

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148404.8 - phase: 0
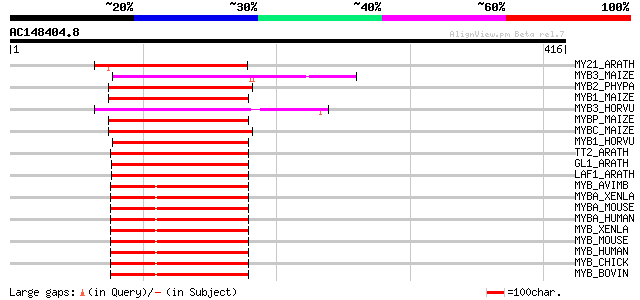
(416 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
MY21_ARATH (Q9LK95) Transcription factor MYB21 (Myb-related prot... 147 6e-35
MYB3_MAIZE (P20025) Myb-related protein Zm38 145 1e-34
MYB2_PHYPA (P80073) Myb-related protein Pp2 145 1e-34
MYB1_MAIZE (P20024) Myb-related protein Zm1 145 2e-34
MYB3_HORVU (P20027) Myb-related protein Hv33 137 5e-32
MYBP_MAIZE (P27898) Myb-related protein P 136 9e-32
MYBC_MAIZE (P10290) Anthocyanin regulatory C1 protein 136 9e-32
MYB1_HORVU (P20026) Myb-related protein Hv1 136 9e-32
TT2_ARATH (Q9FJA2) TRANSPARENT TESTA 2 protein (Myb-related prot... 129 1e-29
GL1_ARATH (P27900) Trichome differentiation protein GL1 (GLABROU... 129 2e-29
LAF1_ARATH (Q9M0K4) Transcription factor LAF1 (Long after far-re... 120 5e-27
MYB_AVIMB (P01104) Transforming protein Myb 119 1e-26
MYBA_XENLA (Q05935) Myb-related protein A (A-Myb) (XAMYB) (MYB-r... 116 1e-25
MYBA_MOUSE (P51960) Myb-related protein A (A-Myb) 115 2e-25
MYBA_HUMAN (P10243) Myb-related protein A (A-Myb) 115 2e-25
MYB_XENLA (Q08759) Myb protein 114 5e-25
MYB_MOUSE (P06876) Myb proto-oncogene protein (C-myb) 113 8e-25
MYB_HUMAN (P10242) Myb proto-oncogene protein (C-myb) 113 8e-25
MYB_CHICK (P01103) Myb proto-oncogene protein (C-myb) 113 8e-25
MYB_BOVIN (P46200) Myb proto-oncogene protein (C-myb) 113 8e-25
>MY21_ARATH (Q9LK95) Transcription factor MYB21 (Myb-related protein
21) (AtMYB21) (Myb homolog 3) (AtMyb3)
Length = 226
Score = 147 bits (370), Expect = 6e-35
Identities = 67/117 (57%), Positives = 87/117 (74%), Gaps = 2/117 (1%)
Query: 64 GDTTGGEGG--EGGLKKGPWTAAEDEILVAHVQKYGEGNWNSVRKCTGLARCGKSCRLRW 121
G ++GG G E ++KGPWT ED IL+ ++ +G+G WNS+ K GL R GKSCRLRW
Sbjct: 6 GGSSGGSGSSAEAEVRKGPWTMEEDLILINYIANHGDGVWNSLAKSAGLKRTGKSCRLRW 65
Query: 122 ANHLRPDLRKGAITAEEERRIIELHHKMGNKWAQMAALLPGRTDNEIKNFWNTRCKK 178
N+LRPD+R+G IT EE+ I+ELH K GN+W+++A LPGRTDNEIKNFW TR +K
Sbjct: 66 LNYLRPDVRRGNITPEEQLIIMELHAKWGNRWSKIAKHLPGRTDNEIKNFWRTRIQK 122
>MYB3_MAIZE (P20025) Myb-related protein Zm38
Length = 255
Score = 145 bits (367), Expect = 1e-34
Identities = 79/190 (41%), Positives = 113/190 (58%), Gaps = 9/190 (4%)
Query: 78 KGPWTAAEDEILVAHVQKYGEGNWNSVRKCTGLARCGKSCRLRWANHLRPDLRKGAITAE 137
+G WT EDE LVA+++ +GEG W S+ K GL RCGKSCRLRW N+LRPDL++G TA+
Sbjct: 14 RGAWTKEEDERLVAYIRAHGEGCWRSLPKAAGLLRCGKSCRLRWINYLRPDLKRGNFTAD 73
Query: 138 EERRIIELHHKMGNKWAQMAALLPGRTDNEIKNFWNTRCKKR--GR-----ANLPIYPED 190
E+ I++LH +GNKW+ +AA LPGRTDNEIKN+WNT +++ GR + PI +
Sbjct: 74 EDDLIVKLHSLLGNKWSLIAARLPGRTDNEIKNYWNTHVRRKLLGRGIDPVTHRPIAADA 133
Query: 191 LSSECLLNQENADMLTNEASQHDEAENKVPEVIFKDYKLRPDILPPCFDKLLAGLLLRPS 250
++ + Q + A++ + K P D L I PPC + + L+PS
Sbjct: 134 VTVTTVSFQPSPSAAAAAAAEAEATAAKAPRC--PDLNLDLCISPPCQQQEEEEVDLKPS 191
Query: 251 KRPWKSDMNL 260
K ++ L
Sbjct: 192 AAVVKREVLL 201
>MYB2_PHYPA (P80073) Myb-related protein Pp2
Length = 421
Score = 145 bits (367), Expect = 1e-34
Identities = 62/108 (57%), Positives = 83/108 (76%)
Query: 75 GLKKGPWTAAEDEILVAHVQKYGEGNWNSVRKCTGLARCGKSCRLRWANHLRPDLRKGAI 134
GL++GPWT+ ED+ LV+H+ G W ++ K GL RCGKSCRLRW N+LRPDL++G
Sbjct: 11 GLRRGPWTSEEDQKLVSHITNNGLSCWRAIPKLAGLLRCGKSCRLRWTNYLRPDLKRGIF 70
Query: 135 TAEEERRIIELHHKMGNKWAQMAALLPGRTDNEIKNFWNTRCKKRGRA 182
+ EE I++LH +GN+W+++AA LPGRTDNEIKN+WNTR KKR R+
Sbjct: 71 SEAEENLILDLHATLGNRWSRIAAQLPGRTDNEIKNYWNTRLKKRLRS 118
>MYB1_MAIZE (P20024) Myb-related protein Zm1
Length = 340
Score = 145 bits (365), Expect = 2e-34
Identities = 64/105 (60%), Positives = 79/105 (74%)
Query: 75 GLKKGPWTAAEDEILVAHVQKYGEGNWNSVRKCTGLARCGKSCRLRWANHLRPDLRKGAI 134
GL +G WT ED L+A++QK+G NW ++ K GL RCGKSCRLRW N+LRPDL++G
Sbjct: 13 GLNRGSWTPQEDMRLIAYIQKHGHTNWRALPKQAGLLRCGKSCRLRWINYLRPDLKRGNF 72
Query: 135 TAEEERRIIELHHKMGNKWAQMAALLPGRTDNEIKNFWNTRCKKR 179
T EEE II LH +GNKW+++AA LPGRTDNEIKN WNT KK+
Sbjct: 73 TDEEEEAIIRLHGLLGNKWSKIAACLPGRTDNEIKNVWNTHLKKK 117
>MYB3_HORVU (P20027) Myb-related protein Hv33
Length = 302
Score = 137 bits (345), Expect = 5e-32
Identities = 76/183 (41%), Positives = 105/183 (56%), Gaps = 13/183 (7%)
Query: 64 GDTTGGEGGEGGLKKGPWTAAEDEILVAHVQKYGEGNWNSVRKCTGLARCGKSCRLRWAN 123
G + G G+ ++KG W+ EDE L H+ ++G G W+SV + L RCGKSCRLRW N
Sbjct: 2 GRPSSGAVGQPKVRKGLWSPEEDEKLYNHIIRHGVGCWSSVPRLAALNRCGKSCRLRWIN 61
Query: 124 HLRPDLRKGAITAEEERRIIELHHKMGNKWAQMAALLPGRTDNEIKNFWNTRCKKRGRAN 183
+LRPDL++G + +EE I+ LH +GN+W+Q+A+ LPGRTDNEIKNFWN+ KK+ R
Sbjct: 62 YLRPDLKRGCFSQQEEDHIVALHQILGNRWSQIASHLPGRTDNEIKNFWNSCIKKKLR-- 119
Query: 184 LPIYPEDLSSECLLNQENADMLTNEASQHD-EAENKVPEVIFKDYKLRP------DILPP 236
+ + +AD T A+ D E E++ P D L P D P
Sbjct: 120 ----QQGIDPATHKPMASADTATAAAALPDAEEEDRKPLCPAVDGSLVPKQPAVFDPFPL 175
Query: 237 CFD 239
C D
Sbjct: 176 CVD 178
>MYBP_MAIZE (P27898) Myb-related protein P
Length = 399
Score = 136 bits (343), Expect = 9e-32
Identities = 58/105 (55%), Positives = 82/105 (77%)
Query: 75 GLKKGPWTAAEDEILVAHVQKYGEGNWNSVRKCTGLARCGKSCRLRWANHLRPDLRKGAI 134
GLK+G WTA ED++L ++ ++GEG+W S+ K GL RCGKSCRLRW N+LR D+++G I
Sbjct: 11 GLKRGRWTAEEDQLLANYIAEHGEGSWRSLPKNAGLLRCGKSCRLRWINYLRADVKRGNI 70
Query: 135 TAEEERRIIELHHKMGNKWAQMAALLPGRTDNEIKNFWNTRCKKR 179
+ EEE II+LH +GN+W+ +A+ LPGRTDNEIKN+WN+ ++
Sbjct: 71 SKEEEDIIIKLHATLGNRWSLIASHLPGRTDNEIKNYWNSHLSRQ 115
>MYBC_MAIZE (P10290) Anthocyanin regulatory C1 protein
Length = 273
Score = 136 bits (343), Expect = 9e-32
Identities = 59/108 (54%), Positives = 80/108 (73%)
Query: 75 GLKKGPWTAAEDEILVAHVQKYGEGNWNSVRKCTGLARCGKSCRLRWANHLRPDLRKGAI 134
G+K+G WT+ ED+ L A+V+ +GEG W V + GL RCGKSCRLRW N+LRP++R+G I
Sbjct: 11 GVKRGAWTSKEDDALAAYVKAHGEGKWREVPQKAGLRRCGKSCRLRWLNYLRPNIRRGNI 70
Query: 135 TAEEERRIIELHHKMGNKWAQMAALLPGRTDNEIKNFWNTRCKKRGRA 182
+ +EE II LH +GN+W+ +A LPGRTDNEIKN+WN+ +R A
Sbjct: 71 SYDEEDLIIRLHRLLGNRWSLIAGRLPGRTDNEIKNYWNSTLGRRAGA 118
>MYB1_HORVU (P20026) Myb-related protein Hv1
Length = 267
Score = 136 bits (343), Expect = 9e-32
Identities = 58/102 (56%), Positives = 77/102 (74%)
Query: 78 KGPWTAAEDEILVAHVQKYGEGNWNSVRKCTGLARCGKSCRLRWANHLRPDLRKGAITAE 137
KG WT ED+ L A+++ +GEG W S+ K GL RCGKSCRLRW N+LRPDL++G + E
Sbjct: 14 KGAWTKEEDDRLTAYIKAHGEGCWRSLPKAAGLLRCGKSCRLRWINYLRPDLKRGNFSHE 73
Query: 138 EERRIIELHHKMGNKWAQMAALLPGRTDNEIKNFWNTRCKKR 179
E+ II+LH +GNKW+ +A LPGRTDNEIKN+WNT +++
Sbjct: 74 EDELIIKLHSLLGNKWSLIAGRLPGRTDNEIKNYWNTHIRRK 115
>TT2_ARATH (Q9FJA2) TRANSPARENT TESTA 2 protein (Myb-related protein
123) (AtMYB123) (Myb-related transcription factor
LBM2-like)
Length = 258
Score = 129 bits (324), Expect = 1e-29
Identities = 55/104 (52%), Positives = 77/104 (73%)
Query: 76 LKKGPWTAAEDEILVAHVQKYGEGNWNSVRKCTGLARCGKSCRLRWANHLRPDLRKGAIT 135
L +G WT ED+IL ++ +GEG W+++ GL RCGKSCRLRW N+LRP +++G I+
Sbjct: 14 LNRGAWTDHEDKILRDYITTHGEGKWSTLPNQAGLKRCGKSCRLRWKNYLRPGIKRGNIS 73
Query: 136 AEEERRIIELHHKMGNKWAQMAALLPGRTDNEIKNFWNTRCKKR 179
++EE II LH+ +GN+W+ +A LPGRTDNEIKN WN+ +KR
Sbjct: 74 SDEEELIIRLHNLLGNRWSLIAGRLPGRTDNEIKNHWNSNLRKR 117
>GL1_ARATH (P27900) Trichome differentiation protein GL1 (GLABROUS1
protein)
Length = 228
Score = 129 bits (323), Expect = 2e-29
Identities = 55/103 (53%), Positives = 73/103 (70%)
Query: 77 KKGPWTAAEDEILVAHVQKYGEGNWNSVRKCTGLARCGKSCRLRWANHLRPDLRKGAITA 136
KKG WT ED IL+ +V +G G WN + + TGL RCGKSCRLRW N+L P++ KG T
Sbjct: 15 KKGLWTVEEDNILMDYVLNHGTGQWNRIVRKTGLKRCGKSCRLRWMNYLSPNVNKGNFTE 74
Query: 137 EEERRIIELHHKMGNKWAQMAALLPGRTDNEIKNFWNTRCKKR 179
+EE II LH +GN+W+ +A +PGRTDN++KN+WNT K+
Sbjct: 75 QEEDLIIRLHKLLGNRWSLIAKRVPGRTDNQVKNYWNTHLSKK 117
>LAF1_ARATH (Q9M0K4) Transcription factor LAF1 (Long after far-red
light protein 1) (Myb-related protein 18) (AtMYB18)
Length = 283
Score = 120 bits (302), Expect = 5e-27
Identities = 54/103 (52%), Positives = 71/103 (68%)
Query: 77 KKGPWTAAEDEILVAHVQKYGEGNWNSVRKCTGLARCGKSCRLRWANHLRPDLRKGAITA 136
+KG W+ EDE L + + YG W +V GL R GKSCRLRW N+LRP L++ I+A
Sbjct: 11 RKGLWSPEEDEKLRSFILSYGHSCWTTVPIKAGLQRNGKSCRLRWINYLRPGLKRDMISA 70
Query: 137 EEERRIIELHHKMGNKWAQMAALLPGRTDNEIKNFWNTRCKKR 179
EEE I+ H +GNKW+Q+A LPGRTDNEIKN+W++ KK+
Sbjct: 71 EEEETILTFHSSLGNKWSQIAKFLPGRTDNEIKNYWHSHLKKK 113
>MYB_AVIMB (P01104) Transforming protein Myb
Length = 382
Score = 119 bits (298), Expect = 1e-26
Identities = 51/104 (49%), Positives = 72/104 (69%), Gaps = 1/104 (0%)
Query: 76 LKKGPWTAAEDEILVAHVQKYGEGNWNSVRKCTGLARCGKSCRLRWANHLRPDLRKGAIT 135
L KGPWT ED+ ++ HVQKYG W+ + K R GK CR RW NHL P+++K + T
Sbjct: 19 LNKGPWTKEEDQRVIEHVQKYGPKRWSDIAKHLK-GRIGKQCRERWHNHLNPEVKKTSWT 77
Query: 136 AEEERRIIELHHKMGNKWAQMAALLPGRTDNEIKNFWNTRCKKR 179
EE+R I + H ++GN+WA++A LLPGRTDN +KN WN+ +++
Sbjct: 78 EEEDRIIYQAHKRLGNRWAEIAKLLPGRTDNAVKNHWNSTMRRK 121
>MYBA_XENLA (Q05935) Myb-related protein A (A-Myb) (XAMYB)
(MYB-related protein 2) (XMYB2)
Length = 728
Score = 116 bits (290), Expect = 1e-25
Identities = 53/104 (50%), Positives = 69/104 (65%), Gaps = 1/104 (0%)
Query: 76 LKKGPWTAAEDEILVAHVQKYGEGNWNSVRKCTGLARCGKSCRLRWANHLRPDLRKGAIT 135
L KGPWT ED+ ++ V KYG W+ + K R GK CR RW NHL PD++K + T
Sbjct: 84 LVKGPWTKEEDQRVIELVHKYGPKKWSIIAKHLK-GRIGKQCRERWHNHLNPDVKKSSWT 142
Query: 136 AEEERRIIELHHKMGNKWAQMAALLPGRTDNEIKNFWNTRCKKR 179
EE+R I H +MGN+WA++A LLPGRTDN IKN WN+ K++
Sbjct: 143 EEEDRIIYSAHKRMGNRWAEIAKLLPGRTDNSIKNHWNSTMKRK 186
>MYBA_MOUSE (P51960) Myb-related protein A (A-Myb)
Length = 751
Score = 115 bits (289), Expect = 2e-25
Identities = 52/104 (50%), Positives = 71/104 (68%), Gaps = 1/104 (0%)
Query: 76 LKKGPWTAAEDEILVAHVQKYGEGNWNSVRKCTGLARCGKSCRLRWANHLRPDLRKGAIT 135
L KGPWT ED+ ++ VQKYG W+ + K R GK CR RW NHL P+++K + T
Sbjct: 85 LIKGPWTKEEDQRVIELVQKYGPKRWSLIAKHLK-GRIGKQCRERWHNHLNPEVKKSSWT 143
Query: 136 AEEERRIIELHHKMGNKWAQMAALLPGRTDNEIKNFWNTRCKKR 179
EE+R I E H ++GN+WA++A LLPGRTDN IKN WN+ +++
Sbjct: 144 EEEDRIIYEAHKRLGNRWAEIAKLLPGRTDNSIKNHWNSTMRRK 187
>MYBA_HUMAN (P10243) Myb-related protein A (A-Myb)
Length = 752
Score = 115 bits (289), Expect = 2e-25
Identities = 52/104 (50%), Positives = 71/104 (68%), Gaps = 1/104 (0%)
Query: 76 LKKGPWTAAEDEILVAHVQKYGEGNWNSVRKCTGLARCGKSCRLRWANHLRPDLRKGAIT 135
L KGPWT ED+ ++ VQKYG W+ + K R GK CR RW NHL P+++K + T
Sbjct: 85 LIKGPWTKEEDQRVIELVQKYGPKRWSLIAKHLK-GRIGKQCRERWHNHLNPEVKKSSWT 143
Query: 136 AEEERRIIELHHKMGNKWAQMAALLPGRTDNEIKNFWNTRCKKR 179
EE+R I E H ++GN+WA++A LLPGRTDN IKN WN+ +++
Sbjct: 144 EEEDRIIYEAHKRLGNRWAEIAKLLPGRTDNSIKNHWNSTMRRK 187
>MYB_XENLA (Q08759) Myb protein
Length = 624
Score = 114 bits (285), Expect = 5e-25
Identities = 51/104 (49%), Positives = 70/104 (67%), Gaps = 1/104 (0%)
Query: 76 LKKGPWTAAEDEILVAHVQKYGEGNWNSVRKCTGLARCGKSCRLRWANHLRPDLRKGAIT 135
L KGPWT ED+ ++ V KYG W+ + K R GK CR RW NHL P+++K + T
Sbjct: 87 LIKGPWTKEEDQRVIELVHKYGPKRWSVIAKHLK-GRIGKQCRERWHNHLNPEVKKSSWT 145
Query: 136 AEEERRIIELHHKMGNKWAQMAALLPGRTDNEIKNFWNTRCKKR 179
EE+R I E H ++GN+WA++A LLPGRTDN IKN WN+ +++
Sbjct: 146 EEEDRTIYEAHKRLGNRWAEIAKLLPGRTDNAIKNHWNSTMRRK 189
>MYB_MOUSE (P06876) Myb proto-oncogene protein (C-myb)
Length = 636
Score = 113 bits (283), Expect = 8e-25
Identities = 51/104 (49%), Positives = 71/104 (68%), Gaps = 1/104 (0%)
Query: 76 LKKGPWTAAEDEILVAHVQKYGEGNWNSVRKCTGLARCGKSCRLRWANHLRPDLRKGAIT 135
L KGPWT ED+ ++ VQKYG W+ + K R GK CR RW NHL P+++K + T
Sbjct: 90 LIKGPWTKEEDQRVIELVQKYGPKRWSVIAKHLK-GRIGKQCRERWHNHLNPEVKKTSWT 148
Query: 136 AEEERRIIELHHKMGNKWAQMAALLPGRTDNEIKNFWNTRCKKR 179
EE+R I + H ++GN+WA++A LLPGRTDN IKN WN+ +++
Sbjct: 149 EEEDRIIYQAHKRLGNRWAEIAKLLPGRTDNAIKNHWNSTMRRK 192
>MYB_HUMAN (P10242) Myb proto-oncogene protein (C-myb)
Length = 640
Score = 113 bits (283), Expect = 8e-25
Identities = 51/104 (49%), Positives = 71/104 (68%), Gaps = 1/104 (0%)
Query: 76 LKKGPWTAAEDEILVAHVQKYGEGNWNSVRKCTGLARCGKSCRLRWANHLRPDLRKGAIT 135
L KGPWT ED+ ++ VQKYG W+ + K R GK CR RW NHL P+++K + T
Sbjct: 90 LIKGPWTKEEDQRVIELVQKYGPKRWSVIAKHLK-GRIGKQCRERWHNHLNPEVKKTSWT 148
Query: 136 AEEERRIIELHHKMGNKWAQMAALLPGRTDNEIKNFWNTRCKKR 179
EE+R I + H ++GN+WA++A LLPGRTDN IKN WN+ +++
Sbjct: 149 EEEDRIIYQAHKRLGNRWAEIAKLLPGRTDNAIKNHWNSTMRRK 192
>MYB_CHICK (P01103) Myb proto-oncogene protein (C-myb)
Length = 641
Score = 113 bits (283), Expect = 8e-25
Identities = 51/104 (49%), Positives = 71/104 (68%), Gaps = 1/104 (0%)
Query: 76 LKKGPWTAAEDEILVAHVQKYGEGNWNSVRKCTGLARCGKSCRLRWANHLRPDLRKGAIT 135
L KGPWT ED+ ++ VQKYG W+ + K R GK CR RW NHL P+++K + T
Sbjct: 90 LIKGPWTKEEDQRVIELVQKYGPKRWSVIAKHLK-GRIGKQCRERWHNHLNPEVKKTSWT 148
Query: 136 AEEERRIIELHHKMGNKWAQMAALLPGRTDNEIKNFWNTRCKKR 179
EE+R I + H ++GN+WA++A LLPGRTDN IKN WN+ +++
Sbjct: 149 EEEDRIIYQAHKRLGNRWAEIAKLLPGRTDNAIKNHWNSTMRRK 192
>MYB_BOVIN (P46200) Myb proto-oncogene protein (C-myb)
Length = 640
Score = 113 bits (283), Expect = 8e-25
Identities = 51/104 (49%), Positives = 71/104 (68%), Gaps = 1/104 (0%)
Query: 76 LKKGPWTAAEDEILVAHVQKYGEGNWNSVRKCTGLARCGKSCRLRWANHLRPDLRKGAIT 135
L KGPWT ED+ ++ VQKYG W+ + K R GK CR RW NHL P+++K + T
Sbjct: 90 LIKGPWTKEEDQRVIELVQKYGPKRWSVIAKHLK-GRIGKQCRERWHNHLNPEVKKTSWT 148
Query: 136 AEEERRIIELHHKMGNKWAQMAALLPGRTDNEIKNFWNTRCKKR 179
EE+R I + H ++GN+WA++A LLPGRTDN IKN WN+ +++
Sbjct: 149 EEEDRIIYQAHKRLGNRWAEIAKLLPGRTDNAIKNHWNSTMRRK 192
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.312 0.131 0.399
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 55,559,551
Number of Sequences: 164201
Number of extensions: 2599044
Number of successful extensions: 5894
Number of sequences better than 10.0: 68
Number of HSP's better than 10.0 without gapping: 46
Number of HSP's successfully gapped in prelim test: 22
Number of HSP's that attempted gapping in prelim test: 5762
Number of HSP's gapped (non-prelim): 99
length of query: 416
length of database: 59,974,054
effective HSP length: 113
effective length of query: 303
effective length of database: 41,419,341
effective search space: 12550060323
effective search space used: 12550060323
T: 11
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 67 (30.4 bits)
Medicago: description of AC148404.8


