

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148347.5 - phase: 0
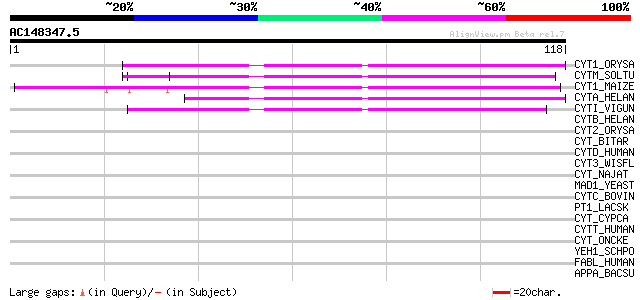
(118 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
CYT1_ORYSA (P09229) Cysteine proteinase inhibitor-I (Oryzacystat... 49 3e-06
CYTM_SOLTU (P37842) Multicystatin (MC) 48 3e-06
CYT1_MAIZE (P31726) Cystatin I precursor (Corn kernel cysteine p... 46 1e-05
CYTA_HELAN (Q10992) Cysteine proteinase inhibitor A (Cystatin A)... 42 3e-04
CYTI_VIGUN (Q06445) Cysteine proteinase inhibitor (Cystatin) 41 6e-04
CYTB_HELAN (Q10993) Cysteine proteinase inhibitor B (Cystatin B)... 39 0.002
CYT2_ORYSA (P20907) Cysteine proteinase inhibitor-II (Oryzacysta... 39 0.002
CYT_BITAR (P08935) Cystatin 36 0.014
CYTD_HUMAN (P28325) Cystatin D precursor 35 0.030
CYT3_WISFL (P19864) Cystatin WCPI-3 (Fragments) 35 0.030
CYT_NAJAT (P81714) Cystatin (Fragment) 33 0.088
MAD1_YEAST (P40957) Spindle assembly checkpoint component MAD1 (... 32 0.20
CYTC_BOVIN (P01035) Cystatin C precursor (Colostrum thiol protei... 32 0.26
PT1_LACSK (O07126) Phosphoenolpyruvate-protein phosphotransferas... 32 0.33
CYT_CYPCA (P35481) Cystatin precursor (Ovarian cystatin) (P12) 32 0.33
CYTT_HUMAN (P09228) Cystatin SA precursor (Cystatin S5) 32 0.33
CYT_ONCKE (Q98967) Cystatin precursor 31 0.44
YEH1_SCHPO (O13944) Oxysterol-binding protein homolog C23H4.01c 31 0.57
FABL_HUMAN (P07148) Fatty acid-binding protein, liver (L-FABP) 30 0.75
APPA_BACSU (P42061) Oligopeptide-binding protein appA precursor 30 0.75
>CYT1_ORYSA (P09229) Cysteine proteinase inhibitor-I
(Oryzacystatin-I)
Length = 102
Score = 48.5 bits (114), Expect = 3e-06
Identities = 30/94 (31%), Positives = 46/94 (48%), Gaps = 4/94 (4%)
Query: 25 IGNLSPINNINDPKVIDVANFAVKEYNNRRRKPEEKLRLWKVIKGESQIVADGVNYRLTL 84
+G + P+ N ND ++D+A FAV E+N +K L K++ + Q+VA G Y T+
Sbjct: 9 LGGVEPVGNENDLHLVDLARFAVTEHN---KKANSLLEFEKLVSVKQQVVA-GTLYYFTI 64
Query: 85 SATKVYTSNTYEAIVLEWSLQHLRNLTSFKLIQA 118
+ YEA V E + L FK + A
Sbjct: 65 EVKEGDAKKLYEAKVWEKPWMDFKELQEFKPVDA 98
>CYTM_SOLTU (P37842) Multicystatin (MC)
Length = 756
Score = 48.1 bits (113), Expect = 3e-06
Identities = 31/92 (33%), Positives = 49/92 (52%), Gaps = 4/92 (4%)
Query: 25 IGNLSPINNINDPKVIDVANFAVKEYNNRRRKPEEKLRLWKVIKGESQIVADGVNYRLTL 84
+G L + N + D+A FAV++YN +K + L KV+ + QIVA G+ Y +T
Sbjct: 4 VGGLVDVPFENKVEFDDLARFAVQDYN---QKNDSSLEFKKVLNVKQQIVA-GIMYYITF 59
Query: 85 SATKVYTSNTYEAIVLEWSLQHLRNLTSFKLI 116
AT+ YEA +L + L+ + FKL+
Sbjct: 60 EATEGGNKKEYEAKILLRKWEDLKKVVGFKLV 91
Score = 43.9 bits (102), Expect = 7e-05
Identities = 27/91 (29%), Positives = 47/91 (50%), Gaps = 4/91 (4%)
Query: 26 GNLSPINNINDPKVIDVANFAVKEYNNRRRKPEEKLRLWKVIKGESQIVADGVNYRLTLS 85
G + + N N P+ D+A FAV++YNN + L + + + Q+V+ G+ Y +TL+
Sbjct: 288 GGIINVPNPNSPEFQDLARFAVQDYNNTQ---NAHLEFVENLNVKEQLVS-GMMYYITLA 343
Query: 86 ATKVYTSNTYEAIVLEWSLQHLRNLTSFKLI 116
AT YEA + + + + FKL+
Sbjct: 344 ATDAGNKKEYEAKIWVKEWEDFKKVIDFKLV 374
Score = 42.4 bits (98), Expect = 2e-04
Identities = 28/92 (30%), Positives = 47/92 (50%), Gaps = 4/92 (4%)
Query: 25 IGNLSPINNINDPKVIDVANFAVKEYNNRRRKPEEKLRLWKVIKGESQIVADGVNYRLTL 84
IG + + N+P+ D+A FAV++YN +K L + + + Q+VA G+ Y +TL
Sbjct: 570 IGGFTDVPFPNNPEFQDLARFAVQDYN---KKENAHLEYVENLNVKEQLVA-GMIYYITL 625
Query: 85 SATKVYTSNTYEAIVLEWSLQHLRNLTSFKLI 116
AT YEA + + + + FKL+
Sbjct: 626 VATDAGKKKIYEAKIWVKEWEDFKKVVEFKLV 657
Score = 42.0 bits (97), Expect = 2e-04
Identities = 25/82 (30%), Positives = 44/82 (53%), Gaps = 4/82 (4%)
Query: 35 NDPKVIDVANFAVKEYNNRRRKPEEKLRLWKVIKGESQIVADGVNYRLTLSATKVYTSNT 94
N P+ D+A FAV+++N +K L + + + Q+VA G+ Y +TL+AT
Sbjct: 674 NSPEFQDLARFAVQDFN---KKENGHLEFVENLNVKEQVVA-GMMYYITLAATDARKKEI 729
Query: 95 YEAIVLEWSLQHLRNLTSFKLI 116
YE +L ++ + + FKL+
Sbjct: 730 YETKILVKEWENFKEVQEFKLV 751
Score = 39.7 bits (91), Expect = 0.001
Identities = 27/92 (29%), Positives = 47/92 (50%), Gaps = 4/92 (4%)
Query: 25 IGNLSPINNINDPKVIDVANFAVKEYNNRRRKPEEKLRLWKVIKGESQIVADGVNYRLTL 84
+G + + N+P+ D+A FAV++YN +K L + + + Q+VA G+ Y +TL
Sbjct: 476 LGGIINVPFPNNPEFQDLARFAVQDYN---KKENAHLEFVENLNVKEQLVA-GMLYYITL 531
Query: 85 SATKVYTSNTYEAIVLEWSLQHLRNLTSFKLI 116
A YEA + ++ + + FKLI
Sbjct: 532 VAIDAGKKKIYEAKIWVKEWENFKKVIEFKLI 563
Score = 38.9 bits (89), Expect = 0.002
Identities = 28/97 (28%), Positives = 51/97 (51%), Gaps = 15/97 (15%)
Query: 26 GNLSPINNINDPKVIDVANFAVKEYNNRRRKPEEKLRLWKVIKGESQIVADGVNYRLTLS 85
G + + N N+ K ++A FA+++YN +K L + + + Q+VA G+ Y +TL+
Sbjct: 99 GGIVNVPNPNNTKFQELARFAIQDYN---KKQNAHLEFVENLNVKEQVVA-GIMYYITLA 154
Query: 86 AT------KVYTSNTYEAIVLEWSLQHLRNLTSFKLI 116
AT K+Y + + V EW + + + FKL+
Sbjct: 155 ATDDAGKKKIYKAKIW---VKEW--EDFKKVVEFKLV 186
Score = 38.5 bits (88), Expect = 0.003
Identities = 24/92 (26%), Positives = 46/92 (49%), Gaps = 4/92 (4%)
Query: 25 IGNLSPINNINDPKVIDVANFAVKEYNNRRRKPEEKLRLWKVIKGESQIVADGVNYRLTL 84
+G ++ + N+P+ D+A FA++ YN +K L + + + Q+VA G+ Y +TL
Sbjct: 193 LGGITDVPFPNNPEFQDLARFAIQVYN---KKENVHLEFVENLNVKQQVVA-GMMYYITL 248
Query: 85 SATKVYTSNTYEAIVLEWSLQHLRNLTSFKLI 116
+A YE + + + + FKL+
Sbjct: 249 AAIDAGKKKIYETKIWVKEWEDFKKVVEFKLV 280
Score = 34.7 bits (78), Expect = 0.040
Identities = 25/106 (23%), Positives = 46/106 (42%), Gaps = 4/106 (3%)
Query: 11 VVLLAFTATKQAIPIGNLSPINNINDPKVIDVANFAVKEYNNRRRKPEEKLRLWKVIKGE 70
V+ A +G + + N P+ D+ FAV +YN + L + + +
Sbjct: 368 VIDFKLVGNDSAKKLGGFTEVPFPNSPEFQDLTRFAVHQYN---KDQNAHLEFVENLNVK 424
Query: 71 SQIVADGVNYRLTLSATKVYTSNTYEAIVLEWSLQHLRNLTSFKLI 116
Q+VA G+ Y +T +AT YE + ++ + + FKL+
Sbjct: 425 KQVVA-GMLYYITFAATDGGKKKIYETKIWVKVWENFKKVVEFKLV 469
>CYT1_MAIZE (P31726) Cystatin I precursor (Corn kernel cysteine
proteinase inhibitor)
Length = 135
Score = 46.2 bits (108), Expect = 1e-05
Identities = 39/130 (30%), Positives = 64/130 (49%), Gaps = 18/130 (13%)
Query: 2 RLESMVLVLVVLLAFTAT-------KQAIP------IGNLSPIN-NINDPKVIDVANFAV 47
R+ S+V L+VLLA A K+++ G + + N ND ++ ++A FAV
Sbjct: 5 RIVSLVAALLVLLALAAVSSTRSTQKESVADNAGMLAGGIKDVPANENDLQLQELARFAV 64
Query: 48 KEYNNRRRKPEEKLRLWKVIKGESQIVADGVNYRLTLSATKVYTSNTYEAIVLEWSLQHL 107
E+N +K L K++K ++Q+VA G Y LT+ YEA V E ++
Sbjct: 65 NEHN---QKANALLGFEKLVKAKTQVVA-GTMYYLTIEVKDGEVKKLYEAKVWEKPWENF 120
Query: 108 RNLTSFKLIQ 117
+ L FK ++
Sbjct: 121 KQLQEFKPVE 130
>CYTA_HELAN (Q10992) Cysteine proteinase inhibitor A (Cystatin A)
(SCA)
Length = 82
Score = 41.6 bits (96), Expect = 3e-04
Identities = 28/81 (34%), Positives = 42/81 (51%), Gaps = 4/81 (4%)
Query: 38 KVIDVANFAVKEYNNRRRKPEEKLRLWKVIKGESQIVADGVNYRLTLSATKVYTSNTYEA 97
++ ++A FAV E+N +K L KV+ + Q+VA G Y +TL AT TYEA
Sbjct: 3 EIDELARFAVDEHN---KKQNALLEFGKVVNTKEQVVA-GKMYYITLEATNGGVKKTYEA 58
Query: 98 IVLEWSLQHLRNLTSFKLIQA 118
V ++ + L FK + A
Sbjct: 59 KVWVKPWENFKELQEFKPVDA 79
>CYTI_VIGUN (Q06445) Cysteine proteinase inhibitor (Cystatin)
Length = 97
Score = 40.8 bits (94), Expect = 6e-04
Identities = 28/89 (31%), Positives = 42/89 (46%), Gaps = 4/89 (4%)
Query: 26 GNLSPINNINDPKVIDVANFAVKEYNNRRRKPEEKLRLWKVIKGESQIVADGVNYRLTLS 85
GN N N ++ +A FAV+E+N +K L +V+ + Q+V+ G Y +TL
Sbjct: 6 GNRDVAGNQNSLEIDSLARFAVEEHN---KKQNALLEFGRVVSAQQQVVS-GTLYTITLE 61
Query: 86 ATKVYTSNTYEAIVLEWSLQHLRNLTSFK 114
A YEA V E + + L FK
Sbjct: 62 AKDGGQKKVYEAKVWEKPWLNFKELQEFK 90
>CYTB_HELAN (Q10993) Cysteine proteinase inhibitor B (Cystatin B)
(SCB)
Length = 101
Score = 39.3 bits (90), Expect = 0.002
Identities = 30/96 (31%), Positives = 48/96 (49%), Gaps = 5/96 (5%)
Query: 23 IPIGNLSPINNINDPKVIDVANFAVKEYNNRRRKPEE---KLRLWKVIKGESQIVADGVN 79
IP G N D ++ + +++V EYN +R + L+ +VI E+Q+VA G
Sbjct: 1 IPGGRTKVKNVKTDTEIQQLGSYSVDEYNRLQRTKKTGAGDLKFSQVIAAETQVVA-GTK 59
Query: 80 YRLTLSA-TKVYTSNTYEAIVLEWSLQHLRNLTSFK 114
Y L + A TK ++A V+ S +H + L FK
Sbjct: 60 YYLKIEAITKGGKMKVFDAEVVVQSWKHSKKLLGFK 95
>CYT2_ORYSA (P20907) Cysteine proteinase inhibitor-II
(Oryzacystatin-II)
Length = 107
Score = 38.9 bits (89), Expect = 0.002
Identities = 28/86 (32%), Positives = 45/86 (51%), Gaps = 5/86 (5%)
Query: 30 PINNINDPKVIDVANFAVKEYNNRRRKPEEKLRLWKVIKGESQIVADGVNYRLTLSATKV 89
P ND +++A FAV E+N+ K L L +V+K Q+V ++Y LT+ +
Sbjct: 19 PAGRENDLTTVELARFAVAEHNS---KANAMLELERVVKVRQQVVGGFMHY-LTVEVKEP 74
Query: 90 YTSN-TYEAIVLEWSLQHLRNLTSFK 114
+N YEA V E + ++ + L FK
Sbjct: 75 GGANKLYEAKVWERAWENFKQLQDFK 100
>CYT_BITAR (P08935) Cystatin
Length = 111
Score = 36.2 bits (82), Expect = 0.014
Identities = 24/69 (34%), Positives = 38/69 (54%), Gaps = 3/69 (4%)
Query: 26 GNLSPINNINDPKVIDVANFAVKEYNNRRRKPEEKLRLWKVIKGESQIVADGVNYRLTLS 85
G LSP ++ DP V + A FAV++Y N K + + +V++ +SQ+V+ GV Y L +
Sbjct: 3 GGLSP-RDVTDPDVQEAAAFAVEKY-NAGSKNDYYFKERRVVEAQSQVVS-GVKYYLMME 59
Query: 86 ATKVYTSNT 94
K T
Sbjct: 60 LLKTTCKKT 68
>CYTD_HUMAN (P28325) Cystatin D precursor
Length = 142
Score = 35.0 bits (79), Expect = 0.030
Identities = 24/75 (32%), Positives = 38/75 (50%), Gaps = 1/75 (1%)
Query: 6 MVLVLVVLLAFTATKQAIPIGNLSPINNINDPKVIDVANFAVKEYNNRRRKPEEKLRLWK 65
++ L+V +A +A+ Q+ + ++ND V +FA+ EYN K E R +
Sbjct: 11 LLTALMVAVAGSASAQSRTLAGGIHATDLNDKSVQCALDFAISEYNKVINKDEYYSRPLQ 70
Query: 66 VIKGESQIVADGVNY 80
V+ QIV GVNY
Sbjct: 71 VMAAYQQIVG-GVNY 84
>CYT3_WISFL (P19864) Cystatin WCPI-3 (Fragments)
Length = 46
Score = 35.0 bits (79), Expect = 0.030
Identities = 21/59 (35%), Positives = 31/59 (51%), Gaps = 18/59 (30%)
Query: 26 GNLSPINNINDPKVIDVANFAVKEYNNRRRKPEEKLRLWKVIKGESQIVADGVNYRLTL 84
G +P+ +I+D V+++AN+AV EYN + S +VA GVNYR L
Sbjct: 5 GGYTPVPDIDDIHVVEIANYAVTEYNKK-----------------SGVVA-GVNYRFVL 45
>CYT_NAJAT (P81714) Cystatin (Fragment)
Length = 99
Score = 33.5 bits (75), Expect = 0.088
Identities = 25/69 (36%), Positives = 35/69 (50%), Gaps = 3/69 (4%)
Query: 26 GNLSPINNINDPKVIDVANFAVKEYNNRRRKPEEKLRLWKVIKGESQIVADGVNYRLTLS 85
G LSP +++DP V A FAV+EYN L +V++ +SQ VA G Y L +
Sbjct: 3 GGLSP-RSVSDPDVQKAAAFAVQEYNAGSANAHYYKEL-RVVEAQSQSVA-GEKYFLMME 59
Query: 86 ATKVYTSNT 94
K + T
Sbjct: 60 LVKTKCAKT 68
>MAD1_YEAST (P40957) Spindle assembly checkpoint component MAD1
(Mitotic arrest deficient protein 1)
Length = 749
Score = 32.3 bits (72), Expect = 0.20
Identities = 25/87 (28%), Positives = 41/87 (46%), Gaps = 5/87 (5%)
Query: 32 NNINDPKVIDVANFAVKEYNNRRRKPEEKLRLWKVIKGESQIVADGVNYRLTLSATKVYT 91
+N ND + N + NN R PEE +R WK+ K E I+ D +N +L L +
Sbjct: 348 DNNNDTSNNNNINNNNRTKNNIRNNPEEIIRDWKLTKKECLILTD-MNDKLRLDNNNLKL 406
Query: 92 SNTYEAI----VLEWSLQHLRNLTSFK 114
N A+ +L+ + + N+ + K
Sbjct: 407 LNDEMALERNQILDLNKNYENNIVNLK 433
>CYTC_BOVIN (P01035) Cystatin C precursor (Colostrum thiol
proteinase inhibitor)
Length = 148
Score = 32.0 bits (71), Expect = 0.26
Identities = 24/85 (28%), Positives = 44/85 (51%), Gaps = 8/85 (9%)
Query: 3 LESMVLVLVVLLAFTATKQAIP-----IGNLSPINNINDPKVIDVANFAVKEYNNRRRKP 57
L S+++ L + LA + P +G L + +N+ V + +FAV E+N R
Sbjct: 12 LASLIVALALALAVSPAAAQGPRKGRLLGGLMEAD-VNEEGVQEALSFAVSEFNKRSNDA 70
Query: 58 EEKLRLWKVIKGESQIVADGVNYRL 82
+ R+ +V++ Q+V+ G+NY L
Sbjct: 71 YQS-RVVRVVRARKQVVS-GMNYFL 93
>PT1_LACSK (O07126) Phosphoenolpyruvate-protein phosphotransferase
(EC 2.7.3.9) (Phosphotransferase system, enzyme I)
Length = 574
Score = 31.6 bits (70), Expect = 0.33
Identities = 15/73 (20%), Positives = 34/73 (46%)
Query: 8 LVLVVLLAFTATKQAIPIGNLSPINNINDPKVIDVANFAVKEYNNRRRKPEEKLRLWKVI 67
L + ++ Q + G+L ++ IN V+D + + EY+ + + ++ W+ +
Sbjct: 198 LEIPAIVGTETVTQDVKAGDLLIVDGINGDVVLDPTDADIAEYDVKAQAFADQKAEWEKL 257
Query: 68 KGESQIVADGVNY 80
K E + DG +
Sbjct: 258 KNEKSVTKDGKTF 270
>CYT_CYPCA (P35481) Cystatin precursor (Ovarian cystatin) (P12)
Length = 129
Score = 31.6 bits (70), Expect = 0.33
Identities = 29/84 (34%), Positives = 42/84 (49%), Gaps = 6/84 (7%)
Query: 1 MRLESMVLVLVVLLAFTATKQAIPIGNLSPINNINDPKVIDVANFAVKEYNNRRRKPEEK 60
M L+ +VL L V L +T IP G + +IND V FAV YN + +
Sbjct: 1 MYLKVIVLFLAVTLVVEST--GIPGGLVDA--DINDKDVQKALRFAVDHYNGQSNDAFVR 56
Query: 61 LRLWKVIKGESQIVADGVNYRLTL 84
++ KVIK + Q+ A G+ Y T+
Sbjct: 57 -KVSKVIKVQQQVAA-GMKYIFTV 78
>CYTT_HUMAN (P09228) Cystatin SA precursor (Cystatin S5)
Length = 141
Score = 31.6 bits (70), Expect = 0.33
Identities = 18/48 (37%), Positives = 28/48 (57%), Gaps = 2/48 (4%)
Query: 33 NINDPKVIDVANFAVKEYNNRRRKPEEKLRLWKVIKGESQIVADGVNY 80
++ND +V +F + EY N+ + E RL +V++ QIV GVNY
Sbjct: 38 DLNDERVQRALHFVISEY-NKATEDEYYRRLLRVLRAREQIVG-GVNY 83
>CYT_ONCKE (Q98967) Cystatin precursor
Length = 130
Score = 31.2 bits (69), Expect = 0.44
Identities = 22/82 (26%), Positives = 42/82 (50%), Gaps = 4/82 (4%)
Query: 3 LESMVLVLVVLLAFTATKQAIPIGNLSPINNINDPKVIDVANFAVKEYNNRRRKPEEKLR 62
+E ++V ++ +AFT + G + N+ND D FAV E+N + + +
Sbjct: 1 MEWKIVVPLLAVAFTVANAGLVGGPMDA--NMNDQGTRDALQFAVVEHNKKTNDMFVR-Q 57
Query: 63 LWKVIKGESQIVADGVNYRLTL 84
+ KV+ + Q+V+ G+ Y T+
Sbjct: 58 VAKVVNAQKQVVS-GMKYIFTV 78
>YEH1_SCHPO (O13944) Oxysterol-binding protein homolog C23H4.01c
Length = 945
Score = 30.8 bits (68), Expect = 0.57
Identities = 16/48 (33%), Positives = 28/48 (58%), Gaps = 1/48 (2%)
Query: 35 NDPKVIDVANFAVKEYNNRRRKPEEKLRLWKVIKGES-QIVADGVNYR 81
+D K+ VA FAV ++N R K +++ + GE+ ++V + NYR
Sbjct: 622 DDIKIFYVAAFAVSNFSNMRHKERSVRKVFSPLLGETFELVREDRNYR 669
>FABL_HUMAN (P07148) Fatty acid-binding protein, liver (L-FABP)
Length = 127
Score = 30.4 bits (67), Expect = 0.75
Identities = 14/39 (35%), Positives = 24/39 (60%)
Query: 57 PEEKLRLWKVIKGESQIVADGVNYRLTLSATKVYTSNTY 95
PEE ++ K IKG S+IV +G +++ T++A N +
Sbjct: 25 PEELIQKGKDIKGVSEIVQNGKHFKFTITAGSKVIQNEF 63
>APPA_BACSU (P42061) Oligopeptide-binding protein appA precursor
Length = 543
Score = 30.4 bits (67), Expect = 0.75
Identities = 21/73 (28%), Positives = 32/73 (43%), Gaps = 3/73 (4%)
Query: 35 NDPKVIDVANFAVKEYNNRRRKPEEKLRLWKVIKGESQIVADGVNYRLTLSATKVYTSNT 94
N PK IDV F EYN ++ K WK G+ + DG + TL +
Sbjct: 341 NYPKDIDVPKF---EYNEKKAKQMLAEAGWKDTNGDGILDKDGKKFSFTLKTNQGNKVRE 397
Query: 95 YEAIVLEWSLQHL 107
A+V++ L+ +
Sbjct: 398 DIAVVVQEQLKKI 410
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.319 0.134 0.371
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 12,235,364
Number of Sequences: 164201
Number of extensions: 431895
Number of successful extensions: 1344
Number of sequences better than 10.0: 41
Number of HSP's better than 10.0 without gapping: 17
Number of HSP's successfully gapped in prelim test: 24
Number of HSP's that attempted gapping in prelim test: 1310
Number of HSP's gapped (non-prelim): 48
length of query: 118
length of database: 59,974,054
effective HSP length: 94
effective length of query: 24
effective length of database: 44,539,160
effective search space: 1068939840
effective search space used: 1068939840
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 58 (26.9 bits)
Medicago: description of AC148347.5


