

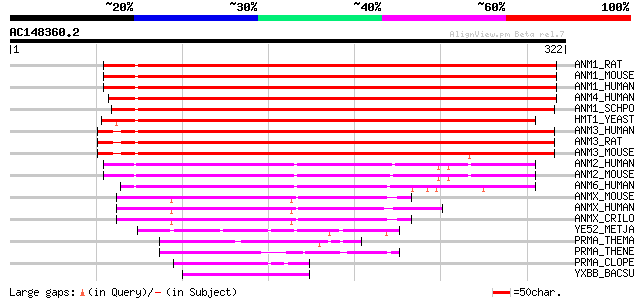
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148360.2 + phase: 0 /pseudo
(322 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
ANM1_RAT (Q63009) Protein arginine N-methyltransferase 1 (EC 2.1... 335 1e-91
ANM1_MOUSE (Q9JIF0) Protein arginine N-methyltransferase 1 (EC 2... 335 1e-91
ANM1_HUMAN (Q99873) Protein arginine N-methyltransferase 1 (EC 2... 333 3e-91
ANM4_HUMAN (Q9NR22) Protein arginine N-methyltransferase 4 (EC 2... 328 1e-89
ANM1_SCHPO (Q9URX7) Probable protein arginine N-methyltransferas... 308 2e-83
HMT1_YEAST (P38074) HNRNP arginine N-methyltransferase (EC 2.1.1... 271 1e-72
ANM3_HUMAN (O60678) Protein arginine N-methyltransferase 3 (EC 2... 244 2e-64
ANM3_RAT (O70467) Protein arginine N-methyltransferase 3 (EC 2.1... 244 3e-64
ANM3_MOUSE (Q922H1) Protein arginine N-methyltransferase 3 (EC 2... 239 9e-63
ANM2_HUMAN (P55345) Protein arginine N-methyltransferase 2 (EC 2... 174 3e-43
ANM2_MOUSE (Q9R144) Protein arginine N-methyltransferase 2 (EC 2... 173 6e-43
ANM6_HUMAN (Q96LA8) Protein arginine N-methyltransferase 6 (EC 2... 164 4e-40
ANMX_MOUSE (Q922X9) Protein arginine N-methyltransferase (EC 2.1... 63 1e-09
ANMX_HUMAN (Q9NVM4) Protein arginine N-methyltransferase (EC 2.1... 62 2e-09
ANMX_CRILO (Q99MI9) Protein arginine N-methyltransferase (EC 2.1... 62 3e-09
YE52_METJA (Q58847) Hypothetical protein MJ1452 57 8e-08
PRMA_THEMA (Q9X0G8) Ribosomal protein L11 methyltransferase (EC ... 47 6e-05
PRMA_THENE (O86951) Ribosomal protein L11 methyltransferase (EC ... 47 8e-05
PRMA_CLOPE (Q8XIT6) Ribosomal protein L11 methyltransferase (EC ... 45 3e-04
YXBB_BACSU (P46326) Hypothetical protein yxbB 44 4e-04
>ANM1_RAT (Q63009) Protein arginine N-methyltransferase 1 (EC
2.1.1.-)
Length = 353
Score = 335 bits (858), Expect = 1e-91
Identities = 160/263 (60%), Positives = 203/263 (76%), Gaps = 1/263 (0%)
Query: 55 DSNDKTSADYYFDSYSHFGNFHKEMLKDTVRTKTYQNVIYQNRFLFKNKVVLDVGAGTGI 114
++ D TS DYYFDSY+HFG H+EMLKD VRT TY+N ++ NR LFK+KVVLDVG+GTGI
Sbjct: 25 NAEDMTSKDYYFDSYAHFG-IHEEMLKDEVRTLTYRNSMFHNRHLFKDKVVLDVGSGTGI 83
Query: 115 LSLFCAKAGAAHVYAVECSHMADRAKEIVETNGYSKVITVLKGKIEELELPVPKVDIIIS 174
L +F AKAGA V +ECS ++D A +IV+ N V+T++KGK+EE+ELPV KVDIIIS
Sbjct: 84 LCMFAAKAGARKVIGIECSSISDYAVKIVKANKLDHVVTIIKGKVEEVELPVEKVDIIIS 143
Query: 175 EWMGYFLLFENMLNSVLFARDKWLVDDGVILPDIASLYLTAIEDKDYKEDKIEFWNNVYG 234
EWMGY L +E+MLN+VL ARDKWL DG+I PD A+LY+TAIED+ YK+ KI +W NVYG
Sbjct: 144 EWMGYCLFYESMLNTVLHARDKWLAPDGLIFPDRATLYVTAIEDRQYKDYKIHWWENVYG 203
Query: 235 FDMSCIKKQALMEPLVDTVDQNQIATNCQLLKSMDISKMSSGDCSFTAPFKLVAARDDFI 294
FDMSCIK A+ EPLVD VD Q+ TN L+K +DI + D +FT+PF L R+D++
Sbjct: 204 FDMSCIKDVAIKEPLVDVVDPKQLVTNACLIKEVDIYTVKVEDLTFTSPFCLQVKRNDYV 263
Query: 295 HAFVAYFDVSFTKCHKLMGFSTA 317
HA VAYF++ FT+CHK GFST+
Sbjct: 264 HALVAYFNIEFTRCHKRTGFSTS 286
>ANM1_MOUSE (Q9JIF0) Protein arginine N-methyltransferase 1 (EC
2.1.1.-)
Length = 371
Score = 335 bits (858), Expect = 1e-91
Identities = 160/263 (60%), Positives = 203/263 (76%), Gaps = 1/263 (0%)
Query: 55 DSNDKTSADYYFDSYSHFGNFHKEMLKDTVRTKTYQNVIYQNRFLFKNKVVLDVGAGTGI 114
++ D TS DYYFDSY+HFG H+EMLKD VRT TY+N ++ NR LFK+KVVLDVG+GTGI
Sbjct: 43 NAEDMTSKDYYFDSYAHFG-IHEEMLKDEVRTLTYRNSMFHNRHLFKDKVVLDVGSGTGI 101
Query: 115 LSLFCAKAGAAHVYAVECSHMADRAKEIVETNGYSKVITVLKGKIEELELPVPKVDIIIS 174
L +F AKAGA V +ECS ++D A +IV+ N V+T++KGK+EE+ELPV KVDIIIS
Sbjct: 102 LCMFAAKAGARKVIGIECSSISDYAVKIVKANKLDHVVTIIKGKVEEVELPVEKVDIIIS 161
Query: 175 EWMGYFLLFENMLNSVLFARDKWLVDDGVILPDIASLYLTAIEDKDYKEDKIEFWNNVYG 234
EWMGY L +E+MLN+VL ARDKWL DG+I PD A+LY+TAIED+ YK+ KI +W NVYG
Sbjct: 162 EWMGYCLFYESMLNTVLHARDKWLAPDGLIFPDRATLYVTAIEDRQYKDYKIHWWENVYG 221
Query: 235 FDMSCIKKQALMEPLVDTVDQNQIATNCQLLKSMDISKMSSGDCSFTAPFKLVAARDDFI 294
FDMSCIK A+ EPLVD VD Q+ TN L+K +DI + D +FT+PF L R+D++
Sbjct: 222 FDMSCIKDVAIKEPLVDVVDPKQLVTNACLIKEVDIYTVKVEDLTFTSPFCLQVKRNDYV 281
Query: 295 HAFVAYFDVSFTKCHKLMGFSTA 317
HA VAYF++ FT+CHK GFST+
Sbjct: 282 HALVAYFNIEFTRCHKRTGFSTS 304
>ANM1_HUMAN (Q99873) Protein arginine N-methyltransferase 1 (EC
2.1.1.-) (Interferon receptor 1-bound protein 4)
Length = 361
Score = 333 bits (855), Expect = 3e-91
Identities = 159/263 (60%), Positives = 203/263 (76%), Gaps = 1/263 (0%)
Query: 55 DSNDKTSADYYFDSYSHFGNFHKEMLKDTVRTKTYQNVIYQNRFLFKNKVVLDVGAGTGI 114
++ D TS DYYFDSY+HFG H+EMLKD VRT TY+N ++ NR LFK+KVVLDVG+GTGI
Sbjct: 33 NAEDMTSKDYYFDSYAHFG-IHEEMLKDEVRTLTYRNSMFHNRHLFKDKVVLDVGSGTGI 91
Query: 115 LSLFCAKAGAAHVYAVECSHMADRAKEIVETNGYSKVITVLKGKIEELELPVPKVDIIIS 174
L +F AKAGA V + CS ++D A +IV+ N V+T++KGK+EE+ELPV KVDIIIS
Sbjct: 92 LCMFAAKAGARKVIGIVCSSISDYAVKIVKANKLDHVVTIIKGKVEEVELPVEKVDIIIS 151
Query: 175 EWMGYFLLFENMLNSVLFARDKWLVDDGVILPDIASLYLTAIEDKDYKEDKIEFWNNVYG 234
EWMGY L +E+MLN+VL+ARDKWL DG+I PD A+LY+TAIED+ YK+ KI +W NVYG
Sbjct: 152 EWMGYCLFYESMLNTVLYARDKWLAPDGLIFPDRATLYVTAIEDRQYKDYKIHWWENVYG 211
Query: 235 FDMSCIKKQALMEPLVDTVDQNQIATNCQLLKSMDISKMSSGDCSFTAPFKLVAARDDFI 294
FDMSCIK A+ EPLVD VD Q+ TN L+K +DI + D +FT+PF L R+D++
Sbjct: 212 FDMSCIKDVAIKEPLVDVVDPKQLVTNACLIKEVDIYTVKVEDLTFTSPFCLQVKRNDYV 271
Query: 295 HAFVAYFDVSFTKCHKLMGFSTA 317
HA VAYF++ FT+CHK GFST+
Sbjct: 272 HALVAYFNIEFTRCHKRTGFSTS 294
>ANM4_HUMAN (Q9NR22) Protein arginine N-methyltransferase 4 (EC
2.1.1.-) (Heterogeneous nuclear ribonucleoprotein
methyltransferase-like protein 4)
Length = 334
Score = 328 bits (840), Expect = 1e-89
Identities = 153/260 (58%), Positives = 202/260 (76%), Gaps = 1/260 (0%)
Query: 58 DKTSADYYFDSYSHFGNFHKEMLKDTVRTKTYQNVIYQNRFLFKNKVVLDVGAGTGILSL 117
+ TS DYYFDSY+HFG H+EMLKD VRT TY+N +Y N+ +FK+KVVLDVG+GTGILS+
Sbjct: 9 EMTSRDYYFDSYAHFG-IHEEMLKDEVRTLTYRNSMYHNKHVFKDKVVLDVGSGTGILSM 67
Query: 118 FCAKAGAAHVYAVECSHMADRAKEIVETNGYSKVITVLKGKIEELELPVPKVDIIISEWM 177
F AKAGA V+ +ECS ++D +++I++ N +IT+ KGK+EE+ELPV KVDIIISEWM
Sbjct: 68 FAAKAGAKKVFGIECSSISDYSEKIIKANHLDNIITIFKGKVEEVELPVEKVDIIISEWM 127
Query: 178 GYFLLFENMLNSVLFARDKWLVDDGVILPDIASLYLTAIEDKDYKEDKIEFWNNVYGFDM 237
GY L +E+MLN+V+FARDKWL G++ PD A+LY+ AIED+ YK+ KI +W NVYGFDM
Sbjct: 128 GYCLFYESMLNTVIFARDKWLKPGGLMFPDRAALYVVAIEDRQYKDFKIHWWENVYGFDM 187
Query: 238 SCIKKQALMEPLVDTVDQNQIATNCQLLKSMDISKMSSGDCSFTAPFKLVAARDDFIHAF 297
+CI+ A+ EPLVD VD Q+ TN L+K +DI + + + SFT+ F L R+D++HA
Sbjct: 188 TCIRDVAMKEPLVDIVDPKQVVTNACLIKEVDIYTVKTEELSFTSAFCLQIQRNDYVHAL 247
Query: 298 VAYFDVSFTKCHKLMGFSTA 317
V YF++ FTKCHK MGFSTA
Sbjct: 248 VTYFNIEFTKCHKKMGFSTA 267
>ANM1_SCHPO (Q9URX7) Probable protein arginine N-methyltransferase
(EC 2.1.1.-)
Length = 339
Score = 308 bits (788), Expect = 2e-83
Identities = 145/257 (56%), Positives = 198/257 (76%), Gaps = 1/257 (0%)
Query: 60 TSADYYFDSYSHFGNFHKEMLKDTVRTKTYQNVIYQNRFLFKNKVVLDVGAGTGILSLFC 119
T+ DYYFDSYSH+G H+EMLKD VRT +Y++ I QN LF++K+VLDVG GTGILS+FC
Sbjct: 14 TAKDYYFDSYSHWG-IHEEMLKDDVRTLSYRDAIMQNPHLFRDKIVLDVGCGTGILSMFC 72
Query: 120 AKAGAAHVYAVECSHMADRAKEIVETNGYSKVITVLKGKIEELELPVPKVDIIISEWMGY 179
A+AGA HVY V+ S + +A +IVE N S IT+++GK+EE++LPV KVDII+SEWMGY
Sbjct: 73 ARAGAKHVYGVDMSEIIHKAVQIVEVNKLSDRITLIQGKMEEIQLPVEKVDIIVSEWMGY 132
Query: 180 FLLFENMLNSVLFARDKWLVDDGVILPDIASLYLTAIEDKDYKEDKIEFWNNVYGFDMSC 239
FLL+E+ML++VL ARD++L DG++ PD A + L AIED DYK +KI FW++VYGFD S
Sbjct: 133 FLLYESMLDTVLVARDRYLAPDGLLFPDRAQIQLAAIEDADYKSEKIGFWDDVYGFDFSP 192
Query: 240 IKKQALMEPLVDTVDQNQIATNCQLLKSMDISKMSSGDCSFTAPFKLVAARDDFIHAFVA 299
IKK EPLVDTVD+ + TN ++ +D+ + D +F++PF++ A R+DF+HAF+A
Sbjct: 193 IKKDVWKEPLVDTVDRIAVNTNSCVILDLDLKTVKKEDLAFSSPFEITATRNDFVHAFLA 252
Query: 300 YFDVSFTKCHKLMGFST 316
+FD+ F+ CHK + FST
Sbjct: 253 WFDIEFSACHKPIKFST 269
>HMT1_YEAST (P38074) HNRNP arginine N-methyltransferase (EC 2.1.1.-)
(ODP1 protein)
Length = 348
Score = 271 bits (694), Expect = 1e-72
Identities = 131/256 (51%), Positives = 183/256 (71%), Gaps = 5/256 (1%)
Query: 54 DDSNDKT----SADYYFDSYSHFGNFHKEMLKDTVRTKTYQNVIYQNRFLFKNKVVLDVG 109
D + +KT S +YF+SY H+G H+EML+DTVRT +Y+N I QN+ LFK+K+VLDVG
Sbjct: 8 DSATEKTKLSESEQHYFNSYDHYG-IHEEMLQDTVRTLSYRNAIIQNKDLFKDKIVLDVG 66
Query: 110 AGTGILSLFCAKAGAAHVYAVECSHMADRAKEIVETNGYSKVITVLKGKIEELELPVPKV 169
GTGILS+F AK GA HV V+ S + + AKE+VE NG+S IT+L+GK+E++ LP PKV
Sbjct: 67 CGTGILSMFAAKHGAKHVIGVDMSSIIEMAKELVELNGFSDKITLLRGKLEDVHLPFPKV 126
Query: 170 DIIISEWMGYFLLFENMLNSVLFARDKWLVDDGVILPDIASLYLTAIEDKDYKEDKIEFW 229
DIIISEWMGYFLL+E+M+++VL+ARD +LV+ G+I PD S++L +ED YK++K+ +W
Sbjct: 127 DIIISEWMGYFLLYESMMDTVLYARDHYLVEGGLIFPDKCSIHLAGLEDSQYKDEKLNYW 186
Query: 230 NNVYGFDMSCIKKQALMEPLVDTVDQNQIATNCQLLKSMDISKMSSGDCSFTAPFKLVAA 289
+VYGFD S L EP+VDTV++N + T L D++ + D +F + FKL A
Sbjct: 187 QDVYGFDYSPFVPLVLHEPIVDTVERNNVNTTSDKLIEFDLNTVKISDLAFKSNFKLTAK 246
Query: 290 RDDFIHAFVAYFDVSF 305
R D I+ V +FD+ F
Sbjct: 247 RQDMINGIVTWFDIVF 262
>ANM3_HUMAN (O60678) Protein arginine N-methyltransferase 3 (EC
2.1.1.-) (Heterogeneous nuclear ribonucleoprotein
methyltransferase-like protein 3)
Length = 531
Score = 244 bits (623), Expect = 2e-64
Identities = 126/266 (47%), Positives = 173/266 (64%), Gaps = 6/266 (2%)
Query: 52 DLDDSNDKTSADYYFDSYSHFGNFHKEMLKDTVRTKTYQNVIYQNRFLFKNKVVLDVGAG 111
DL + D YF SY H+G H+EMLKD +RT++Y++ IYQN +FK+KVVLDVG G
Sbjct: 211 DLQEDEDGV----YFSSYGHYG-IHEEMLKDKIRTESYRDFIYQNPHIFKDKVVLDVGCG 265
Query: 112 TGILSLFCAKAGAAHVYAVECSHMADRAKEIVETNGYSKVITVLKGKIEELELPVPKVDI 171
TGILS+F AKAGA V V+ S + +A +I+ N IT++KGKIEE+ LPV KVD+
Sbjct: 266 TGILSMFAAKAGAKKVLGVDQSEILYQAMDIIRLNKLEDTITLIKGKIEEVHLPVEKVDV 325
Query: 172 IISEWMGYFLLFENMLNSVLFARDKWLVDDGVILPDIASLYLTAIEDKDYKEDKIEFWNN 231
IISEWMGYFLLFE+ML+SVL+A++K+L G + PDI ++ L A+ D + D+I FW++
Sbjct: 326 IISEWMGYFLLFESMLDSVLYAKNKYLAKGGSVYPDICTISLVAVSDVNKHADRIAFWDD 385
Query: 232 VYGFDMSCIKKQALMEPLVDTVDQNQIATNCQLLKSMDISKMSSGDCSFTAPFKLVAARD 291
VYGF MSC+KK + E +V+ +D + + +K +D S D F++ F L R
Sbjct: 386 VYGFKMSCMKKAVIPEAVVEVLDPKTLISEPCGIKHIDCHTTSISDLEFSSDFTLKITRT 445
Query: 292 DFIHAFVAYFDVSFTK-CHKLMGFST 316
A YFD+ F K CH + FST
Sbjct: 446 SMCTAIAGYFDIYFEKNCHNRVVFST 471
>ANM3_RAT (O70467) Protein arginine N-methyltransferase 3 (EC
2.1.1.-) (Heterogeneous nuclear ribonucleoprotein
methyltransferase-like protein 3)
Length = 528
Score = 244 bits (622), Expect = 3e-64
Identities = 127/266 (47%), Positives = 173/266 (64%), Gaps = 6/266 (2%)
Query: 52 DLDDSNDKTSADYYFDSYSHFGNFHKEMLKDTVRTKTYQNVIYQNRFLFKNKVVLDVGAG 111
DL + D YF SY H+G H+EMLKD VRT++Y++ IYQN +FK+KVVLDVG G
Sbjct: 208 DLQEDEDGV----YFSSYGHYG-IHEEMLKDKVRTESYRDFIYQNPHIFKDKVVLDVGCG 262
Query: 112 TGILSLFCAKAGAAHVYAVECSHMADRAKEIVETNGYSKVITVLKGKIEELELPVPKVDI 171
TGILS+F AKAGA V AV+ S + +A +I+ N I ++KGKIEE+ LPV KVD+
Sbjct: 263 TGILSMFAAKAGAKKVIAVDQSEILYQAMDIIRLNKLEDTIVLIKGKIEEVSLPVEKVDV 322
Query: 172 IISEWMGYFLLFENMLNSVLFARDKWLVDDGVILPDIASLYLTAIEDKDYKEDKIEFWNN 231
IISEWMGYFLLFE+ML+SVL+A+ K+L G + PDI ++ L A+ D D+I FW++
Sbjct: 323 IISEWMGYFLLFESMLDSVLYAKSKYLAKGGSVYPDICTISLVAVSDVSKHADRIAFWDD 382
Query: 232 VYGFDMSCIKKQALMEPLVDTVDQNQIATNCQLLKSMDISKMSSGDCSFTAPFKLVAARD 291
VYGF+MSC+KK + E +V+ VD + ++ +K +D S D F++ F L +
Sbjct: 383 VYGFNMSCMKKAVIPEAVVEVVDHKTLISDPCDIKHIDCHTTSISDLEFSSDFTLRTTKT 442
Query: 292 DFIHAFVAYFDVSFTK-CHKLMGFST 316
A YFD+ F K CH + FST
Sbjct: 443 AMCTAVAGYFDIYFEKNCHNRVVFST 468
>ANM3_MOUSE (Q922H1) Protein arginine N-methyltransferase 3 (EC
2.1.1.-) (Heterogeneous nuclear ribonucleoprotein
methyltransferase-like protein 3)
Length = 532
Score = 239 bits (609), Expect = 9e-63
Identities = 127/270 (47%), Positives = 173/270 (64%), Gaps = 10/270 (3%)
Query: 52 DLDDSNDKTSADYYFDSYSHFGNFHKEMLKDTVRTKTYQNVIYQNRFLFKNKVVLDVGAG 111
DL + D YF SY H+G H+EMLKD VRT++Y++ IYQN +FK+KVVLDVG G
Sbjct: 208 DLQEDEDGV----YFSSYGHYG-IHEEMLKDKVRTESYRDFIYQNPHIFKDKVVLDVGCG 262
Query: 112 TGILSLFCAKAGAAHVYAVECSHMADRAKEIVETNGYSKVITVLKGKIEELELPVPKVDI 171
TGILS+F AK GA V AV+ S + +A +I+ N I ++KGKIEE+ LPV KVD+
Sbjct: 263 TGILSMFAAKVGAKKVIAVDQSEILYQAMDIIRLNKLEDTIVLIKGKIEEVSLPVEKVDV 322
Query: 172 IISEWMGYFLLFENMLNSVLFARDKWLVDDGVILPDIASLYLTAIEDKDYKEDKIEFWNN 231
IISEWMGYFLLFE+ML+SVL+A+ K+L G + PDI ++ L A+ D D+I FW++
Sbjct: 323 IISEWMGYFLLFESMLDSVLYAKSKYLAKGGSVYPDICTISLVAVSDVSKHADRIAFWDD 382
Query: 232 VYGFDMSCIKKQALMEPLVDTVDQNQIATN-CQLL---KSMDISKMSSGDCSFTAPFKLV 287
VYGF+MSC+KK + E +V+ VD + ++ C + K +D S D F++ F L
Sbjct: 383 VYGFNMSCMKKAVIPEAVVEVVDHKTLISDPCDIKMDGKHIDCHTTSISDLEFSSDFTLR 442
Query: 288 AARDDFIHAFVAYFDVSFTK-CHKLMGFST 316
+ A YFD+ F K CH + FST
Sbjct: 443 TTKTAMCTAVAGYFDIYFEKNCHNRVVFST 472
>ANM2_HUMAN (P55345) Protein arginine N-methyltransferase 2 (EC
2.1.1.-)
Length = 433
Score = 174 bits (441), Expect = 3e-43
Identities = 105/260 (40%), Positives = 153/260 (58%), Gaps = 13/260 (5%)
Query: 55 DSNDKTSADYYFDSYSHFGNFHKEMLKDTVRTKTYQNVIYQNRFLFKNKVVLDVGAGTGI 114
D D + YF SY H EML D RT Y +VI QN+ +KV+LDVG GTGI
Sbjct: 92 DPEDTWQDEEYFGSYGTL-KLHLEMLADQPRTTKYHSVILQNKESLTDKVILDVGCGTGI 150
Query: 115 LSLFCAK-AGAAHVYAVECSHMADRAKEIVETNGYSKVITVLKGKIEELELPVPKVDIII 173
+SLFCA A VYAVE S MA ++V NG++ +ITV + K+E++ LP KVD+++
Sbjct: 151 ISLFCAHYARPRAVYAVEASEMAQHTGQLVLQNGFADIITVYQQKVEDVVLP-EKVDVLV 209
Query: 174 SEWMGYFLLFENMLNSVLFARDKWLVDDGVILPDIASLYLTAIE-DKDYKEDKIEFWNNV 232
SEWMG LLFE M+ S+L+ARD WL +DGVI P +A+L+L DKDY+ K+ FW+N
Sbjct: 210 SEWMGTCLLFEFMIESILYARDAWLKEDGVIWPTMAALHLVPCSADKDYR-SKVLFWDNA 268
Query: 233 YGFDMSCIKKQALME----PLVDTV--DQNQIATNCQLLKSMDISKMSSGDC-SFTAPFK 285
Y F++S +K A+ E P + + ++ ++ C +L+ +D+ + D + +
Sbjct: 269 YEFNLSALKSLAVKEFFSKPKYNHILKPEDCLSEPCTILQ-LDMRTVQISDLETLRGELR 327
Query: 286 LVAARDDFIHAFVAYFDVSF 305
+ +H F A+F V F
Sbjct: 328 FDIRKAGTLHGFTAWFSVHF 347
>ANM2_MOUSE (Q9R144) Protein arginine N-methyltransferase 2 (EC
2.1.1.-)
Length = 448
Score = 173 bits (438), Expect = 6e-43
Identities = 105/260 (40%), Positives = 151/260 (57%), Gaps = 13/260 (5%)
Query: 55 DSNDKTSADYYFDSYSHFGNFHKEMLKDTVRTKTYQNVIYQNRFLFKNKVVLDVGAGTGI 114
D D + YFDSY H EML D RT Y +VI QN+ K+KV+LDVG GTGI
Sbjct: 104 DPEDTWQDEEYFDSYGTL-KLHLEMLADQPRTTKYHSVILQNKESLKDKVILDVGCGTGI 162
Query: 115 LSLFCAK-AGAAHVYAVECSHMADRAKEIVETNGYSKVITVLKGKIEELELPVPKVDIII 173
+SLFCA A VYAVE S MA ++V NG++ ITV + K+E++ LP KVD+++
Sbjct: 163 ISLFCAHHARPKAVYAVEASDMAQHTSQLVLQNGFADTITVFQQKVEDVVLP-EKVDVLV 221
Query: 174 SEWMGYFLLFENMLNSVLFARDKWLVDDGVILPDIASLYLTAIE-DKDYKEDKIEFWNNV 232
SEWMG LLFE M+ S+L+ARD WL DG+I P A+L+L +KDY K+ FW+N
Sbjct: 222 SEWMGTCLLFEFMIESILYARDTWLKGDGIIWPTTAALHLVPCSAEKDY-HSKVLFWDNA 280
Query: 233 YGFDMSCIKKQALME----PLVDTV--DQNQIATNCQLLKSMDISKMSSGDC-SFTAPFK 285
Y F++S +K A+ E P + + ++ ++ C +L+ +D+ + D + +
Sbjct: 281 YEFNLSALKSLAIKEFFSRPKSNHILKPEDCLSEPCTILQ-LDMRTVQVPDLETMRGELR 339
Query: 286 LVAARDDFIHAFVAYFDVSF 305
+ +H F A+F V F
Sbjct: 340 FDIQKAGTLHGFTAWFSVYF 359
>ANM6_HUMAN (Q96LA8) Protein arginine N-methyltransferase 6 (EC
2.1.1.-) (Heterogeneous nuclear ribonucleoprotein
methyltransferase-like protein 6)
Length = 375
Score = 164 bits (414), Expect = 4e-40
Identities = 101/253 (39%), Positives = 146/253 (56%), Gaps = 15/253 (5%)
Query: 65 YFDSYSHFGNFHKEMLKDTVRTKTYQNVIYQNRFLFKNKVVLDVGAGTGILSLFCAKAGA 124
Y++ YS + H+EM+ D VRT Y+ I +N + K VLDVGAGTGILS+FCA+AGA
Sbjct: 47 YYECYSDV-SVHEEMIADRVRTDAYRLGILRNWAALRGKTVLDVGAGTGILSIFCAQAGA 105
Query: 125 AHVYAVECSHMADRAKEIVETNGYSKVITVLKGKIEELELPVPKVDIIISEWMGYFLLFE 184
VYAVE S + +A+E+V NG + VL G +E +ELP +VD I+SEWMGY LL E
Sbjct: 106 RRVYAVEASAIWQQAREVVRFNGLEDRVHVLPGPVETVELP-EQVDAIVSEWMGYGLLHE 164
Query: 185 NMLNSVLFARDKWLVDDGVILPDIASLYLTAIEDKDYKEDKIEFWNNV---YGFDMSCIK 241
+ML+SVL AR KWL + G++LP A L++ I D+ E ++ FW+ V YG DMSC++
Sbjct: 165 SMLSSVLHARTKWLKEGGLLLPASAELFIAPISDQ-MLEWRLGFWSQVKQHYGVDMSCLE 223
Query: 242 ---KQALM---EPLVDTVDQNQIATNCQLLKSMDISKM---SSGDCSFTAPFKLVAARDD 292
+ LM E +V + + Q +++S+ + F+
Sbjct: 224 GFATRCLMGHSEIVVQGLSGEDVLARPQRFAQLELSRAGLEQELEAGVGGRFRCSCYGSA 283
Query: 293 FIHAFVAYFDVSF 305
+H F +F V+F
Sbjct: 284 PMHGFAIWFQVTF 296
>ANMX_MOUSE (Q922X9) Protein arginine N-methyltransferase (EC
2.1.1.-)
Length = 692
Score = 62.8 bits (151), Expect = 1e-09
Identities = 53/183 (28%), Positives = 91/183 (48%), Gaps = 18/183 (9%)
Query: 63 DYYFDSYSHFG-NFHKEMLKDTVRT-KTYQNV---IYQNRFLFKNKVVLDVGAGTGILSL 117
D ++D + + + +ML D R K YQ + + + + + +VLD+G GTG+LS+
Sbjct: 21 DEHYDYHQEIARSSYADMLHDKDRNIKYYQGIRAAVSRVKDRGQKALVLDIGTGTGLLSM 80
Query: 118 FCAKAGAAHVYAVEC-SHMADRAKEIVETNGYSKVITVLKGKIEEL------ELPVPKVD 170
AGA YA+E MA+ A +IVE NG+S I V+ E+ +LP + +
Sbjct: 81 MAVTAGADFCYAIEVFKPMAEAAVKIVERNGFSDKIKVINKHSTEVTVGPDGDLPC-RAN 139
Query: 171 IIISEWMGYFLLFENMLNSVLFARDKWLVDDGVILPDIASLYLTAIEDKDYKEDKIEFWN 230
I+I+E L+ E L S A + +D +P A++Y +E + ++ WN
Sbjct: 140 ILITELFDTELIGEGALPSYEHAHKHLVQEDCEAVPHRATVYAQLVESR-----RMWSWN 194
Query: 231 NVY 233
++
Sbjct: 195 KLF 197
Score = 35.4 bits (80), Expect = 0.19
Identities = 40/154 (25%), Positives = 64/154 (40%), Gaps = 22/154 (14%)
Query: 80 LKDTVRTKTYQNVIYQNRFLFKNKVVLDVGAGTGILSLFCAKAGAAHVYAVECSHMADRA 139
+ D RT Y + L V L V G+ +LS+ GA V+ VE S + R
Sbjct: 382 INDQDRTDHYAQAL--RTVLLPGSVCLCVSDGS-LLSMLAHHLGAEQVFTVESSVASYRL 438
Query: 140 -KEIVETNGYSKVITVLKGKIEEL---ELPVPKVDIIISE---------WMGYFLLFENM 186
K I + N I+V+ + E L +L KV +++ E W + +
Sbjct: 439 MKRIFKVNHLEDKISVINKRPELLTAADLEGKKVSLLLGEPFFTTSLLPWHNLYFWY--- 495
Query: 187 LNSVLFARDKWLVDDGVILPDIASLYLTAIEDKD 220
V + D+ L V++P ASL+ +E +D
Sbjct: 496 ---VRTSVDQHLAPGAVVMPQAASLHAVIVEFRD 526
>ANMX_HUMAN (Q9NVM4) Protein arginine N-methyltransferase (EC
2.1.1.-)
Length = 692
Score = 62.0 bits (149), Expect = 2e-09
Identities = 54/201 (26%), Positives = 97/201 (47%), Gaps = 18/201 (8%)
Query: 63 DYYFDSYSHFG-NFHKEMLKDTVRT-KTYQNV---IYQNRFLFKNKVVLDVGAGTGILSL 117
D ++D + + + +ML D R K YQ + + + + + +VLD+G GTG+LS+
Sbjct: 21 DEHYDYHQEIARSSYADMLHDKDRNVKYYQGIRAAVSRVKDRGQKALVLDIGTGTGLLSM 80
Query: 118 FCAKAGAAHVYAVEC-SHMADRAKEIVETNGYSKVITVLKGKIEEL------ELPVPKVD 170
AGA YA+E MAD A +IVE NG+S I V+ E+ ++P + +
Sbjct: 81 MAVTAGADFCYAIEVFKPMADAAVKIVEKNGFSDKIKVINKHSTEVTVGPEGDMPC-RAN 139
Query: 171 IIISEWMGYFLLFENMLNSVLFARDKWLVDDGVILPDIASLYLTAIEDKDYKEDKIEFWN 230
I+++E L+ E L S A + ++ +P A++Y +E ++ WN
Sbjct: 140 ILVTELFDTELIGEGALPSYEHAHRHLVEENCEAVPHRATVYAQLVE-----SGRMWSWN 194
Query: 231 NVYGFDMSCIKKQALMEPLVD 251
++ + + ++ P VD
Sbjct: 195 KLFPIHVQTSLGEQVIVPPVD 215
Score = 30.4 bits (67), Expect = 6.2
Identities = 36/154 (23%), Positives = 65/154 (41%), Gaps = 22/154 (14%)
Query: 80 LKDTVRTKTYQNVIYQNRFLFKNKVVLDVGAGTGILSLFCAKAGAAHVYAVECSHMADRA 139
+ D RT Y + L + V L V G+ +LS+ G V+ VE S + +
Sbjct: 382 INDQDRTDRYVQAL--RTVLKPDSVCLCVSDGS-LLSVLAHHLGVEQVFTVESSAASHKL 438
Query: 140 -KEIVETNGYSKVITVLKGKIEEL---ELPVPKVDIIISE---------WMGYFLLFENM 186
++I + N I +++ + E L +L KV +++ E W + +
Sbjct: 439 LRKIFKANHLEDKINIIEKRPELLTNEDLQGRKVSLLLGEPFFTTSLLPWHNLYFWY--- 495
Query: 187 LNSVLFARDKWLVDDGVILPDIASLYLTAIEDKD 220
V A D+ L +++P ASL+ +E +D
Sbjct: 496 ---VRTAVDQHLGPGAMVMPQAASLHAVVVEFRD 526
>ANMX_CRILO (Q99MI9) Protein arginine N-methyltransferase (EC
2.1.1.-)
Length = 729
Score = 61.6 bits (148), Expect = 3e-09
Identities = 52/183 (28%), Positives = 91/183 (49%), Gaps = 18/183 (9%)
Query: 63 DYYFDSYSHFG-NFHKEMLKDTVRT-KTYQNV---IYQNRFLFKNKVVLDVGAGTGILSL 117
D ++D + + + +ML D R K YQ + + + + + +VLD+G GTG+LS+
Sbjct: 58 DEHYDYHQEIARSSYADMLHDKDRNIKYYQGIRAAVSRVKDRGQKALVLDIGTGTGLLSM 117
Query: 118 FCAKAGAAHVYAVEC-SHMADRAKEIVETNGYSKVITVLKGKIEEL------ELPVPKVD 170
AGA YA+E MAD A +IVE NG+S I V+ E+ +LP + +
Sbjct: 118 MAVTAGADFCYAIEVFKPMADAAVKIVEKNGFSDKIKVINKHSTEVTVGPDGDLPC-RAN 176
Query: 171 IIISEWMGYFLLFENMLNSVLFARDKWLVDDGVILPDIASLYLTAIEDKDYKEDKIEFWN 230
I+++E L+ E L S A + ++ +P A++Y +E + ++ WN
Sbjct: 177 ILVTELFDTELIGEGALPSYEHAHRHLVQENCEAVPHKATVYAQLVESR-----RMWSWN 231
Query: 231 NVY 233
++
Sbjct: 232 KLF 234
Score = 38.5 bits (88), Expect = 0.023
Identities = 42/150 (28%), Positives = 68/150 (45%), Gaps = 14/150 (9%)
Query: 80 LKDTVRTKTYQNVIYQNRFLFKNKVVLDVGAGTGILSLFCAKAGAAHVYAVECSHMADRA 139
+ D RT Y + L + L V G+ +LSL GA V+ VE S + R
Sbjct: 419 INDQDRTDQYAQAL--RTVLMPGTICLCVSDGS-LLSLLAHHLGAEQVFTVESSAASYRL 475
Query: 140 -KEIVETNGYSKVITVLKGKIEEL---ELPVPKVDIIISE--WMGYFLLFENMLNSVLFA 193
K I + N ++++K + E L +L KV +++ E + L + N+ +A
Sbjct: 476 MKRIFKANHLEDKVSIIKKRPELLTSADLEGKKVSLLLGEPFFATSLLPWHNLY--FWYA 533
Query: 194 R---DKWLVDDGVILPDIASLYLTAIEDKD 220
R D+ L V++P ASLY +E +D
Sbjct: 534 RTSVDQHLEPGAVVMPQAASLYAMIVEFRD 563
>YE52_METJA (Q58847) Hypothetical protein MJ1452
Length = 259
Score = 56.6 bits (135), Expect = 8e-08
Identities = 47/164 (28%), Positives = 84/164 (50%), Gaps = 19/164 (11%)
Query: 75 FHKEMLKDTVRTKTYQNVIYQNRFLFKNKVVLDVGAGTGILSLFCAKAGAAHVYAVECSH 134
+H +L D R ++N I R + ++ VV D+G G+GIL++ AK A VYA+E
Sbjct: 10 WHYSLLTDYERLAIFKNAI--ERVVDEDDVVFDLGTGSGILAMIAAKK-AKKVYAIELDP 66
Query: 135 MA-DRAKEIVETNGYSKVITVLKGKIEELELPVPKVDIIISEWMGYFLLFE---NMLNSV 190
D AKE ++ NG++ I +++G K D++I+E + L+ E ++NS+
Sbjct: 67 FTYDYAKENIKVNGFNN-IEIIEGDASTYNFK-EKADVVIAELLDTALIIEPQVKVMNSI 124
Query: 191 LFARDKWLVDDGVILPDIASLYLTAIE--------DKDYKEDKI 226
+ +L +D I+P A + +E D+D K +++
Sbjct: 125 I--ERGFLKEDVKIIPAKAISTIQLVEAKMSHIYYDEDIKSEEV 166
>PRMA_THEMA (Q9X0G8) Ribosomal protein L11 methyltransferase (EC
2.1.1.-) (L11 Mtase)
Length = 264
Score = 47.0 bits (110), Expect = 6e-05
Identities = 37/119 (31%), Positives = 61/119 (51%), Gaps = 7/119 (5%)
Query: 88 TYQNVIYQNRFLFKNKVVLDVGAGTGILSLFCAKAGAAHVYAVECSHMADRAKEIVETNG 147
T +V + ++L + VLDVG GTGIL++ K GA+ V AV+ + ++A E+ E N
Sbjct: 116 TRMSVFFLKKYLKEGNTVLDVGCGTGILAIAAKKLGASRVVAVD---VDEQAVEVAEENV 172
Query: 148 YSKVITVLKGKIEELELPVPKVDIIISEWMG--YFLLFENMLNSVLFARDKWLVDDGVI 204
+ VL + L DI++S + + L E+ +N V RD L+ G++
Sbjct: 173 RKNDVDVLVKWSDLLSEVEGTFDIVVSNILAEIHVKLLED-VNRVTH-RDSMLILSGIV 229
>PRMA_THENE (O86951) Ribosomal protein L11 methyltransferase (EC
2.1.1.-) (L11 Mtase)
Length = 264
Score = 46.6 bits (109), Expect = 8e-05
Identities = 43/141 (30%), Positives = 71/141 (49%), Gaps = 22/141 (15%)
Query: 88 TYQNVIYQNRFLFKNKVVLDVGAGTGILSLFCAKAGAAHVYAVECSHMA-DRAKEIVETN 146
T +V++ ++L K V+DVG GTGIL++ K GA++V AV+ A + AKE V+ N
Sbjct: 116 TQMSVLFLKKYLKKGDRVVDVGCGTGILAIVAKKLGASYVLAVDVDEQAVEVAKENVQKN 175
Query: 147 GYSKVITVLKGKIEELELPVPKVDIIISEWMGYF-LLFENMLNSVLFARDKWLVDDGVIL 205
+++ V + D ++SE G F L+ N+L + + L D +
Sbjct: 176 --------------SVDVTVKRSD-LLSEVEGVFDLVVSNILAEIHL---RLLEDVSRVT 217
Query: 206 PDIASLYLTAIEDKDYKEDKI 226
+ + L L+ I D KED +
Sbjct: 218 HEKSILILSGIVDT--KEDMV 236
>PRMA_CLOPE (Q8XIT6) Ribosomal protein L11 methyltransferase (EC
2.1.1.-) (L11 Mtase)
Length = 313
Score = 44.7 bits (104), Expect = 3e-04
Identities = 28/80 (35%), Positives = 46/80 (57%), Gaps = 4/80 (5%)
Query: 96 NRFLFKNKVVLDVGAGTGILSLFCAKAGAAHVYAVECSHMA-DRAKEIVETNGYSKVITV 154
++++ + V D+G G+GIL+L +K GA HV V+ +A D AKE + N I V
Sbjct: 172 DKYVKADTTVFDIGTGSGILALVASKLGAKHVVGVDLDPVAVDSAKENISFNNVDN-IEV 230
Query: 155 LKGKIEELELPVPKVDIIIS 174
L G + L++ K DI+++
Sbjct: 231 LYGNL--LDVVDGKADIVVA 248
>YXBB_BACSU (P46326) Hypothetical protein yxbB
Length = 244
Score = 44.3 bits (103), Expect = 4e-04
Identities = 27/75 (36%), Positives = 42/75 (56%), Gaps = 1/75 (1%)
Query: 101 KNKVVLDVGAGTGILSLFCAKAGAAHVYAVECS-HMADRAKEIVETNGYSKVITVLKGKI 159
KNKV++D+G G G LS+ AK AHV+AV+ + M + A+E + +G S +I+ +
Sbjct: 36 KNKVIIDMGTGPGYLSIQLAKRTNAHVHAVDINPAMHEIAQEEAKKSGVSSLISFDLEDV 95
Query: 160 EELELPVPKVDIIIS 174
L D I+S
Sbjct: 96 HHLSYADQYADFIVS 110
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.319 0.135 0.399
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 39,448,823
Number of Sequences: 164201
Number of extensions: 1728527
Number of successful extensions: 12107
Number of sequences better than 10.0: 270
Number of HSP's better than 10.0 without gapping: 194
Number of HSP's successfully gapped in prelim test: 77
Number of HSP's that attempted gapping in prelim test: 7341
Number of HSP's gapped (non-prelim): 1850
length of query: 322
length of database: 59,974,054
effective HSP length: 110
effective length of query: 212
effective length of database: 41,911,944
effective search space: 8885332128
effective search space used: 8885332128
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 66 (30.0 bits)
Medicago: description of AC148360.2


