = 11/210 (5%)
Query: 118 SEFSDYFDAVYESEGKSYGTLCNSFHELEGDYENLYKS-TMGIKAWSVGPVSAWLKKEQN 176
++F + A YE + G + N+ +EG Y L + G K W++GP + L E+
Sbjct: 184 AQFKGFRTAQYEFRKFNNGDIYNTSRVIEGPYVELLELFNGGKKVWALGPFNP-LAVEKK 242
Query: 177 EDVIVESELLNWLNSKPNDSVLYVSFGSLTRLSHSQIVEIAHGLENSGHNFIWVVRKKD- 235
+ + + WL+ + SV+Y+SFG+ T L QI +IA GLE S FIWV+R+ D
Sbjct: 243 DSIGFRHPCMEWLDKQEPSSVIYISFGTTTALRDEQIQQIATGLEQSKQKFIWVLREADK 302
Query: 236 ------GEGDEDGFLDDFKQRMKENKKGYIIWNWAPQLLILGHPATAGVVTHCGWNSILE 289
E F++R++ G ++ +WAPQL IL H +T G ++HCGWNS LE
Sbjct: 303 GDIFAGSEAKRYELPKGFEERVEG--MGLVVRDWAPQLEILSHSSTGGFMSHCGWNSCLE 360
Query: 290 SLSVSLPIITWPMFAEQFYNEKLLVFVLKI 319
S+++ +PI TWPM ++Q N L+ VLK+
Sbjct: 361 SITMGVPIATWPMHSDQPRNAVLVTEVLKV 390
>ZOX_PHAVU (P56725) Zeatin O-xylosyltransferase (EC 2.4.2.40)
(Zeatin O-beta-D-xylosyltransferase)
Length = 454
Score = 132 bits (331), Expect = 2e-30
Identities = 76/210 (36%), Positives = 118/210 (56%), Gaps = 11/210 (5%)
Query: 118 SEFSDYFDAVYESEGKSYGTLCNSFHELEGDY-ENLYKSTMGIKAWSVGPVSAWLKKEQN 176
++F+D+ A E + G + N+ +EG Y E L + G + W++GP + L E+
Sbjct: 179 AQFTDFLTAQNEFRKFNNGDIYNTSRVIEGPYVELLERFNGGKEVWALGPFTP-LAVEKK 237
Query: 177 EDVIVESELLNWLNSKPNDSVLYVSFGSLTRLSHSQIVEIAHGLENSGHNFIWVVRKKDG 236
+ + + WL+ + SV+YVSFG+ T L QI E+A GLE S FIWV+R D
Sbjct: 238 DSIGFSHPCMEWLDKQEPSSVIYVSFGTTTALRDEQIQELATGLEQSKQKFIWVLRDADK 297
Query: 237 EGDEDG-------FLDDFKQRMKENKKGYIIWNWAPQLLILGHPATAGVVTHCGWNSILE 289
DG + F++R++ G ++ +WAPQ+ IL H +T G ++HCGWNS LE
Sbjct: 298 GDIFDGSEAKRYELPEGFEERVEG--MGLVVRDWAPQMEILSHSSTGGFMSHCGWNSCLE 355
Query: 290 SLSVSLPIITWPMFAEQFYNEKLLVFVLKI 319
SL+ +P+ TW M ++Q N L+ VLK+
Sbjct: 356 SLTRGVPMATWAMHSDQPRNAVLVTDVLKV 385
>UFO2_MANES (Q40285) Flavonol 3-O-glucosyltransferase 2 (EC
2.4.1.91) (UDP-glucose flavonoid 3-O-glucosyltransferase
2) (Fragment)
Length = 346
Score = 129 bits (323), Expect = 1e-29
Identities = 75/224 (33%), Positives = 117/224 (51%), Gaps = 24/224 (10%)
Query: 126 AVYESEGKSYGTLCNSFHELEGDYENLYKSTMGIKAWSVGPVSAWLKKEQNEDVIVESEL 185
A+ + ++ G + N+F ELE +K + VGP+ +N E+
Sbjct: 83 AIAKKFRQAKGIIVNTFLELESRAIESFKVP---PLYHVGPILDVKSDGRN----THPEI 135
Query: 186 LNWLNSKPNDSVLYVSFGSLTRLSHSQIVEIAHGLENSGHNFIWVVRKK----------D 235
+ WL+ +P SV+++ FGS+ S Q+ EIA+ LENSGH F+W +R+ D
Sbjct: 136 MQWLDDQPEGSVVFLCFGSMGSFSEDQLKEIAYALENSGHRFLWSIRRPPPPDKIASPTD 195
Query: 236 GEGDEDGFLDDFKQRMKENKKGYIIWNWAPQLLILGHPATAGVVTHCGWNSILESLSVSL 295
E D + F +R K + WAPQ+ +L HPA G V+HCGWNS+LESL +
Sbjct: 196 YEDPRDVLPEGFLERTVAVGK---VIGWAPQVAVLAHPAIGGFVSHCGWNSVLESLWFGV 252
Query: 296 PIITWPMFAEQFYNEKLLVFVLKIVVSVGSKVNTFWSNEGEVAV 339
PI TWPM+AEQ +N F + + + +G +++ + E + V
Sbjct: 253 PIATWPMYAEQQFN----AFEMVVELGLGVEIDMGYRKESGIIV 292
>UFO1_MANES (Q40284) Flavonol 3-O-glucosyltransferase 1 (EC
2.4.1.91) (UDP-glucose flavonoid 3-O-glucosyltransferase
1)
Length = 449
Score = 127 bits (320), Expect = 3e-29
Identities = 86/290 (29%), Positives = 141/290 (47%), Gaps = 37/290 (12%)
Query: 53 AAQVGLPDGVENIKDATSREMLAHFIKKQ-----KPHENLVSDSQKFSIPGLPHNIEITS 107
A + G+P + A + H K P E SD + +PGL ++ +
Sbjct: 113 ANEFGVPSYIFYTSGAAFLNFMLHVQKIHDEENFNPTEFNASDGE-LQVPGLVNSFPSKA 171
Query: 108 LQLQEYVREWSEFSDYFDAVYESE---GKSYGTLCNSFHELEGDYENLYKSTMGIKAWSV 164
+ ++W F + E+ G++ G + N+F ELE +K + V
Sbjct: 172 MPTAILSKQW------FPPLLENTRRYGEAKGVIINTFFELESHAIESFKDP---PIYPV 222
Query: 165 GPVSAWLKKEQNEDVIVESELLNWLNSKPNDSVLYVSFGSLTRLSHSQIVEIAHGLENSG 224
GP+ +N + E++ WL+ +P SV+++ FGS S Q+ EIA LE+SG
Sbjct: 223 GPILDVRSNGRNTN----QEIMQWLDDQPPSSVVFLCFGSNGSFSKDQVKEIACALEDSG 278
Query: 225 HNFIWVVRKKDGEGDEDGFLD---DFKQRMKENKKGYI--------IWNWAPQLLILGHP 273
H F+W + + GFL+ D++ + +G++ + WAPQ+ +L HP
Sbjct: 279 HRFLWSL----ADHRAPGFLESPSDYEDLQEVLPEGFLERTSGIEKVIGWAPQVAVLAHP 334
Query: 274 ATAGVVTHCGWNSILESLSVSLPIITWPMFAEQFYNEKLLVFVLKIVVSV 323
AT G+V+H GWNSILES+ +P+ TWPM+AEQ +N +V L + V +
Sbjct: 335 ATGGLVSHSGWNSILESIWFGVPVATWPMYAEQQFNAFQMVIELGLAVEI 384
>HQGT_ARATH (Q9M156) Probable hydroquinone glucosyltransferase (EC
2.4.1.218) (Arbutin synthase)
Length = 480
Score = 125 bits (314), Expect = 2e-28
Identities = 77/192 (40%), Positives = 106/192 (55%), Gaps = 18/192 (9%)
Query: 136 GTLCNSFHELEGDYENLYKSTMGIK--AWSVGPVSAWLKKEQNEDVIVESELLNWLNSKP 193
G L N+F ELE + + K + VGP+ K+E + ESE L WL+++P
Sbjct: 209 GILVNTFFELEPNAIKALQEPGLDKPPVYPVGPLVNIGKQEAKQTE--ESECLKWLDNQP 266
Query: 194 NDSVLYVSFGSLTRLSHSQIVEIAHGLENSGHNFIWVVRKKDGEGDEDGFLDD------- 246
SVLYVSFGS L+ Q+ E+A GL +S F+WV+R G + F
Sbjct: 267 LGSVLYVSFGSGGTLTCEQLNELALGLADSEQRFLWVIRSPSGIANSSYFDSHSQTDPLT 326
Query: 247 -----FKQRMKENKKGYIIWNWAPQLLILGHPATAGVVTHCGWNSILESLSVSLPIITWP 301
F +R K K+G++I WAPQ +L HP+T G +THCGWNS LES+ +P+I WP
Sbjct: 327 FLPPGFLERTK--KRGFVIPFWAPQAQVLAHPSTGGFLTHCGWNSTLESVVSGIPLIAWP 384
Query: 302 MFAEQFYNEKLL 313
++AEQ N LL
Sbjct: 385 LYAEQKMNAVLL 396
>HQGT_RAUSE (Q9AR73) Hydroquinone glucosyltransferase (EC 2.4.1.218)
(Arbutin synthase)
Length = 470
Score = 124 bits (310), Expect = 4e-28
Identities = 72/199 (36%), Positives = 112/199 (56%), Gaps = 17/199 (8%)
Query: 136 GTLCNSFHELE-GDYENLYKSTMGIK-AWSVGPVSAWLKKEQNEDVIVESELLNWLNSKP 193
G + N+F++LE G + L + G + +GP L + + + + E L WL+ +P
Sbjct: 206 GIMVNTFNDLEPGPLKALQEEDQGKPPVYPIGP----LIRADSSSKVDDCECLKWLDDQP 261
Query: 194 NDSVLYVSFGSLTRLSHSQIVEIAHGLENSGHNFIWVVRKKDGE---------GDEDGFL 244
SVL++SFGS +SH+Q +E+A GLE S F+WVVR + + +++ L
Sbjct: 262 RGSVLFISFGSGGAVSHNQFIELALGLEMSEQRFLWVVRSPNDKIANATYFSIQNQNDAL 321
Query: 245 DDFKQRMKENKKG--YIIWNWAPQLLILGHPATAGVVTHCGWNSILESLSVSLPIITWPM 302
+ E KG ++ +WAPQ IL H +T G +THCGWNSILES+ +P+I WP+
Sbjct: 322 AYLPEGFLERTKGRCLLVPSWAPQTEILSHGSTGGFLTHCGWNSILESVVNGVPLIAWPL 381
Query: 303 FAEQFYNEKLLVFVLKIVV 321
+AEQ N +L LK+ +
Sbjct: 382 YAEQKMNAVMLTEGLKVAL 400
>UFO6_MANES (Q40288) Flavonol 3-O-glucosyltransferase 6 (EC
2.4.1.91) (UDP-glucose flavonoid 3-O-glucosyltransferase
6) (Fragment)
Length = 394
Score = 123 bits (308), Expect = 8e-28
Identities = 76/197 (38%), Positives = 104/197 (52%), Gaps = 10/197 (5%)
Query: 136 GTLCNSFHELEGDYENLYKSTMGIKAWSVGPVSAWLK-KEQNEDVIVE-SELLNWLNSKP 193
G + N+F ELE N K K + PV LK Q DV E SE++ WL+ +P
Sbjct: 131 GIMVNTFMELESHALNSLKDDQS-KIPPIYPVGPILKLSNQENDVGPEGSEIIEWLDDQP 189
Query: 194 NDSVLYVSFGSLTRLSHSQIVEIAHGLENSGHNFIWVVRKKDGEG------DEDGFLDDF 247
SV+++ FGS+ Q EIA LE S H F+W +R+ +G D + +
Sbjct: 190 PSSVVFLCFGSMGGFDMDQAKEIACALEQSRHRFLWSLRRPPPKGKIETSTDYENLQEIL 249
Query: 248 KQRMKENKKGY-IIWNWAPQLLILGHPATAGVVTHCGWNSILESLSVSLPIITWPMFAEQ 306
E G + WAPQ+ IL HPA G V+HCGWNSILES+ S+PI TWP++AEQ
Sbjct: 250 PVGFSERTAGMGKVVGWAPQVAILEHPAIGGFVSHCGWNSILESIWFSVPIATWPLYAEQ 309
Query: 307 FYNEKLLVFVLKIVVSV 323
+N +V L + V +
Sbjct: 310 QFNAFTMVTELGLAVEI 326
>UFO5_MANES (Q40287) Flavonol 3-O-glucosyltransferase 5 (EC
2.4.1.91) (UDP-glucose flavonoid 3-O-glucosyltransferase
5)
Length = 487
Score = 122 bits (307), Expect = 1e-27
Identities = 64/152 (42%), Positives = 92/152 (60%), Gaps = 16/152 (10%)
Query: 184 ELLNWLNSKPNDSVLYVSFGSLTRLSHSQIVEIAHGLENSGHNFIWVVRKK--------- 234
ELL+WL+ +P +SV+YVSFGS LS Q++E+A GLE S FIWVVR+
Sbjct: 260 ELLDWLDQQPKESVVYVSFGSGGTLSLEQMIELAWGLERSQQRFIWVVRQPTVKTGDAAF 319
Query: 235 ----DGEGDEDGFLDD-FKQRMKENKKGYIIWNWAPQLLILGHPATAGVVTHCGWNSILE 289
DG D G+ + F R++ G ++ W+PQ+ I+ HP+ ++HCGWNS+LE
Sbjct: 320 FTQGDGADDMSGYFPEGFLTRIQN--VGLVVPQWSPQIHIMSHPSVGVFLSHCGWNSVLE 377
Query: 290 SLSVSLPIITWPMFAEQFYNEKLLVFVLKIVV 321
S++ +PII WP++AEQ N LL L + V
Sbjct: 378 SITAGVPIIAWPIYAEQRMNATLLTEELGVAV 409
>LGT_CITUN (Q9MB73) Limonoid UDP-glucosyltransferase (EC 2.4.1.210)
(Limonoid glucosyltransferase) (Limonoid GTase) (LGTase)
Length = 511
Score = 120 bits (300), Expect = 6e-27
Identities = 71/200 (35%), Positives = 109/200 (54%), Gaps = 18/200 (9%)
Query: 128 YESEGKSYGTLCNSFHELEGDYENLYKSTMGIKAWSVGPVSAWLKKEQ---NEDVIVESE 184
YE+ GK + L ++F+ELE + + IK VGP+ K +D + E
Sbjct: 206 YENLGKPFCILLDTFYELEKEIIDYMAKICPIKP--VGPLFKNPKAPTLTVRDDCMKPDE 263
Query: 185 LLNWLNSKPNDSVLYVSFGSLTRLSHSQIVEIAHGLENSGHNFIWVVRKKDGEGD----- 239
++WL+ KP SV+Y+SFG++ L Q+ EI + L NSG +F+WV++ +
Sbjct: 264 CIDWLDKKPPSSVVYISFGTVVYLKQEQVEEIGYALLNSGISFLWVMKPPPEDSGVKIVD 323
Query: 240 -EDGFLDDFKQRMKENKKGYIIWNWAPQLLILGHPATAGVVTHCGWNSILESLSVSLPII 298
DGFL+ K KG ++ W+PQ +L HP+ A VTHCGWNS +ESL+ +P+I
Sbjct: 324 LPDGFLE------KVGDKGKVV-QWSPQEKVLAHPSVACFVTHCGWNSTMESLASGVPVI 376
Query: 299 TWPMFAEQFYNEKLLVFVLK 318
T+P + +Q + L V K
Sbjct: 377 TFPQWGDQVTDAMYLCDVFK 396
>UFOG_SOLME (Q43641) Flavonol 3-O-glucosyltransferase (EC 2.4.1.91)
(UDP-glucose flavonoid 3-O-glucosyltransferase)
Length = 433
Score = 109 bits (272), Expect = 1e-23
Identities = 71/200 (35%), Positives = 108/200 (53%), Gaps = 17/200 (8%)
Query: 133 KSYGTLCNSFHELEGD---YENLYKSTMGIKAWSVGPVSAWLKKEQNEDVIVESELLNWL 189
K+ + NSF EL+ D ++L K+ K +++GP+ Q+ + ES + WL
Sbjct: 200 KADAVVLNSFQELDRDPLINKDLQKNLQ--KVFNIGPLVL-----QSSRKLDESGCIQWL 252
Query: 190 NSKPNDSVLYVSFGSLTRLSHSQIVEIAHGLENSGHNFIWVVRKKDGEGDEDGFLDDFKQ 249
+ + SV+Y+SFG++T L ++I IA LE FIW +R + GFL+
Sbjct: 253 DKQKEKSVVYLSFGTVTTLPPNEIGSIAEALETKKTPFIWSLRNNGVKNLPKGFLE---- 308
Query: 250 RMKENKKGYIIWNWAPQLLILGHPATAGVVTHCGWNSILESLSVSLPIITWPMFAEQFYN 309
R KE K I +WAPQL IL H + VTHCGWNSILE +S +P+I P F +Q N
Sbjct: 309 RTKEFGK---IVSWAPQLEILAHKSVGVFVTHCGWNSILEGISFGVPMICRPFFGDQKLN 365
Query: 310 EKLLVFVLKIVVSVGSKVNT 329
+++ V +I + + + T
Sbjct: 366 SRMVESVWEIGLQIEGGIFT 385
>UFOG_GENTR (Q96493) Flavonol 3-O-glucosyltransferase (EC 2.4.1.91)
(UDP-glucose flavonoid 3-O-glucosyltransferase)
Length = 453
Score = 102 bits (255), Expect = 1e-21
Identities = 64/197 (32%), Positives = 103/197 (51%), Gaps = 11/197 (5%)
Query: 133 KSYGTLCNSFHELEGDYENLYKSTMGIKAWSVGPVSAWLKKEQNEDVIVESELLNWLNSK 192
K+ NSF E++ N +ST + ++GP+ ED +E L WL ++
Sbjct: 211 KATAVAVNSFEEIDPIITNHLRSTNQLNILNIGPLQTLSSSIPPED----NECLKWLQTQ 266
Query: 193 PNDSVLYVSFGSLTRLSHSQIVEIAHGLENSGHNFIWVVRKKDGEGDEDGFLDDFKQRMK 252
SV+Y+SFG++ +++ +A LE+ F+W +R + + + F+D +
Sbjct: 267 KESSVVYLSFGTVINPPPNEMAALASTLESRKIPFLWSLRDEARKHLPENFID------R 320
Query: 253 ENKKGYIIWNWAPQLLILGHPATAGVVTHCGWNSILESLSVSLPIITWPMFAEQFYNEKL 312
+ G I+ +WAPQL +L +PA VTHCGWNS LES+ +P+I P F +Q N ++
Sbjct: 321 TSTFGKIV-SWAPQLHVLENPAIGVFVTHCGWNSTLESIFCRVPVIGRPFFGDQKVNARM 379
Query: 313 LVFVLKIVVSVGSKVNT 329
+ V KI V V V T
Sbjct: 380 VEDVWKIGVGVKGGVFT 396
>IAAG_MAIZE (Q41819) Indole-3-acetate beta-glucosyltransferase (EC
2.4.1.121) (IAA-Glu synthetase) ((Uridine
5'-diphosphate-glucose:indol-3-ylacetyl)-beta-D-glucosyl
transferase)
Length = 471
Score = 100 bits (250), Expect = 4e-21
Identities = 69/191 (36%), Positives = 96/191 (50%), Gaps = 18/191 (9%)
Query: 132 GKSYGTLCNSFHELEGDYENLYKSTMGIKAWSVGPV----SAWLKKEQNEDVIVESELLN 187
GK L NSF ELE E L T +KA ++GP +A N + + L+
Sbjct: 206 GKDDWVLFNSFEELET--EVLAGLTKYLKARAIGPCVPLPTAGRTAGANGRITYGANLVK 263
Query: 188 -------WLNSKPNDSVLYVSFGSLTRLSHSQIVEIAHGLENSGHNFIWVVRKKDGEGDE 240
WL++KP+ SV YVSFGSL L ++Q E+A GL +G F+WVVR D
Sbjct: 264 PEDACTKWLDTKPDRSVAYVSFGSLASLGNAQKEELARGLLAAGKPFLWVVRASDEHQVP 323
Query: 241 DGFLDDFKQRMKENKKGYIIWNWAPQLLILGHPATAGVVTHCGWNSILESLSVSLPIITW 300
L + ++ W PQL +L HPA VTHCGWNS LE+LS +P++
Sbjct: 324 RYLLAEATAT-----GAAMVVPWCPQLDVLAHPAVGCFVTHCGWNSTLEALSFGVPMVAM 378
Query: 301 PMFAEQFYNEK 311
++ +Q N +
Sbjct: 379 ALWTDQPTNAR 389
>UFO7_MANES (Q40289) Flavonol 3-O-glucosyltransferase 7 (EC
2.4.1.91) (UDP-glucose flavonoid 3-O-glucosyltransferase
7) (Fragment)
Length = 287
Score = 92.0 bits (227), Expect = 2e-18
Identities = 70/234 (29%), Positives = 104/234 (43%), Gaps = 19/234 (8%)
Query: 86 NLVSDSQKFSIPGLPHNIEITSLQLQEYVREWSEFSDYFDAVYESEGKSYGTLCNSFHEL 145
NL+ K I LP + +L+ S FS + ++ L NSF EL
Sbjct: 3 NLIPGMSKIQIRDLPEGVLFGNLE--------SLFSQMLHNMGRMLPRAAAVLMNSFEEL 54
Query: 146 EGDYENLYKSTMGIKAWSVGPVSAWLKKEQNEDVIVESELLNWLNSKPNDSVLYVSFGSL 205
+ + S +GP + D + WL+ + SV Y+SFGS+
Sbjct: 55 DPTIVSDLNSKFN-NILCIGPFNLVSPPPPVPDTY---GCMAWLDKQKPASVAYISFGSV 110
Query: 206 TRLSHSQIVEIAHGLENSGHNFIWVVRKKDGEGDEDGFLDDFKQRMKENKKGYIIWNWAP 265
++V +A LE S F+W ++ +GFLD K I+ +WAP
Sbjct: 111 ATPPPHELVALAEALEASKVPFLWSLKDHSKVHLPNGFLD-------RTKSHGIVLSWAP 163
Query: 266 QLLILGHPATAGVVTHCGWNSILESLSVSLPIITWPMFAEQFYNEKLLVFVLKI 319
Q+ IL H A VTHCGWNSILES+ +P+I P F +Q N +++ V +I
Sbjct: 164 QVEILEHAALGVFVTHCGWNSILESIVGGVPMICRPFFGDQRLNGRMVEDVWEI 217
>UFO1_MAIZE (P16166) Flavonol 3-O-glucosyltransferase (EC 2.4.1.91)
(UDP-glucose flavonoid 3-O-glucosyltransferase)
(Bronze-1) (Bz-McC allele)
Length = 471
Score = 89.4 bits (220), Expect = 1e-17
Identities = 49/131 (37%), Positives = 70/131 (53%), Gaps = 5/131 (3%)
Query: 186 LNWLNSKPNDSVLYVSFGSLTRLSHSQIVEIAHGLENSGHNFIWVVRKKDGEGDEDGFLD 245
L WL +P V YVSFG++ ++ E+A GLE+SG F+W +R+ GFLD
Sbjct: 279 LAWLGRQPARGVAYVSFGTVACPRPDELRELAAGLEDSGAPFLWSLREDSWPHLPPGFLD 338
Query: 246 DFKQRMKENKKGYIIWNWAPQLLILGHPATAGVVTHCGWNSILESLSVSLPIITWPMFAE 305
R G ++ WAPQ+ +L HP+ VTH GW S+LE LS +P+ P F +
Sbjct: 339 ----RAAGTGSGLVV-PWAPQVAVLRHPSVGAFVTHAGWASVLEGLSSGVPMACRPFFGD 393
Query: 306 QFYNEKLLVFV 316
Q N + + V
Sbjct: 394 QRMNARSVAHV 404
>UFO3_MAIZE (P16167) Flavonol 3-O-glucosyltransferase (EC 2.4.1.91)
(UDP-glucose flavonoid 3-O-glucosyltransferase)
(Bronze-1) (Bz-W22 allele)
Length = 471
Score = 86.3 bits (212), Expect = 1e-16
Identities = 48/131 (36%), Positives = 69/131 (52%), Gaps = 5/131 (3%)
Query: 186 LNWLNSKPNDSVLYVSFGSLTRLSHSQIVEIAHGLENSGHNFIWVVRKKDGEGDEDGFLD 245
L WL +P V YVSFG++ ++ E+A GLE SG F+W +R+ GFLD
Sbjct: 279 LAWLGRQPARGVAYVSFGTVACPRPDELRELAAGLEASGAPFLWSLREDSWTLLPPGFLD 338
Query: 246 DFKQRMKENKKGYIIWNWAPQLLILGHPATAGVVTHCGWNSILESLSVSLPIITWPMFAE 305
R G ++ WAPQ+ +L HP+ VTH GW S+LE +S +P+ P F +
Sbjct: 339 ----RAAGTGSGLVV-PWAPQVAVLRHPSVGAFVTHAGWASVLEGVSSGVPMACRPFFGD 393
Query: 306 QFYNEKLLVFV 316
Q N + + V
Sbjct: 394 QRMNARSVAHV 404
>UFO2_MAIZE (P16165) Flavonol 3-O-glucosyltransferase (EC 2.4.1.91)
(UDP-glucose flavonoid 3-O-glucosyltransferase)
(Bronze-1) (Bz-Mc2 allele)
Length = 471
Score = 84.0 bits (206), Expect = 5e-16
Identities = 47/131 (35%), Positives = 68/131 (51%), Gaps = 5/131 (3%)
Query: 186 LNWLNSKPNDSVLYVSFGSLTRLSHSQIVEIAHGLENSGHNFIWVVRKKDGEGDEDGFLD 245
L WL +P V YVSFG++ ++ E+A GLE S F+W +R+ GFLD
Sbjct: 279 LAWLGRQPARGVAYVSFGTVACPRPDELRELAAGLEASAAPFLWSLREDSWTLLPPGFLD 338
Query: 246 DFKQRMKENKKGYIIWNWAPQLLILGHPATAGVVTHCGWNSILESLSVSLPIITWPMFAE 305
R G ++ WAPQ+ +L HP+ VTH GW S+LE +S +P+ P F +
Sbjct: 339 ----RAAGTGSGLVV-PWAPQVAVLRHPSVGAFVTHAGWASVLEGVSSGVPMACRPFFGD 393
Query: 306 QFYNEKLLVFV 316
Q N + + V
Sbjct: 394 QRMNARSVAHV 404
>UFOG_HORVU (P14726) Flavonol 3-O-glucosyltransferase (EC 2.4.1.91)
(UDP-glucose flavonoid 3-O-glucosyltransferase)
(Bronze-1)
Length = 455
Score = 79.7 bits (195), Expect = 1e-14
Identities = 47/131 (35%), Positives = 67/131 (50%), Gaps = 10/131 (7%)
Query: 186 LNWLNSKPNDSVLYVSFGSLTRLSHSQIVEIAHGLENSGHNFIWVVRKKDGEGDEDGFLD 245
L WL+ +P SV YVSFG+ ++ E+A GLE SG F+W +R GFL
Sbjct: 268 LAWLDRRPARSVAYVSFGTNATARPDELQELAAGLEASGAPFLWSLRGVVAAAPR-GFL- 325
Query: 246 DFKQRMKENKKGYIIWNWAPQLLILGHPATAGVVTHCGWNSILESLSVSLPIITWPMFAE 305
E G ++ WAPQ+ +L H A VTH GW S++E +S +P+ P F +
Sbjct: 326 -------ERAPGLVV-PWAPQVGVLRHAAVGAFVTHAGWASVMEGVSSGVPMACRPFFGD 377
Query: 306 QFYNEKLLVFV 316
Q N + + V
Sbjct: 378 QTMNARSVASV 388
>UFOG_PETHY (Q43716) Flavonol 3-O-glucosyltransferase (EC 2.4.1.91)
(UDP-glucose flavonoid 3-O-glucosyltransferase)
(Anthocyanin rhamnosyl transferase)
Length = 473
Score = 73.6 bits (179), Expect = 7e-13
Identities = 86/377 (22%), Positives = 149/377 (38%), Gaps = 72/377 (19%)
Query: 1 MNPMIDTARLFAKHGVDVTIITTQANALLFKKPIDNDLFSGYSIKACVIQFPAAQVGLPD 60
++P + A + +GV V+ T NA K ++ S + + P + GLP
Sbjct: 25 ISPFVQLANKLSSYGVKVSFFTASGNASRVKSMLN----SAPTTHIVPLTLPHVE-GLPP 79
Query: 61 GVENIKDAT--SREMLA-----------HFIKKQKPHENLVSDSQKFSIPGLPHNIEITS 107
G E+ + T S E+L + KPH L +Q++ +P + + + I +
Sbjct: 80 GAESTAELTPASAELLKVALDLMQPQIKTLLSHLKPHFVLFDFAQEW-LPKMANGLGIKT 138
Query: 108 LQLQEYV----------------REWSEFSDY-----------FDAVYESEGKSYGTLCN 140
+ V +++ D +V E + + +
Sbjct: 139 VYYSVVVALSTAFLTCPARVLEPKKYPSLEDMKKPPLGFPQTSVTSVRTFEARDFLYVFK 198
Query: 141 SFHELEGDYENLYKSTMGIKAWSV-------GPVSAWLKKEQNEDVIV------------ 181
SFH Y+ + G A GP +++ + N+ V +
Sbjct: 199 SFHNGPTLYDRIQSGLRGCSAILAKTCSQMEGPYIKYVEAQFNKPVFLIGPVVPDPPSGK 258
Query: 182 -ESELLNWLNSKPNDSVLYVSFGSLTRLSHSQIVEIAHGLENSGHNFIWVVR---KKDGE 237
E + WLN +V+Y SFGS T L+ Q+ E+A GLE +G F V+ D
Sbjct: 259 LEEKWATWLNKFEGGTVIYCSFGSETFLTDDQVKELALGLEQTGLPFFLVLNFPANVDVS 318
Query: 238 GDEDGFL-DDFKQRMKENKKGYIIWNWAPQLLILGHPATAGVVTHCGWNSILESLSVSLP 296
+ + L + F +R+K+ KG I W Q IL H + V H G++S++E+L
Sbjct: 319 AELNRALPEGFLERVKD--KGIIHSGWVQQQNILAHSSVGCYVCHAGFSSVIEALVNDCQ 376
Query: 297 IITWPMFAEQFYNEKLL 313
++ P +Q N KL+
Sbjct: 377 VVMLPQKGDQILNAKLV 393
>UFOG_VITVI (P51094) Flavonol 3-O-glucosyltransferase (EC 2.4.1.91)
(UDP-glucose flavonoid 3-O-glucosyltransferase)
(Fragment)
Length = 154
Score = 57.0 bits (136), Expect = 7e-08
Identities = 30/78 (38%), Positives = 45/78 (57%), Gaps = 1/78 (1%)
Query: 253 ENKKGY-IIWNWAPQLLILGHPATAGVVTHCGWNSILESLSVSLPIITWPMFAEQFYNEK 311
E +GY ++ WAPQ +L VTHCGWNS+ ES++ +P+I P F +Q N +
Sbjct: 19 EKTRGYGMVVPWAPQAEVLALRQFGAFVTHCGWNSLWESVAGGVPLICRPFFGDQRLNGR 78
Query: 312 LLVFVLKIVVSVGSKVNT 329
++ VL+I V + V T
Sbjct: 79 MVEDVLEIGVRIEGGVFT 96
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.317 0.135 0.408
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 42,197,502
Number of Sequences: 164201
Number of extensions: 1819197
Number of successful extensions: 4847
Number of sequences better than 10.0: 97
Number of HSP's better than 10.0 without gapping: 85
Number of HSP's successfully gapped in prelim test: 12
Number of HSP's that attempted gapping in prelim test: 4724
Number of HSP's gapped (non-prelim): 112
length of query: 339
length of database: 59,974,054
effective HSP length: 111
effective length of query: 228
effective length of database: 41,747,743
effective search space: 9518485404
effective search space used: 9518485404
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 66 (30.0 bits)
Medicago: description of AC148481.8


