

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148395.5 - phase: 0
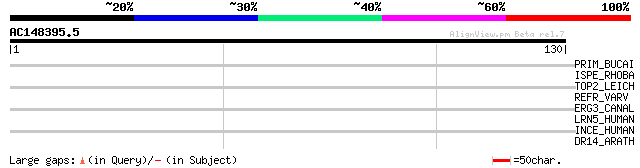
(130 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
PRIM_BUCAI (P57164) DNA primase (EC 2.7.7.-) 28 2.7
ISPE_RHOBA (Q7UEV3) 4-diphosphocytidyl-2-C-methyl-D-erythritol k... 28 2.7
TOP2_LEICH (O61078) DNA topoisomerase II (EC 5.99.1.3) 28 4.6
REFR_VARV (P33810) Rifampicin resistance protein (62 kDa protein) 28 4.6
ERG3_CANAL (O93875) C-5 sterol desaturase (EC 1.3.3.-) (Sterol-C... 28 4.6
LRN5_HUMAN (O75325) Leucine rich repeat neuronal protein 5 precu... 27 7.9
INCE_HUMAN (Q9NQS7) Inner centromere protein 27 7.9
DR14_ARATH (Q9SI85) Probable disease resistance protein At1g6263... 27 7.9
>PRIM_BUCAI (P57164) DNA primase (EC 2.7.7.-)
Length = 577
Score = 28.5 bits (62), Expect = 2.7
Identities = 28/101 (27%), Positives = 44/101 (42%), Gaps = 24/101 (23%)
Query: 1 MAEEHTLKEYATPSTEEPQTVI-----------VYPTIDANNFDIKPALLNIVQQNQFFE 49
++++ TLK P+ E+P T+I + I + F K L NI
Sbjct: 325 ISDKKTLKFILLPNQEDPDTIIRKEGREKFQKRIDNAITMSKFFFKNILKNI-------N 377
Query: 50 LPTEDPNLHISTF-LRLSGTLKSNGVTPETIRLHLFPFLLR 89
L ++D H+S L L T+ S +TIR++L L R
Sbjct: 378 LSSDDDKFHLSVHALPLINTISS-----DTIRIYLRQILAR 413
>ISPE_RHOBA (Q7UEV3) 4-diphosphocytidyl-2-C-methyl-D-erythritol
kinase (EC 2.7.1.148) (CMK)
(4-(cytidine-5'-diphospho)-2-C-methyl-D-erythritol
kinase)
Length = 329
Score = 28.5 bits (62), Expect = 2.7
Identities = 13/37 (35%), Positives = 22/37 (59%)
Query: 37 ALLNIVQQNQFFELPTEDPNLHISTFLRLSGTLKSNG 73
A L + + N+ +PT+D NL + RL+ TL+ +G
Sbjct: 70 AELQVDEDNELLLVPTDDKNLVVRALNRLTETLRLDG 106
>TOP2_LEICH (O61078) DNA topoisomerase II (EC 5.99.1.3)
Length = 1236
Score = 27.7 bits (60), Expect = 4.6
Identities = 22/77 (28%), Positives = 38/77 (48%), Gaps = 2/77 (2%)
Query: 8 KEYATPSTEEPQTVIVYPTIDANNFDIKPALLNIVQQNQFFELPTEDPNLHISTFLRLSG 67
KEY +P+ Q ++V DA+ IK ++N ++ + +L +P +IS F
Sbjct: 499 KEYRSPAELRYQRLLVMTDQDADGSHIKGLVINAF-ESLWPKLLQNNPG-YISLFSTPIV 556
Query: 68 TLKSNGVTPETIRLHLF 84
+K +G + E I H F
Sbjct: 557 KIKVSGKSKEVIAFHSF 573
>REFR_VARV (P33810) Rifampicin resistance protein (62 kDa protein)
Length = 551
Score = 27.7 bits (60), Expect = 4.6
Identities = 10/38 (26%), Positives = 24/38 (62%)
Query: 42 VQQNQFFELPTEDPNLHISTFLRLSGTLKSNGVTPETI 79
++++ F + ++ P L++ ++ LSG + +NG +TI
Sbjct: 16 IKRSNVFAVDSQIPTLYMPQYISLSGVMTNNGPDNQTI 53
>ERG3_CANAL (O93875) C-5 sterol desaturase (EC 1.3.3.-)
(Sterol-C5-desaturase) (Ergosterol delta 5,6 desaturase)
Length = 386
Score = 27.7 bits (60), Expect = 4.6
Identities = 26/97 (26%), Positives = 43/97 (43%), Gaps = 2/97 (2%)
Query: 23 VYPTIDANNFDIKPALLNIVQQNQFFELPTEDPNLHISTFLRLSGTLKSNGVTPETIRLH 82
V+P A + +KPA+ + Q F LP D + ++ L S + V P TI +
Sbjct: 20 VFPKDGAVHEFLKPAIQSF-SQIDFPSLPNLD-SFDTNSTLISSNNFNISNVNPATIPSY 77
Query: 83 LFPFLLRDRASAWFHSLPNGSITPWDQLRQAFLARSS 119
LF + + + + L D + +FLARS+
Sbjct: 78 LFSKIASYQDKSEIYGLAPKFFPATDFINTSFLARSN 114
>LRN5_HUMAN (O75325) Leucine rich repeat neuronal protein 5
precursor (Glioma amplified on chromosome 1 protein)
Length = 713
Score = 26.9 bits (58), Expect = 7.9
Identities = 13/23 (56%), Positives = 14/23 (60%)
Query: 79 IRLHLFPFLLRDRASAWFHSLPN 101
+RLHL LLR S WF LPN
Sbjct: 168 LRLHLNSNLLRAIDSRWFEMLPN 190
>INCE_HUMAN (Q9NQS7) Inner centromere protein
Length = 919
Score = 26.9 bits (58), Expect = 7.9
Identities = 19/70 (27%), Positives = 31/70 (44%), Gaps = 9/70 (12%)
Query: 12 TPSTEEPQTVIVYPTIDANNFDIKPALLNIVQQNQFFELPTEDPNLHISTFLRLSGTLKS 71
TPS+ P + +V P L+ VQ+NQ PT P + +F++ + L+
Sbjct: 478 TPSSPCPASKVVRPL---------RTFLHTVQRNQMLMTPTSAPRSVMKSFIKRNTPLRM 528
Query: 72 NGVTPETIRL 81
+ E RL
Sbjct: 529 DPKEKERQRL 538
>DR14_ARATH (Q9SI85) Probable disease resistance protein At1g62630
(pNd4)
Length = 893
Score = 26.9 bits (58), Expect = 7.9
Identities = 17/45 (37%), Positives = 23/45 (50%), Gaps = 2/45 (4%)
Query: 26 TIDANNFDIKP--ALLNIVQQNQFFELPTEDPNLHISTFLRLSGT 68
TI + F+ P A+L++ FELP E NL +L LS T
Sbjct: 560 TISSEFFNCMPKLAVLDLSHNQSLFELPEEISNLVSLKYLNLSHT 604
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.318 0.134 0.402
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 16,024,485
Number of Sequences: 164201
Number of extensions: 624329
Number of successful extensions: 1101
Number of sequences better than 10.0: 8
Number of HSP's better than 10.0 without gapping: 3
Number of HSP's successfully gapped in prelim test: 5
Number of HSP's that attempted gapping in prelim test: 1098
Number of HSP's gapped (non-prelim): 8
length of query: 130
length of database: 59,974,054
effective HSP length: 106
effective length of query: 24
effective length of database: 42,568,748
effective search space: 1021649952
effective search space used: 1021649952
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 58 (26.9 bits)
Medicago: description of AC148395.5


