

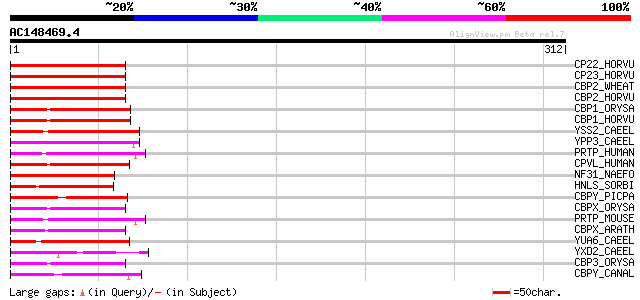
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148469.4 - phase: 0 /pseudo
(312 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
CP22_HORVU (P55748) Serine carboxypeptidase II-2 precursor (EC 3... 72 2e-12
CP23_HORVU (P52711) Serine carboxypeptidase II-3 precursor (EC 3... 71 3e-12
CBP2_WHEAT (P08819) Serine carboxypeptidase II chains A and B (E... 68 3e-11
CBP2_HORVU (P08818) Serine carboxypeptidase II precursor (EC 3.4... 67 7e-11
CBP1_ORYSA (P37890) Serine carboxypeptidase I precursor (EC 3.4.... 60 5e-09
CBP1_HORVU (P07519) Serine carboxypeptidase I precursor (EC 3.4.... 60 7e-09
YSS2_CAEEL (Q09991) Putative serine carboxypeptidase K10B2.2 pre... 59 1e-08
YPP3_CAEEL (P52716) Putative serine carboxypeptidase F32A5.3 pre... 58 3e-08
PRTP_HUMAN (P10619) Lysosomal protective protein precursor (EC 3... 58 3e-08
CPVL_HUMAN (Q9H3G5) Probable serine carboxypeptidase CPVL precur... 57 8e-08
NF31_NAEFO (P42661) Virulence-related protein Nf314 (EC 3.4.16.-) 54 5e-07
HNLS_SORBI (P52708) P-(S)-hydroxymandelonitrile lyase precursor ... 53 9e-07
CBPY_PICPA (P52710) Carboxypeptidase Y precursor (EC 3.4.16.5) (... 53 1e-06
CBPX_ORYSA (P52712) Serine carboxypeptidase-like precursor (EC 3... 53 1e-06
PRTP_MOUSE (P16675) Lysosomal protective protein precursor (EC 3... 52 1e-06
CBPX_ARATH (P32826) Serine carboxypeptidase precursor (EC 3.4.16.-) 50 6e-06
YUA6_CAEEL (P52715) Putative serine carboxypeptidase F13S12.6 pr... 49 1e-05
YXD2_CAEEL (P52714) Putative serine carboxypeptidase C08H9.1 (EC... 49 2e-05
CBP3_ORYSA (P37891) Serine carboxypeptidase III precursor (EC 3.... 48 3e-05
CBPY_CANAL (P30574) Carboxypeptidase Y precursor (EC 3.4.16.5) (... 48 4e-05
>CP22_HORVU (P55748) Serine carboxypeptidase II-2 precursor (EC
3.4.16.6) (CP-MII.2) (Fragment)
Length = 436
Score = 71.6 bits (174), Expect = 2e-12
Identities = 31/65 (47%), Positives = 47/65 (71%)
Query: 1 MLYLESPAGVGFSYSANTSDYFMVTDERTARDVLIFLQGWVTKFQKYQNSDFFITGESYA 60
+L+L+SP GVG+SYS ++D DERTA+D L+FL W+ +F +Y+ +F++TGESYA
Sbjct: 92 ILFLDSPVGVGYSYSNTSADILSNGDERTAKDSLVFLTKWLERFPQYKEREFYLTGESYA 151
Query: 61 VWNLP 65
+P
Sbjct: 152 GHYVP 156
>CP23_HORVU (P52711) Serine carboxypeptidase II-3 precursor (EC
3.4.16.6) (CP-MII.3)
Length = 516
Score = 71.2 bits (173), Expect = 3e-12
Identities = 33/65 (50%), Positives = 44/65 (66%)
Query: 1 MLYLESPAGVGFSYSANTSDYFMVTDERTARDVLIFLQGWVTKFQKYQNSDFFITGESYA 60
+L+LESPAGVG+SYS T+DY D TA D FL W+ +F +Y+ +F+ITGESYA
Sbjct: 179 VLFLESPAGVGYSYSNTTADYGRSGDNGTAEDAYQFLDNWLERFPEYKGREFYITGESYA 238
Query: 61 VWNLP 65
+P
Sbjct: 239 GHYVP 243
>CBP2_WHEAT (P08819) Serine carboxypeptidase II chains A and B (EC
3.4.16.6) (Carboxypeptidase D) (CPDW-II) (CP-WII)
Length = 423
Score = 67.8 bits (164), Expect = 3e-11
Identities = 32/65 (49%), Positives = 42/65 (64%)
Query: 1 MLYLESPAGVGFSYSANTSDYFMVTDERTARDVLIFLQGWVTKFQKYQNSDFFITGESYA 60
+L+L+SPAGVGFSY+ +SD + D RTA D FL W +F Y+ DF+I GESYA
Sbjct: 101 VLFLDSPAGVGFSYTNTSSDIYTSGDNRTAHDSYAFLAKWFERFPHYKYRDFYIAGESYA 160
Query: 61 VWNLP 65
+P
Sbjct: 161 GHYVP 165
>CBP2_HORVU (P08818) Serine carboxypeptidase II precursor (EC
3.4.16.6) (Carboxypeptidase D) (CP-MII) [Contains:
Serine carboxypeptidase II chain A; Serine
carboxypeptidase II chain B]
Length = 476
Score = 66.6 bits (161), Expect = 7e-11
Identities = 30/65 (46%), Positives = 42/65 (64%)
Query: 1 MLYLESPAGVGFSYSANTSDYFMVTDERTARDVLIFLQGWVTKFQKYQNSDFFITGESYA 60
+L+L+SPAGVGFSY+ +SD + D RTA D FL W +F Y+ +F++ GESYA
Sbjct: 133 VLFLDSPAGVGFSYTNTSSDIYTSGDNRTAHDSYAFLAAWFERFPHYKYREFYVAGESYA 192
Query: 61 VWNLP 65
+P
Sbjct: 193 GHYVP 197
>CBP1_ORYSA (P37890) Serine carboxypeptidase I precursor (EC
3.4.16.5) (Carboxypeptidase C)
Length = 510
Score = 60.5 bits (145), Expect = 5e-09
Identities = 31/68 (45%), Positives = 44/68 (64%), Gaps = 1/68 (1%)
Query: 1 MLYLESPAGVGFSYSANTSDYFMVTDERTARDVLIFLQGWVTKFQKYQNSDFFITGESYA 60
++YL+SPAGVG SYS NTSDY D +TA D FL W + ++ ++ F+I GESYA
Sbjct: 138 VIYLDSPAGVGLSYSKNTSDY-NTGDLKTAADSHTFLLKWFQLYPEFLSNPFYIAGESYA 196
Query: 61 VWNLPLIT 68
+P ++
Sbjct: 197 GVYVPTLS 204
>CBP1_HORVU (P07519) Serine carboxypeptidase I precursor (EC
3.4.16.5) (Carboxypeptidase C) (CP-MI)
Length = 499
Score = 60.1 bits (144), Expect = 7e-09
Identities = 31/68 (45%), Positives = 43/68 (62%), Gaps = 1/68 (1%)
Query: 1 MLYLESPAGVGFSYSANTSDYFMVTDERTARDVLIFLQGWVTKFQKYQNSDFFITGESYA 60
M+YL+SPAGVG SYS N SDY D +TA D FL W + ++ ++ F+I GESYA
Sbjct: 132 MIYLDSPAGVGLSYSKNVSDY-ETGDLKTATDSHTFLLKWFQLYPEFLSNPFYIAGESYA 190
Query: 61 VWNLPLIT 68
+P ++
Sbjct: 191 GVYVPTLS 198
>YSS2_CAEEL (Q09991) Putative serine carboxypeptidase K10B2.2
precursor (EC 3.4.16.-)
Length = 470
Score = 59.3 bits (142), Expect = 1e-08
Identities = 31/73 (42%), Positives = 48/73 (65%), Gaps = 2/73 (2%)
Query: 1 MLYLESPAGVGFSYSANTSDYFMVTDERTARDVLIFLQGWVTKFQKYQNSDFFITGESYA 60
+L+LESPAGVG+SYS N + V+D+ + + L +++KF +Y+ DF+ITGESYA
Sbjct: 114 VLFLESPAGVGYSYSTNFN--LTVSDDEVSLHNYMALLDFLSKFPEYKGRDFWITGESYA 171
Query: 61 VWNLPLITIPGLN 73
+P + + LN
Sbjct: 172 GVYIPTLAVRILN 184
>YPP3_CAEEL (P52716) Putative serine carboxypeptidase F32A5.3
precursor (EC 3.4.16.-)
Length = 574
Score = 57.8 bits (138), Expect = 3e-08
Identities = 31/77 (40%), Positives = 45/77 (58%), Gaps = 4/77 (5%)
Query: 1 MLYLESPAGVGFSYSANTSDYFMVTDERTARDVLIFLQGWVTKFQ-KYQNSDFFITGESY 59
+LYLESP GVG+SY T YF D+++A L + Q KY N F+++GESY
Sbjct: 114 VLYLESPIGVGYSYDTTTPGYFQANDDQSAAQNYQALTNFFNVAQPKYTNRTFYLSGESY 173
Query: 60 AVWNLPLIT---IPGLN 73
A +P++T + G+N
Sbjct: 174 AGIYIPMLTDLIVQGIN 190
>PRTP_HUMAN (P10619) Lysosomal protective protein precursor (EC
3.4.16.5) (Cathepsin A) (Carboxypeptidase C) (Protective
protein for beta-galactosidase)
Length = 480
Score = 57.8 bits (138), Expect = 3e-08
Identities = 32/81 (39%), Positives = 46/81 (56%), Gaps = 7/81 (8%)
Query: 1 MLYLESPAGVGFSYSANTSDYFMVTDERTARDVLIFLQGWVTKFQKYQNSDFFITGESYA 60
+LYLESPAGVGFSYS + ++ D A+ LQ + F +Y+N+ F+TGESYA
Sbjct: 123 VLYLESPAGVGFSYSDDK--FYATNDTEVAQSNFEALQDFFRLFPEYKNNKLFLTGESYA 180
Query: 61 VWNLPLITI-----PGLNFYG 76
+P + + P +N G
Sbjct: 181 GIYIPTLAVLVMQDPSMNLQG 201
>CPVL_HUMAN (Q9H3G5) Probable serine carboxypeptidase CPVL precursor
(EC 3.4.16.-) (Carboxypeptidase, vitellogenic-like)
(Vitellogenic carboxypeptidase-like protein) (VCP-like
protein)
Length = 476
Score = 56.6 bits (135), Expect = 8e-08
Identities = 28/67 (41%), Positives = 44/67 (64%), Gaps = 1/67 (1%)
Query: 1 MLYLESPAGVGFSYSANTSDYFMVTDERTARDVLIFLQGWVTKFQKYQNSDFFITGESYA 60
MLY+++P G GFS++ +T Y V ++ ARD+ L + F +Y+N+DF++TGESYA
Sbjct: 148 MLYIDNPVGTGFSFTDDTHGY-AVNEDDVARDLYSALIQFFQIFPEYKNNDFYVTGESYA 206
Query: 61 VWNLPLI 67
+P I
Sbjct: 207 GKYVPAI 213
>NF31_NAEFO (P42661) Virulence-related protein Nf314 (EC 3.4.16.-)
Length = 482
Score = 53.9 bits (128), Expect = 5e-07
Identities = 24/59 (40%), Positives = 39/59 (65%)
Query: 1 MLYLESPAGVGFSYSANTSDYFMVTDERTARDVLIFLQGWVTKFQKYQNSDFFITGESY 59
+LY+E P GVGFSYS +T DY + D + A D+ L+ ++T+F ++ + ++ GESY
Sbjct: 106 ILYIEQPVGVGFSYSNSTDDYQNLNDVQAASDMNNALRDFLTRFPQFIGRETYLAGESY 164
>HNLS_SORBI (P52708) P-(S)-hydroxymandelonitrile lyase precursor
(EC 4.1.2.11) (Hydroxynitrile lyase) (HNL) (Fragment)
Length = 366
Score = 53.1 bits (126), Expect = 9e-07
Identities = 27/58 (46%), Positives = 36/58 (61%), Gaps = 1/58 (1%)
Query: 1 MLYLESPAGVGFSYSANTSDYFMVTDERTARDVLIFLQGWVTKFQKYQNSDFFITGES 58
+L+ ESPAGV FSYS NTS + D++ A+D FL W +F Y +F+I GES
Sbjct: 13 ILFAESPAGVVFSYS-NTSSDLSMGDDKMAQDTYTFLVKWFERFPHYNYREFYIAGES 69
>CBPY_PICPA (P52710) Carboxypeptidase Y precursor (EC 3.4.16.5)
(Carboxypeptidase YSCY)
Length = 523
Score = 52.8 bits (125), Expect = 1e-06
Identities = 28/66 (42%), Positives = 40/66 (60%), Gaps = 4/66 (6%)
Query: 1 MLYLESPAGVGFSYSANTSDYFMVTDERTARDVLIFLQGWVTKFQKYQNSDFFITGESYA 60
++YL+ P VGFSYS+++ +V E DV FLQ + F +YQ +DF I GESYA
Sbjct: 196 IIYLDQPVNVGFSYSSSSVSNTVVAGE----DVYAFLQLFFQHFPEYQTNDFHIAGESYA 251
Query: 61 VWNLPL 66
+P+
Sbjct: 252 GHYIPV 257
>CBPX_ORYSA (P52712) Serine carboxypeptidase-like precursor (EC
3.4.16.-)
Length = 429
Score = 52.8 bits (125), Expect = 1e-06
Identities = 25/65 (38%), Positives = 39/65 (59%), Gaps = 1/65 (1%)
Query: 1 MLYLESPAGVGFSYSANTSDYFMVTDERTARDVLIFLQGWVTKFQKYQNSDFFITGESYA 60
++Y++ P G GFSYS+N D + + D+ FLQ + T+ + +DF+ITGESYA
Sbjct: 92 LIYVDQPTGTGFSYSSNPRDT-RHDEAGVSNDLYAFLQAFFTEHPNFAKNDFYITGESYA 150
Query: 61 VWNLP 65
+P
Sbjct: 151 GHYIP 155
>PRTP_MOUSE (P16675) Lysosomal protective protein precursor (EC
3.4.16.5) (Cathepsin A) (Carboxypeptidase C) (Protective
protein for beta-galactosidase)
Length = 474
Score = 52.4 bits (124), Expect = 1e-06
Identities = 29/81 (35%), Positives = 46/81 (55%), Gaps = 7/81 (8%)
Query: 1 MLYLESPAGVGFSYSANTSDYFMVTDERTARDVLIFLQGWVTKFQKYQNSDFFITGESYA 60
+LY+ESPAGVGFSYS + ++ D A + L+ + F +Y+++ F+TGESYA
Sbjct: 118 VLYIESPAGVGFSYSDDKM--YVTNDTEVAENNYEALKDFFRLFPEYKDNKLFLTGESYA 175
Query: 61 VWNLPLITI-----PGLNFYG 76
+P + + P +N G
Sbjct: 176 GIYIPTLAVLVMQDPSMNLQG 196
>CBPX_ARATH (P32826) Serine carboxypeptidase precursor (EC 3.4.16.-)
Length = 516
Score = 50.4 bits (119), Expect = 6e-06
Identities = 25/65 (38%), Positives = 38/65 (58%), Gaps = 1/65 (1%)
Query: 1 MLYLESPAGVGFSYSANTSDYFMVTDERTARDVLIFLQGWVTKFQKYQNSDFFITGESYA 60
+LY++ P G GFSY+ + SD + + D+ FLQ + + K +DF+ITGESYA
Sbjct: 173 LLYVDQPVGTGFSYTTDKSD-IRHDETGVSNDLYDFLQAFFAEHPKLAKNDFYITGESYA 231
Query: 61 VWNLP 65
+P
Sbjct: 232 GHYIP 236
>YUA6_CAEEL (P52715) Putative serine carboxypeptidase F13S12.6
precursor (EC 3.4.16.-)
Length = 454
Score = 49.3 bits (116), Expect = 1e-05
Identities = 25/67 (37%), Positives = 42/67 (62%), Gaps = 2/67 (2%)
Query: 1 MLYLESPAGVGFSYSANTSDYFMVTDERTARDVLIFLQGWVTKFQKYQNSDFFITGESYA 60
+L LE+PAGVG+SY+ T + D++TA + L + +F +Y+ +DF++TGESY
Sbjct: 113 ILTLEAPAGVGYSYA--TDNNIATGDDQTASENWEALVAFFNEFPQYKGNDFYVTGESYG 170
Query: 61 VWNLPLI 67
+P +
Sbjct: 171 GIYVPTL 177
>YXD2_CAEEL (P52714) Putative serine carboxypeptidase C08H9.1 (EC
3.4.16.-)
Length = 505
Score = 48.9 bits (115), Expect = 2e-05
Identities = 32/82 (39%), Positives = 38/82 (46%), Gaps = 19/82 (23%)
Query: 1 MLYLESPAGVGFSYSANTSDYFMVTD----ERTARDVLIFLQGWVTKFQKYQNSDFFITG 56
+L+LESP GVGFSY S++ V D E+ V+ F Q K Y N DFFI
Sbjct: 114 ILFLESPIGVGFSYDTEQSNFTKVNDDSIAEQNFNSVIDFFQ---RKHSSYVNHDFFIAA 170
Query: 57 ESYAVWNLPLITIPGLNFYGPM 78
ESY YGPM
Sbjct: 171 ESYG------------GVYGPM 180
>CBP3_ORYSA (P37891) Serine carboxypeptidase III precursor (EC
3.4.16.5)
Length = 500
Score = 48.1 bits (113), Expect = 3e-05
Identities = 24/65 (36%), Positives = 39/65 (59%), Gaps = 1/65 (1%)
Query: 1 MLYLESPAGVGFSYSANTSDYFMVTDERTARDVLIFLQGWVTKFQKYQNSDFFITGESYA 60
+++++ P G GFSYS++ D + + D+ FLQ + K ++ +DFFITGESYA
Sbjct: 160 IIFVDQPTGTGFSYSSDDRDT-RHDETGVSNDLYSFLQVFFKKHPEFAKNDFFITGESYA 218
Query: 61 VWNLP 65
+P
Sbjct: 219 GHYIP 223
>CBPY_CANAL (P30574) Carboxypeptidase Y precursor (EC 3.4.16.5)
(Carboxypeptidase YSCY)
Length = 542
Score = 47.8 bits (112), Expect = 4e-05
Identities = 28/79 (35%), Positives = 42/79 (52%), Gaps = 9/79 (11%)
Query: 1 MLYLESPAGVGFSYSANTSDYFMVTDERTARDVLIFLQGWVTKFQKYQNSDFFITGESYA 60
+++L+ P VG+SYS+ + + +DV FLQ + F +Y N DF I GESYA
Sbjct: 216 VIFLDQPINVGYSYSSQSVSNTIAA----GKDVYAFLQLFFKNFPEYANLDFHIAGESYA 271
Query: 61 VWNLP-----LITIPGLNF 74
+P ++T P NF
Sbjct: 272 GHYIPAFASEILTHPERNF 290
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.340 0.150 0.525
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 35,654,177
Number of Sequences: 164201
Number of extensions: 1401018
Number of successful extensions: 3551
Number of sequences better than 10.0: 41
Number of HSP's better than 10.0 without gapping: 27
Number of HSP's successfully gapped in prelim test: 14
Number of HSP's that attempted gapping in prelim test: 3490
Number of HSP's gapped (non-prelim): 46
length of query: 312
length of database: 59,974,054
effective HSP length: 110
effective length of query: 202
effective length of database: 41,911,944
effective search space: 8466212688
effective search space used: 8466212688
T: 11
A: 40
X1: 15 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 39 (21.9 bits)
S2: 66 (30.0 bits)
Medicago: description of AC148469.4


