

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148349.13 - phase: 0
(348 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
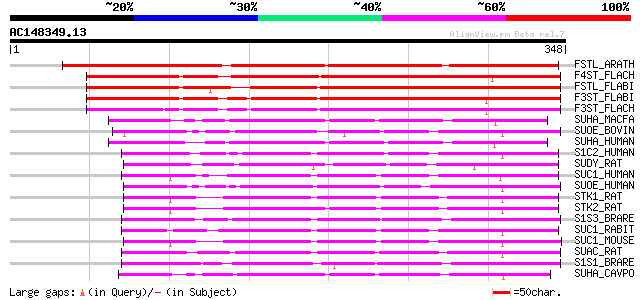
Score E
Sequences producing significant alignments: (bits) Value
FSTL_ARATH (P52839) Flavonol sulfotransferase-like (EC 2.8.2.-) ... 272 1e-72
F4ST_FLACH (P52837) Flavonol 4'-sulfotransferase (EC 2.8.2.-) (F... 249 6e-66
FSTL_FLABI (P52838) Flavonol sulfotransferase-like (EC 2.8.2.-) 246 5e-65
F3ST_FLABI (P52835) Flavonol 3-sulfotransferase (EC 2.8.2.-) (F3... 241 2e-63
F3ST_FLACH (P52836) Flavonol 3-sulfotransferase (EC 2.8.2.-) (F3... 224 2e-58
SUHA_MACFA (P52842) Alcohol sulfotransferase (EC 2.8.2.2) (Hydro... 128 2e-29
SUOE_BOVIN (P19217) Estrogen sulfotransferase (EC 2.8.2.4) (Sulf... 128 2e-29
SUHA_HUMAN (Q06520) Alcohol sulfotransferase (EC 2.8.2.2) (Hydro... 127 4e-29
S1C2_HUMAN (O75897) Sulfotransferase 1C2 (EC 2.8.2.-) (SULT1C) (... 127 5e-29
SUDY_RAT (P52847) DOPA/tyrosine sulfotransferase (EC 2.8.1.-) 125 1e-28
SUC1_HUMAN (O00338) Sulfotransferase 1C1 (EC 2.8.2.-) (SULT1C#1)... 125 2e-28
SUOE_HUMAN (P49888) Estrogen sulfotransferase (EC 2.8.2.4) (Sulf... 124 3e-28
STK1_RAT (Q9WUW8) Sulfotransferase K1 (EC 2.8.2.-) (rSULT1C2) 124 5e-28
STK2_RAT (Q9WUW9) Sulfotransferase K2 (EC 2.8.2.-) (rSULT1C2A) 123 8e-28
S1S3_BRARE (Q7T2V2) Cytosolic sulfotransferase 3 (EC 2.8.2.-) (S... 122 1e-27
SUC1_RABIT (O46503) Sulfotransferase 1C1 (EC 2.8.2.-) (rabSULT1C2) 122 2e-27
SUC1_MOUSE (Q9D939) Sulfotransferase 1C1 (EC 2.8.2.-) 122 2e-27
SUAC_RAT (P50237) N-hydroxyarylamine sulfotransferase (EC 2.8.2.... 122 2e-27
S1S1_BRARE (Q6PH37) Cytosolic sulfotransferase 1 (EC 2.8.2.-) (S... 120 4e-27
SUHA_CAVPO (P50234) 3-alpha-hydroxysteroid sulfotransferase (EC ... 119 9e-27
>FSTL_ARATH (P52839) Flavonol sulfotransferase-like (EC 2.8.2.-)
(RaRO47)
Length = 326
Score = 272 bits (695), Expect = 1e-72
Identities = 141/312 (45%), Positives = 192/312 (61%), Gaps = 11/312 (3%)
Query: 34 LILSLPRENGCETQYMYFFHGFWCPSTLIQSVNSFQNNFHAKDSDIVVASIPKSGTTWLK 93
LI SLP+E G +Y F G W ++Q + Q F AKDSDI++ + PKSGTTWLK
Sbjct: 23 LISSLPKEKGWLVSEIYEFQGLWHTQAILQGILICQKRFEAKDSDIILVTNPKSGTTWLK 82
Query: 94 GLAYAIVNRQHF-TSLENNHPLLLFNPHELVPQFEVNLYGDKDGPLPQIDVSNMTEPRLF 152
L +A++NR F S NHPLL+ NPH LVP E Y P D S++ PRL
Sbjct: 83 ALVFALLNRHKFPVSSSGNHPLLVTNPHLLVPFLEGVYYES-----PDFDFSSLPSPRLM 137
Query: 153 GTHMPFPSLPKSVKESNCKIIYICRNPFDTFVSYWIFINKIRLRKSLTELTLEESFERYC 212
TH+ SLP+SVK S+CKI+Y CRNP D FVS W F K+ ++ + +E++ E +C
Sbjct: 138 NTHISHLSLPESVKSSSCKIVYCCRNPKDMFVSLWHFGKKLAPEET-ADYPIEKAVEAFC 196
Query: 213 KGICLFGPFWDNMLGYLKESIERPDRVLFLKYEDLKEDVNFHTKRIAEFVGIPFTQEEEN 272
+G + GPFWD++L Y S E P++VLF+ YE+LK+ KRIAEF+ F +EEE
Sbjct: 197 EGKFIGGPFWDHILEYWYASRENPNKVLFVTYEELKKQTEVEMKRIAEFLECGFIEEEE- 255
Query: 273 NGVIENIIKLCSFESMKEIEGNQSGTISGDIEKEFYFRKGEIGDWANYLSSSMVEKLSKV 332
+ I+KLCSFES+ +E N+ G + IE + +FRKGEIG W + LS S+ E++ +
Sbjct: 256 ---VREIVKLCSFESLSNLEVNKEGKLPNGIETKTFFRKGEIGGWRDTLSESLAEEIDRT 312
Query: 333 MEEKLNGSSLSF 344
+EEK GS L F
Sbjct: 313 IEEKFKGSGLKF 324
>F4ST_FLACH (P52837) Flavonol 4'-sulfotransferase (EC 2.8.2.-)
(F4-ST)
Length = 320
Score = 249 bits (637), Expect = 6e-66
Identities = 133/299 (44%), Positives = 182/299 (60%), Gaps = 12/299 (4%)
Query: 49 MYFFHGFWCPSTLIQSVNSFQNNFHAKDSDIVVASIPKSGTTWLKGLAYAIVNRQHFTSL 108
+Y + FW I+ Q +F A+ D+ + S PKSGTTWLK LAYAIV R+ F
Sbjct: 32 LYKYQDFWGLQNNIEGAILAQQSFKARPDDVFLCSYPKSGTTWLKALAYAIVTREKFD-- 89
Query: 109 ENNHPLLLFNPHELVPQFEVNLYGDKDGPLPQIDVSNMTEPRLFGTHMPFPSLPKSVKES 168
E PLL PH +P E +L ++ N + THMP+ LPKS+
Sbjct: 90 EFTSPLLTNIPHNCIPYIEKDLK-------KIVENQNNSCFTPMATHMPYHVLPKSILAL 142
Query: 169 NCKIIYICRNPFDTFVSYWIFINKIRLRKSLTELTLEESFERYCKGICLFGPFWDNMLGY 228
NCK++YI RN D VS++ F +I + L + EE+F+ + GI FGP+WD++LGY
Sbjct: 143 NCKMVYIYRNIKDVIVSFYHFGREIT-KLPLEDAPFEEAFDEFYHGISQFGPYWDHLLGY 201
Query: 229 LKESIERPDRVLFLKYEDLKEDVNFHTKRIAEFVGIPFTQEEENNGVIENIIKLCSFESM 288
K S+ERP+ +LFLKYED+K+D + KR+AEF+G PFT EEE GVIE+IIKLCSFE++
Sbjct: 202 WKASLERPEVILFLKYEDVKKDPTSNVKRLAEFIGYPFTFEEEKEGVIESIIKLCSFENL 261
Query: 289 KEIEGNQSGTISG--DIEKEFYFRKGEIGDWANYLSSSMVEKLSKVMEEKLNGSSLSFK 345
+E N+SG G IE YFRK + GDW NY + M EK+ K+++EKL+ + L K
Sbjct: 262 SNLEVNKSGNSKGFLPIENRLYFRKAKDGDWKNYFTDEMTEKIDKLIDEKLSATGLVLK 320
>FSTL_FLABI (P52838) Flavonol sulfotransferase-like (EC 2.8.2.-)
Length = 309
Score = 246 bits (629), Expect = 5e-65
Identities = 129/302 (42%), Positives = 187/302 (61%), Gaps = 19/302 (6%)
Query: 49 MYFFHGFWCPSTLIQSVNSFQNNFHAKDSDIVVASIPKSGTTWLKGLAYAIVNRQHFTSL 108
MY + FW L++ Q +F A+ SD+ + S PK+GTTWLK LA+AIV R++F
Sbjct: 22 MYKYQDFWTSKQLLEGTLMAQQSFKAEPSDVFLCSAPKTGTTWLKALAFAIVTRENFD-- 79
Query: 109 ENNHPLLLFNPHELVP----QFEVNLYGDKDGPLPQIDVSNMTEPRLFGTHMPFPSLPKS 164
E+ PLL HE VP Q E + + LP L TH+P+ SLP+S
Sbjct: 80 ESTSPLLKKLVHECVPFLERQVEEIEHNRESSSLP-----------LVATHLPYASLPES 128
Query: 165 VKESNCKIIYICRNPFDTFVSYWIFINKIRLRKSLTELTLEESFERYCKGICLFGPFWDN 224
V SNC+++YI RN D VS + F+ + + S+ + EE+FE + G +GP+WD+
Sbjct: 129 VIASNCRMVYIYRNIKDVIVSNYHFLREA-FKLSMEDAPFEETFEDFYNGNSSYGPYWDH 187
Query: 225 MLGYLKESIERPDRVLFLKYEDLKEDVNFHTKRIAEFVGIPFTQEEENNGVIENIIKLCS 284
+LGY K S++ PD++LFLKYEDLK + + KR+AEF+G PF+ +EE GVIENII +CS
Sbjct: 188 ILGYRKASLDMPDKILFLKYEDLKSEPISNVKRLAEFIGYPFSNDEEKAGVIENIINMCS 247
Query: 285 FESMKEIEGNQSGTISGD-IEKEFYFRKGEIGDWANYLSSSMVEKLSKVMEEKLNGSSLS 343
FE++ +E N++ G +E Y+RKG+ GDW NY ++ M EK+ K+M+EKL+G+ L
Sbjct: 248 FENLSSLEVNKTRKPKGGMLENRLYYRKGQDGDWKNYFTNEMKEKIDKIMDEKLSGTGLI 307
Query: 344 FK 345
K
Sbjct: 308 LK 309
>F3ST_FLABI (P52835) Flavonol 3-sulfotransferase (EC 2.8.2.-)
(F3-ST)
Length = 312
Score = 241 bits (615), Expect = 2e-63
Identities = 125/301 (41%), Positives = 186/301 (61%), Gaps = 14/301 (4%)
Query: 49 MYFFHGFWCPSTLIQSVNSFQNNFHAKDSDIVVASIPKSGTTWLKGLAYAIVNRQHFTSL 108
+Y + W ++ + F A +D+ +AS PKSGTTWLK LA+AI+ R+ F
Sbjct: 22 LYKYQDVWNHQEFLEGRMLSEQTFKAHPNDVFLASYPKSGTTWLKALAFAIITREKFD-- 79
Query: 109 ENNHPLLLFNPHELVPQFEVNLYGDKDGPLPQIDVSNMTEPRLFGTHMPFPSLPKSVKES 168
++ PLL PH+ +P E +L + + +++ P TH + SLP+S + S
Sbjct: 80 DSTSPLLTTMPHDCIPLLEKDLE-----KIQENQRNSLYTP--ISTHFHYKSLPESARTS 132
Query: 169 NCKIIYICRNPFDTFVSYWIFINKIRLRKSLTELTLEESFERYCKGICLFGPFWDNMLGY 228
NCKI+YI RN D VSY+ F+ +I ++ S+ E EE+ + +C+GI GP+W+++LGY
Sbjct: 133 NCKIVYIYRNMKDVIVSYYHFLRQI-VKLSVEEAPFEEAVDEFCQGISSCGPYWEHILGY 191
Query: 229 LKESIERPDRVLFLKYEDLKEDVNFHTKRIAEFVGIPFTQEEENNGVIENIIKLCSFESM 288
K S+E+P+ LFLKYED+K+D K++A+F+G PFT +EE GVIENIIKLCSFE +
Sbjct: 192 WKASLEKPEIFLFLKYEDMKKDPVPSVKKLADFIGHPFTPKEEEAGVIENIIKLCSFEKL 251
Query: 289 KEIEGNQSG----TISGDIEKEFYFRKGEIGDWANYLSSSMVEKLSKVMEEKLNGSSLSF 344
+E N+SG + IE YFRKG+ GDW NY + M+EK+ K+++EKL + L
Sbjct: 252 SSLEVNKSGMHRPEEAHSIENRLYFRKGKDGDWKNYFTDEMIEKIDKLIDEKLGATGLVL 311
Query: 345 K 345
K
Sbjct: 312 K 312
>F3ST_FLACH (P52836) Flavonol 3-sulfotransferase (EC 2.8.2.-)
(F3-ST)
Length = 311
Score = 224 bits (571), Expect = 2e-58
Identities = 119/301 (39%), Positives = 181/301 (59%), Gaps = 15/301 (4%)
Query: 49 MYFFHGFWCPSTLIQSVNSFQNNFHAKDSDIVVASIPKSGTTWLKGLAYAIVNRQHFTSL 108
+Y + W ++ + F A +D+ +AS PKSGTTWLK I+ R+ F
Sbjct: 22 LYKYKDAWNHQEFLEGRILSEQKFKAHPNDVFLASYPKSGTTWLKAWI-CIITREKFD-- 78
Query: 109 ENNHPLLLFNPHELVPQFEVNLYGDKDGPLPQIDVSNMTEPRLFGTHMPFPSLPKSVKES 168
++ PLL PH+ +P E +L + + +++ P TH + SLP+S + S
Sbjct: 79 DSTSPLLTTMPHDCIPLLEKDLE-----KIQENQRNSLYTP--ISTHFHYKSLPESARTS 131
Query: 169 NCKIIYICRNPFDTFVSYWIFINKIRLRKSLTELTLEESFERYCKGICLFGPFWDNMLGY 228
NCKI+YI RN D VSY+ F+ +I ++ S+ E EE+F+ +C+GI GP+W+++ GY
Sbjct: 132 NCKIVYIYRNMKDVIVSYYHFLRQI-VKLSVEEAPFEEAFDEFCQGISSCGPYWEHIKGY 190
Query: 229 LKESIERPDRVLFLKYEDLKEDVNFHTKRIAEFVGIPFTQEEENNGVIENIIKLCSFESM 288
K S+E+P+ LFLKYED+K+D K++A+F+G PFT +EE GVIE+I+KLCSFE +
Sbjct: 191 WKASLEKPEIFLFLKYEDMKKDPVPSVKKLADFIGHPFTPKEEEAGVIEDIVKLCSFEKL 250
Query: 289 KEIEGNQSG----TISGDIEKEFYFRKGEIGDWANYLSSSMVEKLSKVMEEKLNGSSLSF 344
+E N+SG + IE YFRKG+ GDW NY + M +K+ K+++EKL + L
Sbjct: 251 SSLEVNKSGMHRPEEAHSIENRLYFRKGKDGDWKNYFTDEMTQKIDKLIDEKLGATGLVL 310
Query: 345 K 345
K
Sbjct: 311 K 311
>SUHA_MACFA (P52842) Alcohol sulfotransferase (EC 2.8.2.2)
(Hydroxysteroid sulfotransferase) (HST)
Length = 284
Score = 128 bits (322), Expect = 2e-29
Identities = 85/277 (30%), Positives = 143/277 (50%), Gaps = 25/277 (9%)
Query: 63 QSVNSFQNNFHAKDSDIVVASIPKSGTTWLKGLAYAIVNRQHFTSLENNHPLLLFNPHEL 122
+++ ++ F KD D+++ + PKSGT WL + I + N P + +
Sbjct: 20 ETLRKVRDEFVIKDEDVIILTYPKSGTNWLIEILCLIHS--------NGDPKWI----QS 67
Query: 123 VPQFEVNLYGDKDGPLPQIDVSNMTEPRLFGTHMPFPSLPKSVKESNCKIIYICRNPFDT 182
VP +E + + + + + +S PRLF +H+P PKS S K+IY+ RNP D
Sbjct: 68 VPIWERSPWVETE--MGYKLLSEEEGPRLFSSHLPIQLFPKSFFSSKAKVIYLMRNPRDV 125
Query: 183 FVSYWIFINKIRLRKSLTELTLEESFERYCKGICLFGPFWDNMLGYLKESIERPDRVLFL 242
FVS + F N ++ K + ++ FE +C+G ++G ++D++ G++ + L L
Sbjct: 126 FVSGYFFWNSVKFVKK--PKSWQQYFEWFCQGNVIYGSWFDHIHGWM--PMREKKNFLLL 181
Query: 243 KYEDLKEDVNFHTKRIAEFVGIPFTQEEENNGVIENIIKLCSFESMKEIEGNQSGTISGD 302
YE+LK+D ++I +F+G EE N I+K SF+SMKE + + +S D
Sbjct: 182 SYEELKQDTRRTVEKICQFLGKTLEPEELN-----LILKNSSFQSMKENKMSNFSLLSVD 236
Query: 303 I--EKEFYFRKGEIGDWANYLSSSMVEKLSKVMEEKL 337
EK RKG GDW N+L+ + E K+ +EK+
Sbjct: 237 FVEEKAQLLRKGISGDWKNHLTVAQAEAFDKLFQEKM 273
>SUOE_BOVIN (P19217) Estrogen sulfotransferase (EC 2.8.2.4)
(Sulfotransferase, estrogen-preferring) (ST1E1)
Length = 295
Score = 128 bits (321), Expect = 2e-29
Identities = 85/290 (29%), Positives = 147/290 (50%), Gaps = 31/290 (10%)
Query: 65 VNSFQN--NFHAKDSDIVVASIPKSGTTWLKGLAYAIVNRQHFTSLENNHPLLLFNPHEL 122
+ F N F A+ D+V+ + PKSGTTWL + I N + + ++FN
Sbjct: 25 IEQFHNVEEFEARPDDLVIVTYPKSGTTWLSEIICMIYNNGDVEKCKED---VIFNR--- 78
Query: 123 VPQFEVNLYGDKDGPLPQIDVSNMTEPRLFGTHMPFPSLPKSVKESNCKIIYICRNPFDT 182
VP E + G ++ M PR+ +H+P LP S E NCKIIY+ RN D
Sbjct: 79 VPYLECSTEHVMKGVK---QLNEMASPRIVKSHLPVKLLPVSFWEKNCKIIYLSRNAKDV 135
Query: 183 FVSYWIFINKIRLRKSLTELTLEESF----ERYCKGICLFGPFWDNMLGYLKESIERPDR 238
VSY+ I + T + +SF E++ G +G ++++ + ++S + +
Sbjct: 136 VVSYYFLILMV------TAIPDPDSFQDFVEKFMDGEVPYGSWFEHTKSWWEKS--KNPQ 187
Query: 239 VLFLKYEDLKEDVNFHTKRIAEFVGIPFTQEEENNGVIENIIKLCSFESMKEIEGNQSGT 298
VLFL YED+KE++ ++ EF+G + ++ +++ IIK SF+ MK T
Sbjct: 188 VLFLFYEDMKENIRKEVMKLLEFLG-----RKASDELVDKIIKHTSFQEMKNNPSTNYTT 242
Query: 299 ISGDIEKEF---YFRKGEIGDWANYLSSSMVEKLSKVMEEKLNGSSLSFK 345
+ ++ + + RKG++GDW N+ + ++ EK E+++ GS+L F+
Sbjct: 243 LPDEVMNQKVSPFMRKGDVGDWKNHFTVALNEKFDMHYEQQMKGSTLKFR 292
>SUHA_HUMAN (Q06520) Alcohol sulfotransferase (EC 2.8.2.2)
(Hydroxysteroid Sulfotransferase) (HST)
(Dehydroepiandrosterone sulfotransferase) (DHEA-ST)
(ST2) (ST2A3)
Length = 284
Score = 127 bits (319), Expect = 4e-29
Identities = 81/277 (29%), Positives = 142/277 (51%), Gaps = 25/277 (9%)
Query: 63 QSVNSFQNNFHAKDSDIVVASIPKSGTTWLKGLAYAIVNRQHFTSLENNHPLLLFNPHEL 122
+++ ++ F +D D+++ + PKSGT WL + + ++ +++
Sbjct: 20 ETLRKVRDEFVIRDEDVIILTYPKSGTNWLAEILCLMHSKGDAKWIQS------------ 67
Query: 123 VPQFEVNLYGDKDGPLPQIDVSNMTEPRLFGTHMPFPSLPKSVKESNCKIIYICRNPFDT 182
VP +E + + + + + +S PRLF +H+P PKS S K+IY+ RNP D
Sbjct: 68 VPIWERSPWVESE--IGYTALSETESPRLFSSHLPIQLFPKSFFSSKAKVIYLMRNPRDV 125
Query: 183 FVSYWIFINKIRLRKSLTELTLEESFERYCKGICLFGPFWDNMLGYLKESIERPDRVLFL 242
VS + F ++ K + EE FE +C+G L+G ++D++ G++ E+ L L
Sbjct: 126 LVSGYFFWKNMKFIKK--PKSWEEYFEWFCQGTVLYGSWFDHIHGWMPMREEK--NFLLL 181
Query: 243 KYEDLKEDVNFHTKRIAEFVGIPFTQEEENNGVIENIIKLCSFESMKEIEGNQSGTISGD 302
YE+LK+D ++I +F+G EE N I+K SF+SMKE + + +S D
Sbjct: 182 SYEELKQDTGRTIEKICQFLGKTLEPEELN-----LILKNSSFQSMKENKMSNYSLLSVD 236
Query: 303 --IEKEFYFRKGEIGDWANYLSSSMVEKLSKVMEEKL 337
++K RKG GDW N+ + + E K+ +EK+
Sbjct: 237 YVVDKAQLLRKGVSGDWKNHFTVAQAEDFDKLFQEKM 273
>S1C2_HUMAN (O75897) Sulfotransferase 1C2 (EC 2.8.2.-) (SULT1C)
(SULT1C#2)
Length = 302
Score = 127 bits (318), Expect = 5e-29
Identities = 84/278 (30%), Positives = 144/278 (51%), Gaps = 23/278 (8%)
Query: 71 NFHAKDSDIVVASIPKSGTTWLKGLAYAIVNRQHFTSLENNHPLLLFNPHELVPQFEVNL 130
NF AK D+++++ PK+GTTW + + I N + H+ P E+ +
Sbjct: 40 NFQAKPDDLLISTYPKAGTTWTQEIVELIQNEGDVEKSKRAPT------HQRFPFLEMKI 93
Query: 131 YGDKDGPLPQIDVSNMTEPRLFGTHMPFPSLPKSVKESNCKIIYICRNPFDTFVSYWIFI 190
G L Q M PR+ TH+PF LP S+ E NCKIIY+ RNP D VSY+ F
Sbjct: 94 PSLGSG-LEQAHA--MPSPRILKTHLPFHLLPPSLLEKNCKIIYVARNPKDNMVSYYHF- 149
Query: 191 NKIRLRKSL-TELTLEESFERYCKGICLFGPFWDNMLGYLKESIERPDRVLFLKYEDLKE 249
R+ K+L T EE FE + G +G + +++ G+ + + R+L+L YED+K+
Sbjct: 150 --QRMNKALPAPGTWEEYFETFLAGKVCWGSWHEHVKGWWEAKDKH--RILYLFYEDMKK 205
Query: 250 DVNFHTKRIAEFVGIPFTQEEENNGVIENIIKLCSFESMKEIEGNQSGTISGDIEKEF-- 307
+ +++AEF+G ++ ++ V++ I+ SF+ MK+ +I +I
Sbjct: 206 NPKHEIQKLAEFIG-----KKLDDKVLDKIVHYTSFDVMKQNPMANYSSIPAEIMDHSIS 260
Query: 308 -YFRKGEIGDWANYLSSSMVEKLSKVMEEKLNGSSLSF 344
+ RKG +GDW + + + E+ + ++K+ + L+F
Sbjct: 261 PFMRKGAVGDWKKHFTVAQNERFDEDYKKKMTDTRLTF 298
>SUDY_RAT (P52847) DOPA/tyrosine sulfotransferase (EC 2.8.1.-)
Length = 299
Score = 125 bits (315), Expect = 1e-28
Identities = 85/278 (30%), Positives = 139/278 (49%), Gaps = 24/278 (8%)
Query: 72 FHAKDSDIVVASIPKSGTTWLKGLAYAIVNRQHFTSLENNHPLLLFNPHELVPQFEVNLY 131
F ++ DIV+ + PKSGTTWL + ++N + + + VP E N+
Sbjct: 34 FQSRPCDIVIPTYPKSGTTWLSEIVDMVLNDGNVEKCKRDVIT------SKVPMLEQNVP 87
Query: 132 GDKDGPLPQIDVSNMTEPRLFGTHMPFPSLPKSVKESNCKIIYICRNPFDTFVSYWIF-- 189
G + + + PR+ TH+P LPKS ++ CK+IY+ RN D VSY+ F
Sbjct: 88 GARRSGVELL--KKTPSPRIIKTHLPIDLLPKSFWDNKCKMIYLARNGKDVAVSYYHFDL 145
Query: 190 INKIRLRKSLTELTLEESFERYCKGICLFGPFWDNMLGYLKESIERPDRVLFLKYEDLKE 249
+N I+ T EE E++ G +G ++D++ + ++ P +LFL YEDLK+
Sbjct: 146 MNNIQPLPG----TWEEYLEKFLAGNVAYGSWFDHVKSWWEKREGHP--ILFLYYEDLKK 199
Query: 250 DVNFHTKRIAEFVGIPFTQEEENNGVIENIIKLCSFESMKE---IEGNQSGTISGDIEKE 306
+ K+IA F+ + + +E I+ SFE MK+ + T D K
Sbjct: 200 NPKKEIKKIANFL-----DKTLDEHTLERIVHHTSFEVMKDNPLVNYTHLPTEIMDHSKS 254
Query: 307 FYFRKGEIGDWANYLSSSMVEKLSKVMEEKLNGSSLSF 344
+ RKG +GDW NY + + EK + ++KL+G++L F
Sbjct: 255 PFMRKGVVGDWKNYFTMTQSEKFDAIYKKKLSGTTLEF 292
>SUC1_HUMAN (O00338) Sulfotransferase 1C1 (EC 2.8.2.-) (SULT1C#1)
(ST1C2) (humSULTC2)
Length = 296
Score = 125 bits (313), Expect = 2e-28
Identities = 81/283 (28%), Positives = 144/283 (50%), Gaps = 33/283 (11%)
Query: 71 NFHAKDSDIVVASIPKSGTTWLKGLAYAI-----VNRQHFTSLENNHPLLLFNPHELVPQ 125
+F AK D+++ + PK+GTTW++ + I V + +++ HP + + PQ
Sbjct: 34 SFEAKPDDLLICTYPKAGTTWIQEIVDMIEQNGDVEKCQRAIIQHRHPFIEW---ARPPQ 90
Query: 126 FEVNLYGDKDGPLPQIDVSNMTEPRLFGTHMPFPSLPKSVKESNCKIIYICRNPFDTFVS 185
P M PR+ TH+ LP S E+NCK +Y+ RN D VS
Sbjct: 91 -----------PSGVEKAKAMPSPRILKTHLSTQLLPPSFWENNCKFLYVARNAKDCMVS 139
Query: 186 YWIFINKIRLRKSLTEL-TLEESFERYCKGICLFGPFWDNMLGYLKESIERPDRVLFLKY 244
Y+ F R+ L + T EE FE + G ++G ++D++ G+ + ++ ++LFL Y
Sbjct: 140 YYHF---QRMNHMLPDPGTWEEYFETFINGKVVWGSWFDHVKGWWE--MKDRHQILFLFY 194
Query: 245 EDLKEDVNFHTKRIAEFVGIPFTQEEENNGVIENIIKLCSFESMKEIEGNQSGTISGDIE 304
ED+K D +++ +F+G ++ + V++ I++ SFE MKE T+S I
Sbjct: 195 EDIKRDPKHEIRKVMQFMG-----KKVDETVLDKIVQETSFEKMKENPMTNRSTVSKSIL 249
Query: 305 KE---FYFRKGEIGDWANYLSSSMVEKLSKVMEEKLNGSSLSF 344
+ + RKG +GDW N+ + + E+ ++ K+ G+S++F
Sbjct: 250 DQSISSFMRKGTVGDWKNHFTVAQNERFDEIYRRKMEGTSINF 292
>SUOE_HUMAN (P49888) Estrogen sulfotransferase (EC 2.8.2.4)
(Sulfotransferase, estrogen-preferring) (EST-1)
Length = 294
Score = 124 bits (312), Expect = 3e-28
Identities = 83/277 (29%), Positives = 141/277 (49%), Gaps = 21/277 (7%)
Query: 72 FHAKDSDIVVASIPKSGTTWLKGLAYAIVNRQHFTSLENNHPLLLFNPHELVPQFEVNLY 131
F A+ D+V+A+ PKSGTTW+ + Y I + + ++FN +P E
Sbjct: 33 FQARPDDLVIATYPKSGTTWVSEIVYMIYKEGDVEKCKED---VIFNR---IPFLECRKE 86
Query: 132 GDKDGPLPQIDVSNMTEPRLFGTHMPFPSLPKSVKESNCKIIYICRNPFDTFVSYWIFIN 191
+G + Q+D M PR+ TH+P LP S E +CKIIY+CRN D VS++ F
Sbjct: 87 NLMNG-VKQLD--EMNSPRIVKTHLPPELLPASFWEKDCKIIYLCRNAKDVAVSFYYFF- 142
Query: 192 KIRLRKSLTELTLEESFERYCKGICLFGPFWDNMLGYLKESIERPDRVLFLKYEDLKEDV 251
+ + + E E++ +G +G ++ ++ + ++ + RVLFL YEDLKED+
Sbjct: 143 -LMVAGHPNPGSFPEFVEKFMQGQVPYGSWYKHVKSWWEKG--KSPRVLFLFYEDLKEDI 199
Query: 252 NFHTKRIAEFVGIPFTQEEENNGVIENIIKLCSFESMKEIEGNQSGTISGDIEKEF---Y 308
++ I F + + + +++ II SF+ MK T+ +I + +
Sbjct: 200 RKEVIKL-----IHFLERKPSEELVDRIIHHTSFQEMKNNPSTNYTTLPDEIMNQKLSPF 254
Query: 309 FRKGEIGDWANYLSSSMVEKLSKVMEEKLNGSSLSFK 345
RKG GDW N+ + ++ EK K E+++ S+L F+
Sbjct: 255 MRKGITGDWKNHFTVALNEKFDKHYEQQMKESTLKFR 291
>STK1_RAT (Q9WUW8) Sulfotransferase K1 (EC 2.8.2.-) (rSULT1C2)
Length = 296
Score = 124 bits (310), Expect = 5e-28
Identities = 83/284 (29%), Positives = 144/284 (50%), Gaps = 37/284 (13%)
Query: 72 FHAKDSDIVVASIPKSGTTWLKGLAYAI-----VNRQHFTSLENNHPLLLFNPHELVPQF 126
F AK D+++ + PKSGTTW++ + I V + T +++ HP + +
Sbjct: 35 FKAKPDDLLICTYPKSGTTWIQEIVDMIEQNGDVEKCQRTIIQHRHPFIEW--------- 85
Query: 127 EVNLYGDKDGPLPQ-IDVSN-MTEPRLFGTHMPFPSLPKSVKESNCKIIYICRNPFDTFV 184
P P +D +N M PR+ TH+P LP S +NCK +Y+ RN D V
Sbjct: 86 -------ARPPQPSGVDKANAMPAPRILRTHLPTQLLPPSFWTNNCKFLYVARNAKDCMV 138
Query: 185 SYWIFINKIRLRKSLTEL-TLEESFERYCKGICLFGPFWDNMLGYLKESIERPDRVLFLK 243
SY+ F R+ + L + T E FE + G +G ++D++ G+ + I ++LFL
Sbjct: 139 SYYHF---YRMSQVLPDPGTWNEYFETFINGKVSWGSWFDHVKGWWE--IRDRYQILFLF 193
Query: 244 YEDLKEDVNFHTKRIAEFVGIPFTQEEENNGVIENIIKLCSFESMKEIEGNQSGTISGDI 303
YED+K D +++ +F+G +E V++ I+ SFE MKE T+ +
Sbjct: 194 YEDVKRDPKREIQKVMQFMGKNLDEE-----VVDKIVLETSFEKMKENPMTNRSTVPKSV 248
Query: 304 EKEF---YFRKGEIGDWANYLSSSMVEKLSKVMEEKLNGSSLSF 344
+ + RKG +GDW N+ + + ++ ++ ++K+ G+SL+F
Sbjct: 249 LDQSISPFMRKGTVGDWKNHFTVAQNDRFDEIYKQKMGGTSLNF 292
>STK2_RAT (Q9WUW9) Sulfotransferase K2 (EC 2.8.2.-) (rSULT1C2A)
Length = 296
Score = 123 bits (308), Expect = 8e-28
Identities = 83/283 (29%), Positives = 142/283 (49%), Gaps = 35/283 (12%)
Query: 72 FHAKDSDIVVASIPKSGTTWLKGLAYAI-----VNRQHFTSLENNHPLLLFNPHELVPQF 126
F AK D+++ + PKSGTTW++ + I V + T +++ HP + +
Sbjct: 35 FKAKPDDLLICTYPKSGTTWIQEIVNMIEQNGDVEKCQRTIIQHRHPFIEW--------- 85
Query: 127 EVNLYGDKDGPLPQ-IDVSN-MTEPRLFGTHMPFPSLPKSVKESNCKIIYICRNPFDTFV 184
P P +D +N M PR+ TH+P LP S +NCK +Y+ RN D V
Sbjct: 86 -------ARPPQPSGVDKANAMPAPRILRTHLPIQLLPPSFWTNNCKYLYVARNAKDCMV 138
Query: 185 SYWIFINKIRLRKSLTELTLEESFERYCKGICLFGPFWDNMLGYLKESIERPDRVLFLKY 244
S++ F ++ + T E FE + G G ++D++ G+ + I ++LFL Y
Sbjct: 139 SFYHFYRMCQVLPN--PGTWNEYFETFINGKVSCGSWFDHVKGWWE--IRDRYQILFLFY 194
Query: 245 EDLKEDVNFHTKRIAEFVGIPFTQEEENNGVIENIIKLCSFESMKEIEGNQSGTISGDIE 304
ED+K D +++ +F+G +E V++ I+ SFE MK+ TI I
Sbjct: 195 EDMKRDPKREIQKVMQFMGKNLDEE-----VVDKIVLETSFEKMKDNPLTNFSTIPKTIM 249
Query: 305 KEF---YFRKGEIGDWANYLSSSMVEKLSKVMEEKLNGSSLSF 344
+ + RKG +GDW N+ + + E+ ++ E+K++G+SL+F
Sbjct: 250 DQSISPFMRKGIVGDWKNHFTVAQNERFDEIYEQKMDGTSLNF 292
>S1S3_BRARE (Q7T2V2) Cytosolic sulfotransferase 3 (EC 2.8.2.-)
(SULT1 ST3)
Length = 301
Score = 122 bits (307), Expect = 1e-27
Identities = 79/277 (28%), Positives = 143/277 (51%), Gaps = 18/277 (6%)
Query: 71 NFHAKDSDIVVASIPKSGTTWLKGLAYAIVNRQHFTSLENNHPLLLFNPHELVPQFEVNL 130
NF A+ DI++A+ PK+GTTW+ + + + + P+ + VP E
Sbjct: 38 NFQARPDDILIATYPKAGTTWVSYILDLLYFGNESPERQTSQPIYM-----RVPFLEACF 92
Query: 131 YGDKDGPLPQIDVSNMTEPRLFGTHMPFPSLPKSVKESNCKIIYICRNPFDTFVSYWIFI 190
G G ++ + T PRL TH+P +PKS E N K++Y+ RN D VSY+ F
Sbjct: 93 EGIPFGT--ELADNLPTSPRLIKTHLPVQLVPKSFWEQNSKVVYVARNAKDNAVSYFHF- 149
Query: 191 NKIRLRKSLTELTLEESF-ERYCKGICLFGPFWDNMLGYLKESIERPDRVLFLKYEDLKE 249
R+ E +F +++ +G +FGP++D++ GY K+ + +L++ YED+ E
Sbjct: 150 --DRMNMGQPEPGDWNTFLQKFMEGRNVFGPWYDHVNGYWKKK-QTYSNILYMFYEDMVE 206
Query: 250 DVNFHTKRIAEFVGIPFTQEEENNGVIENIIKLCSFESMKEIEGNQSGTIS-GDIEKEFY 308
+ +R+ F+G+ + E E I K F++MK+ + TI D + +
Sbjct: 207 NTGREVERLCSFLGLSTSAAER-----ERITKGVQFDAMKQNKMTNYSTIPVMDFKISPF 261
Query: 309 FRKGEIGDWANYLSSSMVEKLSKVMEEKLNGSSLSFK 345
RKG++GDW N+ + + E+ +V ++K+ +++ F+
Sbjct: 262 MRKGKVGDWRNHFTVAQNEQFDEVYKQKMKNTTVKFR 298
>SUC1_RABIT (O46503) Sulfotransferase 1C1 (EC 2.8.2.-) (rabSULT1C2)
Length = 296
Score = 122 bits (305), Expect = 2e-27
Identities = 79/278 (28%), Positives = 142/278 (50%), Gaps = 23/278 (8%)
Query: 71 NFHAKDSDIVVASIPKSGTTWLKGLAYAIVNRQHFTSLENNHPLLLFNPHELVPQFEVNL 130
+F AK D+++ + PKSGTTW++ + I + +E L+ + H +
Sbjct: 34 SFEAKPDDLLICTYPKSGTTWIQEIVDMI---EQNGDVEKCQRALIQHRHPFIE------ 84
Query: 131 YGDKDGPLPQIDVSNMTEPRLFGTHMPFPSLPKSVKESNCKIIYICRNPFDTFVSYWIFI 190
+ P M PR+ TH+P LP S E+NCK +Y+ RN D VSY+ F
Sbjct: 85 WARPPQPSGVEKAQAMPSPRILRTHLPTRLLPPSFWENNCKFLYVARNVKDCMVSYYHF- 143
Query: 191 NKIRLRKSLTEL-TLEESFERYCKGICLFGPFWDNMLGYLKESIERPDRVLFLKYEDLKE 249
R+ + L + T EE FE + G +G +++++ G+ + ++ ++LFL YED+K+
Sbjct: 144 --QRMNQVLPDPGTWEEYFETFINGKVAWGSWFEHVKGWWE--VKGRYQILFLFYEDIKK 199
Query: 250 DVNFHTKRIAEFVGIPFTQEEENNGVIENIIKLCSFESMKEIEGNQSGTISGDIEKEF-- 307
D +++A+F+G + + V++ I++ SFE MK+ T+ I +
Sbjct: 200 DPKCEIRKVAQFMG-----KHLDETVLDKIVQETSFEKMKDNPMINRSTVPKSIMDQSIS 254
Query: 308 -YFRKGEIGDWANYLSSSMVEKLSKVMEEKLNGSSLSF 344
+ RKG +GDW N+ + + +L ++ +K+ G S+ F
Sbjct: 255 PFMRKGTVGDWKNHFTVAQSHRLDELYRKKMEGVSIDF 292
>SUC1_MOUSE (Q9D939) Sulfotransferase 1C1 (EC 2.8.2.-)
Length = 296
Score = 122 bits (305), Expect = 2e-27
Identities = 83/286 (29%), Positives = 144/286 (50%), Gaps = 37/286 (12%)
Query: 72 FHAKDSDIVVASIPKSGTTWLKGLAYAI-----VNRQHFTSLENNHPLLLFNPHELVPQF 126
F AK D+++ + PKSGTTW++ + I V + T +++ HP + +
Sbjct: 35 FEAKPDDLLICTYPKSGTTWIQEIVDMIEQNGDVEKCRRTIIQHRHPFIEW--------- 85
Query: 127 EVNLYGDKDGPLPQ-IDVSN-MTEPRLFGTHMPFPSLPKSVKESNCKIIYICRNPFDTFV 184
P P +D +N M PR+ TH+P LP S +NCK +Y+ RN D V
Sbjct: 86 -------ARPPQPSGVDKANEMPAPRILRTHLPTQLLPPSFWTNNCKFLYVARNAKDCMV 138
Query: 185 SYWIFINKIRLRKSLTEL-TLEESFERYCKGICLFGPFWDNMLGYLKESIERPDRVLFLK 243
SY+ F R+ + L E T +E FE + G +G ++D++ G+ + I ++LFL
Sbjct: 139 SYYHF---YRMSQVLPEPGTWDEYFETFINGKVSWGSWFDHVKGWWE--IRDKYQILFLF 193
Query: 244 YEDLKEDVNFHTKRIAEFVGIPFTQEEENNGVIENIIKLCSFESMKEIEGNQSGTISGDI 303
YED+K + +++ +F+G + + V++ I+ SFE MKE T I
Sbjct: 194 YEDMKRNPKHEIQKVMQFMG-----KNLDEDVVDKIVLETSFEKMKENPMTNRSTAPKSI 248
Query: 304 EKEF---YFRKGEIGDWANYLSSSMVEKLSKVMEEKLNGSSLSFKV 346
+ + RKG +GDW N+ + + E+ ++ ++K+ +SL+F +
Sbjct: 249 LDQSISPFMRKGTVGDWKNHFTVAQNERFDEIYKQKMGRTSLNFSM 294
>SUAC_RAT (P50237) N-hydroxyarylamine sulfotransferase (EC 2.8.2.-)
(HAST-I)
Length = 304
Score = 122 bits (305), Expect = 2e-27
Identities = 90/280 (32%), Positives = 139/280 (49%), Gaps = 24/280 (8%)
Query: 71 NFHAKDSDIVVASIPKSGTTWLKGLAYAIVNRQHFTSLENNHPLLLFNPHELVPQFEVNL 130
NF AK D+++A+ K+GTTW + + I N + N ++ P E L
Sbjct: 41 NFQAKPDDLLIATYAKAGTTWTQEIVDMIQNDGDVQKCQRA------NTYDRHPFIEWTL 94
Query: 131 YGDKDGPLPQIDVSN-MTEPRLFGTHMPFPSLPKSVKESNCKIIYICRNPFDTFVSYWIF 189
+ L D++N M PR TH+P LP S + N KIIY+ RN D VSY+ F
Sbjct: 95 PSPLNSGL---DLANKMPSPRTLKTHLPVHMLPPSFWKENSKIIYVARNAKDCLVSYYYF 151
Query: 190 INKIRLRKSLTEL-TLEESFERYCKGICLFGPFWDNMLGYLKESIERPDRVLFLKYEDLK 248
R+ K L + TL E E++ G L+G ++D++ G+ ++ R+L+L YED+K
Sbjct: 152 ---SRMNKMLPDPGTLGEYIEQFKAGKVLWGSWYDHVKGWW--DVKDQHRILYLFYEDMK 206
Query: 249 EDVNFHTKRIAEFVGIPFTQEEENNGVIENIIKLCSFESMKEIEGNQSGTISGDIEKEF- 307
ED K+IA+F+ ++E V+ II SF+ MKE T+ I
Sbjct: 207 EDPKREIKKIAKFLEKDISEE-----VLNKIIYHTSFDVMKENPMANYTTLPSSIMDHSI 261
Query: 308 --YFRKGEIGDWANYLSSSMVEKLSKVMEEKLNGSSLSFK 345
+ RKG GDW NY + + E + K+ GS+++F+
Sbjct: 262 SPFMRKGMPGDWKNYFTVAQSEDFDEDYRRKMAGSNITFR 301
>S1S1_BRARE (Q6PH37) Cytosolic sulfotransferase 1 (EC 2.8.2.-)
(SULT1 ST1)
Length = 299
Score = 120 bits (302), Expect = 4e-27
Identities = 80/279 (28%), Positives = 140/279 (49%), Gaps = 22/279 (7%)
Query: 71 NFHAKDSDIVVASIPKSGTTWLKGLAYAIVNRQHFTSLENNHPLLLFNPHELVPQFEVNL 130
NF A+ DI++A+ PK+GTTW+ + + ++ + P+ + P L F+V
Sbjct: 36 NFQARPDDILIATYPKAGTTWVSYILDLLYFGENAPEEHTSQPIYMRVPF-LESCFKVIA 94
Query: 131 YGDKDGPLPQIDVSNMTEPRLFGTHMPFPSLPKSVKESNCKIIYICRNPFDTFVSYWIFI 190
G ++ + T PRL TH+P +PKS E N +++Y+ RN D VSY+ F
Sbjct: 95 SGT------ELADNMTTSPRLIKTHLPVQLIPKSFWEQNSRVVYVARNAKDNVVSYFHFD 148
Query: 191 NKIRLRKSLTEL---TLEESFERYCKGICLFGPFWDNMLGYLKESIERPDRVLFLKYEDL 247
R ++ E R+ G +FGP++D++ GY E + +L+L YEDL
Sbjct: 149 -----RMNIVEPDPGDWNTFLHRFMDGKSVFGPWYDHVNGYW-EKKQTYSNLLYLFYEDL 202
Query: 248 KEDVNFHTKRIAEFVGIPFTQEEENNGVIENIIKLCSFESMKEIEGNQSGTIS-GDIEKE 306
ED R+ F+G+ + + E I K F++MK+ + T+ D +
Sbjct: 203 VEDTGREVDRLCSFLGLSTSVSDR-----EKITKDVQFDAMKQNKMTNYSTLPVMDFKIS 257
Query: 307 FYFRKGEIGDWANYLSSSMVEKLSKVMEEKLNGSSLSFK 345
+ RKG++GDW N+ + + E+ +V +EK+ +++ F+
Sbjct: 258 PFMRKGKVGDWKNHFTVAQNEQFDEVYKEKMKNATVKFR 296
>SUHA_CAVPO (P50234) 3-alpha-hydroxysteroid sulfotransferase (EC
2.8.2.2) (Alcohol sulfotransferase) (HST1)
Length = 286
Score = 119 bits (299), Expect = 9e-27
Identities = 78/273 (28%), Positives = 138/273 (49%), Gaps = 25/273 (9%)
Query: 69 QNNFHAKDSDIVVASIPKSGTTWLKGLAYAIVNRQHFTSLENNHPLLLFNPHELVPQFEV 128
++ F KD D + + PKSGT WL + I+++ P L+ + VP ++
Sbjct: 26 RDKFLVKDEDTITVTYPKSGTNWLNEIVCLILSK--------GDPKLV----QSVPNWDR 73
Query: 129 NLYGDKDGPLPQIDVSNMTEPRLFGTHMPFPSLPKSVKESNCKIIYICRNPFDTFVSYWI 188
+ + + G V +PR++ +H+P PKS S K+IY RNP D VS +
Sbjct: 74 SPWIEFTGGYEL--VKGQKDPRVYTSHLPLHLFPKSFFSSKAKVIYCIRNPRDALVSGYF 131
Query: 189 FINKIRLRKSLTELTLEESFERYCKGICLFGPFWDNMLGYLKESIERPDRVLFLKYEDLK 248
F++K+ + + TL++ E + +G ++G +++++ G+L S+ + VL L YEDL
Sbjct: 132 FLSKMNVTEK--PETLQQYMEWFLQGNVIYGSWFEHVRGWL--SMREMENVLVLSYEDLI 187
Query: 249 EDVNFHTKRIAEFVGIPFTQEEENNGVIENIIKLCSFESMKEIEGNQSGTISGDIEKEFY 308
+D ++I +F+G EE + ++K SF+ MKE E + + E +
Sbjct: 188 KDTRSTVEKICQFLGKKLKPEE-----TDLVLKYSSFQFMKENEMSNFTLLPHAYTTEGF 242
Query: 309 --FRKGEIGDWANYLSSSMVEKLSKVMEEKLNG 339
RKG +GDW N+ + + E K+ +EK+ G
Sbjct: 243 TLLRKGTVGDWKNHFTVAQAEAFDKIYQEKMAG 275
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.319 0.137 0.411
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 42,928,463
Number of Sequences: 164201
Number of extensions: 1926004
Number of successful extensions: 4624
Number of sequences better than 10.0: 57
Number of HSP's better than 10.0 without gapping: 44
Number of HSP's successfully gapped in prelim test: 13
Number of HSP's that attempted gapping in prelim test: 4429
Number of HSP's gapped (non-prelim): 64
length of query: 348
length of database: 59,974,054
effective HSP length: 111
effective length of query: 237
effective length of database: 41,747,743
effective search space: 9894215091
effective search space used: 9894215091
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 66 (30.0 bits)
Medicago: description of AC148349.13


