

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148345.14 - phase: 0 /pseudo
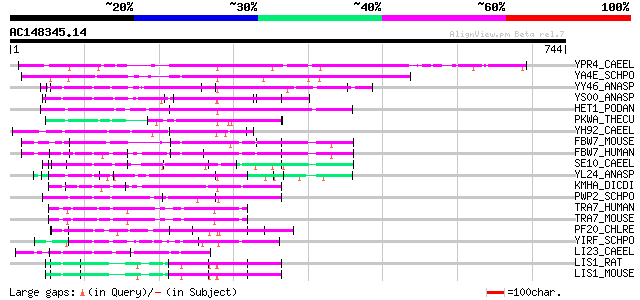
(744 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
YPR4_CAEEL (Q20059) Hypothetical WD-repeat protein F35G12.4 in c... 241 4e-63
YA4E_SCHPO (Q09731) Hypothetical WD-repeat protein C31A2.14 in c... 202 3e-51
YY46_ANASP (Q8YRI1) Hypothetical WD-repeat protein alr3466 100 2e-20
YS00_ANASP (Q8YTC2) Hypothetical WD-repeat protein alr2800 99 3e-20
HET1_PODAN (Q00808) Vegetatible incompatibility protein HET-E-1 94 2e-18
PKWA_THECU (P49695) Probable serine/threonine-protein kinase pkw... 91 1e-17
YH92_CAEEL (Q23256) Hypothetical WD-repeat protein ZC302.2 in ch... 79 4e-14
FBW7_MOUSE (Q8VBV4) F-box/WD-repeat protein 7 (F-box and WD-40 d... 78 1e-13
FBW7_HUMAN (Q969H0) F-box/WD-repeat protein 7 (F-box and WD-40 d... 77 2e-13
SE10_CAEEL (Q93794) F-box/WD-repeat protein sel-10 (Suppressor/e... 75 8e-13
YL24_ANASP (Q8YV57) Hypothetical WD-repeat protein all2124 74 1e-12
KMHA_DICDI (P42527) Myosin heavy chain kinase A (EC 2.7.1.129) (... 72 4e-12
PWP2_SCHPO (Q9C1X1) Periodic tryptophan protein 2 homolog 70 2e-11
TRA7_HUMAN (Q6Q0C0) E3 ubiquitin protein ligase TRAF7 (EC 6.3.2.... 69 4e-11
TRA7_MOUSE (Q922B6) E3 ubiquitin protein ligase TRAF7 (EC 6.3.2.... 68 8e-11
PF20_CHLRE (P93107) Flagellar WD-repeat protein PF20 68 8e-11
YIRF_SCHPO (Q9P7I3) Hypothetical WD-repeat protein C664.15 in ch... 66 3e-10
LI23_CAEEL (Q09990) F-box/WD-repeat protein lin-23 (Abnormal cel... 66 3e-10
LIS1_RAT (P63004) Platelet-activating factor acetylhydrolase IB ... 66 4e-10
LIS1_MOUSE (P63005) Platelet-activating factor acetylhydrolase I... 66 4e-10
>YPR4_CAEEL (Q20059) Hypothetical WD-repeat protein F35G12.4 in
chromosome III
Length = 683
Score = 241 bits (616), Expect = 4e-63
Identities = 202/731 (27%), Positives = 328/731 (44%), Gaps = 143/731 (19%)
Query: 12 NSTRPRKEKRLTYVLNDSDDTKHCAGINCLALLTSTISDGSDYLFTGSRDGRLKRWALAE 71
N+++ +K++++++ D + + + ++ L + LFTG D ++ W++
Sbjct: 6 NTSQTGPKKKISFIIRDEHEYSNRSAVSALQYDAQ-----NGRLFTGGSDTIIRTWSVPH 60
Query: 72 DAATCSA-----------------TFESHVDWVNDAVLVGDNT-LVSCSSDTTLKTWNAL 113
SA + E H DWVND +L G L+S S+DTT+K WN
Sbjct: 61 HKDAFSARGGVRSPGKNSPVQYQGSLEQHTDWVNDMILCGHGKILISASNDTTVKVWNIE 120
Query: 114 SAGT-----CVRTHRQHTDYVTCLAAAGKNSNIVASGGLGGEVFVWDIEAALAPVSKCND 168
C+RTH+ DYV+CLA A V S +FV+DI A
Sbjct: 121 RDNKHGFIDCIRTHK---DYVSCLAYAPIVEKAV-SASFDHNIFVYDINA---------- 166
Query: 169 AMVDESSNGINGSANLLPSTNLRTISSSHSISLHTAQTQGYIPIAAKGHKESVYALAMDE 228
N +T+++ G K+S+Y+LA
Sbjct: 167 --------------------NFKTVNN------------------LIGCKDSIYSLATTP 188
Query: 229 GGSILVSGGTEKVVRVWDTRSGSKILKLKGHTDNIRALLLDSTG-RVLFI---RIIRFYD 284
S+++ GTEK +R++D R+ KI+KL+GHTDN+RAL+++ G R L IR +D
Sbjct: 189 NLSLVLGAGTEKCIRLFDPRTNEKIMKLRGHTDNVRALVVNDDGTRALSAGSDATIRLWD 248
Query: 285 KGQQRCVHSYAVHTDSVWALASTPTFSHVYSGGRDFSLYLTDLQT-RESSLLCTGEHPIR 343
GQQRC+ + H + VW L +F+ VYS G+D + T L +S LL E P++
Sbjct: 249 IGQQRCIATCIAHEEGVWTLQVDSSFTTVYSAGKDKMVVKTPLYDFTKSQLLFKEEAPVK 308
Query: 344 QLALHND----SIWVASTDSSVHRWPAEGCDPQKIFQRGNSFLAGNLSFSRARVSLEGST 399
+L L S+WV + S + RW P G + S + VS S
Sbjct: 309 KLLLSEKDNPVSLWVGTWKSDIKRWSIR---PSAQLSIGGDEDGPSTSNANHSVSASSSP 365
Query: 400 PCF*IMNF**VPIYKE------PTLTIRGTPGIVQHEVLNNKRHVLTKDTSDSVKLWEIT 453
P + F + + + P L I G P I +H +L++KRHVLT+D+ +V L+++
Sbjct: 366 P----VTFKYIRVKDQKGQQSTPELVIPGAPAIKKHAMLSDKRHVLTRDSDGNVALYDVL 421
Query: 454 KGVVVEDYGKVSFKEKKEELFEMVSVPGWFTVDTRLGTLSVHLDTQHCFNAEMYSADLNI 513
++DYGK F+E +E V +P WF VD++ G L + LD ++ + S D
Sbjct: 422 AARKIKDYGKRIFEEVVDENSRQVYIPSWFVVDSKSGMLQITLDELDALSSWLSSKDAGF 481
Query: 514 VGKPEDDKVNLGRETLKGLLTHWLRKRKQRMGSPAPANGELSGKDVASRSLIHSRAEVDV 573
+ K+N G L+ L W P +++ D A + +A ++
Sbjct: 482 DDNDRETKLNYGGMMLRSLFERW---------PPC----KMTNVDAADADDV-QKATLNF 527
Query: 574 SSENDATVYPPFEFSVVSPPSIVTEGTHGGPWRKKITDLDGTEDEKD-----FPWWCLDC 628
S + T P I+ EG +G P + + G E E + P W +D
Sbjct: 528 ISLPEHT------------PLIICEG-NGRPLYRLLVGDAGKEFEANELAQIAPMWVIDA 574
Query: 629 VLNNRLTPRENTKCSFYLQPCEGSSVQILTQGKLSAPRILRIHKVINYVVEKLVLDNK-- 686
+ N+L P+ N K FYL P ++ + + +LSA +L++ KV+ +V EK++ N
Sbjct: 575 IERNQL-PKFN-KMPFYLLPHPSTNPKQPKKDRLSATEMLQVKKVMEHVYEKILSTNDDI 632
Query: 687 -----PLDNLH 692
PL+ +H
Sbjct: 633 TVGSIPLNQIH 643
>YA4E_SCHPO (Q09731) Hypothetical WD-repeat protein C31A2.14 in
chromosome I
Length = 962
Score = 202 bits (514), Expect = 3e-51
Identities = 164/564 (29%), Positives = 258/564 (45%), Gaps = 86/564 (15%)
Query: 17 RKEKRLTYVLND-SDDTKHCAGINCLALLTSTISDGSD----YLFTGSRDGRLKRWALAE 71
RK ++TYVL+ +D H NCL L GS L++G RDG+L W +
Sbjct: 4 RKRYKVTYVLDSCNDQLGHRLDANCLVLGRYFSEQGSAPVTRALYSGGRDGQLFGWDIGY 63
Query: 72 DAATCS---ATFESHVDWVNDAVLVGDNT-LVSCSSDTTLKTW--NALSAGTCVRTHRQH 125
D S A ++H WVND L D+ ++SCSSD+T+K W + L+A +C+ T +H
Sbjct: 64 DYGKASQPLAKIQAHSAWVNDIALTHDSEGVISCSSDSTVKLWKPHVLNA-SCLSTIGEH 122
Query: 126 TDYVTCLAAAG-KNSNIVASGGLGGEVFVWDIEAALAPVSKCNDAMVDESSNGINGSANL 184
TDYV ++ S +VASGGL + VWD + M E
Sbjct: 123 TDYVKRVSIPKYSKSPLVASGGLDKRIIVWDYNVG-------QEVMRFEQ---------- 165
Query: 185 LPSTNLRTISSSHSISLHTAQTQGYIPIAAKGHKESVYALAMDEGGSILVSGGTEKVVRV 244
LP +L G + VY+LA + +I+ +GG +K +++
Sbjct: 166 LPDCSL-----------------------VVGPRSGVYSLAANN--NIIANGGLQKDIQL 200
Query: 245 WDTRSGSKILKLKGHTDNIRALLLDSTGRVLFI----RIIRFYDKGQQRCVHSYAVHTDS 300
WD S +I L GHTDN+R +L+ GR + I+ + Q+C+ S+ H+DS
Sbjct: 201 WDCVSKKRITDLVGHTDNVRDILISDDGRTILTASSDATIKLWSLRAQKCLFSFIAHSDS 260
Query: 301 VWALAST-PTFSHVYSGGRDFSLYLTDLQTRESSLLCTG----EHPIRQLALHNDSIWVA 355
VWAL S P Y+G R + TD++ + ++ CT + P+ + IW
Sbjct: 261 VWALYSEHPDLKVFYAGDRSGLITRTDIRNQPNNQSCTAICKQDAPVSDIVARQSFIWST 320
Query: 356 STDSSVHRWPAEGCDPQKIFQRGNSFLAGNLSFSRARVSLEGST----PCF*IMNF**VP 411
S D S+ RW E Q + + + +LS S S S C + +
Sbjct: 321 SRDGSILRWKDEPLFNQDVGAALSKHTSSHLSVSSDCPSRHSSDIRNHSCPTLYHDDAED 380
Query: 412 IY---------------KEPTLTIRGTPGIVQHEVLNNKRHVLTKDTSDSVKLWEITKGV 456
IY K P I G G++++ +L ++RHVLT+D + LW+I
Sbjct: 381 IYYDLHHTESYSNINLTKTPDYVIHGGIGLLKYRMLGDRRHVLTEDAVGNKCLWDILACK 440
Query: 457 VVEDYGK-VSFKEKKEELFEMVSVPGWFTVDTRLGTLSVHLDTQHCFNAEMYSADLNI-- 513
++ K F++ + L + ++P W +V+ LG L+V LD H +AE+Y+ + +
Sbjct: 441 QAGEFDKSEDFEKIVQSLDTVQAIPRWASVNCLLGILAVTLDENHYMDAEIYADECPLLK 500
Query: 514 VGKPEDDKVNLGRETLKGLLTHWL 537
V P D ++NLG LK L ++
Sbjct: 501 VDSPSDKRINLGVWILKNLFREFI 524
Score = 36.2 bits (82), Expect = 0.32
Identities = 22/76 (28%), Positives = 36/76 (46%), Gaps = 2/76 (2%)
Query: 618 EKDFPWWCLDCVLNNRLTPRENTKCSFYLQPCEGSSVQILT--QGKLSAPRILRIHKVIN 675
E P+W +L N + +F LQP GS + ++ +LSA +LR K+++
Sbjct: 835 ENIMPFWLGRLLLLNEFPSKTAPTVNFTLQPFPGSGLPLIVNENTRLSASAMLRAQKIMD 894
Query: 676 YVVEKLVLDNKPLDNL 691
Y KL K + +L
Sbjct: 895 YSYSKLSQQRKDVSSL 910
>YY46_ANASP (Q8YRI1) Hypothetical WD-repeat protein alr3466
Length = 1526
Score = 100 bits (248), Expect = 2e-20
Identities = 106/436 (24%), Positives = 181/436 (41%), Gaps = 53/436 (12%)
Query: 42 ALLTSTISDGSDYLFTGSRDGRLKRW--ALAEDAATCSATFESHVDWVNDAVLVGDNT-L 98
++LT S TG G ++ W A ++ TC + H WVN D L
Sbjct: 866 SVLTVAFSPDGKLFATGDSGGIVRFWEAATGKELLTC----KGHNSWVNSVGFSQDGKML 921
Query: 99 VSCSSDTTLKTWNALSAGTCVRTHRQHTDYVTCLAAAGKNSNIVASGGLGGEVFVWDIEA 158
S S D T++ W+ +S+G C++T + HT V + + NS ++ASG V +WDI +
Sbjct: 922 ASGSDDQTVRLWD-ISSGQCLKTFKGHTSRVRSVVFS-PNSLMLASGSSDQTVRLWDISS 979
Query: 159 ALAPVSKCNDAMVDESSNG-INGSANLLPSTNLRTISSSHSISLHTAQTQGYIPIAAKGH 217
+C + + G + A L + L T S ++ L + I +GH
Sbjct: 980 G-----EC--LYIFQGHTGWVYSVAFNLDGSMLATGSGDQTVRLWDISSSQCFYIF-QGH 1031
Query: 218 KESVYALAMDEGGSILVSGGTEKVVRVWDTRSGSKILKLKGHTDNIRALLLDSTGRVLFI 277
V ++ G++L SG ++ VR+WD SG+ + L+GHT +R+++ G +L
Sbjct: 1032 TSCVRSVVFSSDGAMLASGSDDQTVRLWDISSGNCLYTLQGHTSCVRSVVFSPDGAMLAS 1091
Query: 278 ----RIIRFYDKGQQRCVHSYAVHTDSVWALASTPTFSHVYSGGRDFSLYLTDLQTRESS 333
+I+R +D C+++ +T V L +P + +G D + L D+ +++
Sbjct: 1092 GGDDQIVRLWDISSGNCLYTLQGYTSWVRFLVFSPNGVTLANGSSDQIVRLWDISSKKCL 1151
Query: 334 LLCTGE-HPIRQLALHNDSIWVA--STDSSVHRWPAEGCD------------PQKIFQRG 378
G + + +A D +A S D +V W +F
Sbjct: 1152 YTLQGHTNWVNAVAFSPDGATLASGSGDQTVRLWDISSSKCLYILQGHTSWVNSVVFNPD 1211
Query: 379 NSFLAGNLSFSRARVSLEGSTPCF*IMNF**VPIYKEPTLTIRGTPGIVQHEVLNNKRHV 438
S LA S R+ S+ C T +G V V N +
Sbjct: 1212 GSTLASGSSDQTVRLWEINSSKCL---------------CTFQGHTSWVNSVVFNPDGSM 1256
Query: 439 LTKDTSD-SVKLWEIT 453
L +SD +V+LW+I+
Sbjct: 1257 LASGSSDKTVRLWDIS 1272
Score = 90.5 bits (223), Expect = 1e-17
Identities = 101/443 (22%), Positives = 193/443 (42%), Gaps = 31/443 (6%)
Query: 55 LFTGSRDGRLKRWALAEDAATCSATFESHVDWVNDAVLVGDN-TLVSCSSDTTLKTWNAL 113
L +G D ++ W ++ + C T + + WV V + TL + SSD ++ W+ +
Sbjct: 1089 LASGGDDQIVRLWDIS--SGNCLYTLQGYTSWVRFLVFSPNGVTLANGSSDQIVRLWD-I 1145
Query: 114 SAGTCVRTHRQHTDYVTCLAAAGKNSNIVASGGLGGEVFVWDIEAALAPVSKCNDAMVDE 173
S+ C+ T + HT++V +A + + + ASG V +WDI + SKC +
Sbjct: 1146 SSKKCLYTLQGHTNWVNAVAFSPDGATL-ASGSGDQTVRLWDISS-----SKCLYILQGH 1199
Query: 174 SSNGINGSANLLPSTNLRTISSSHSISLHTAQTQGYIPIAAKGHKESVYALAMDEGGSIL 233
+S +N + L + SS ++ L + + +GH V ++ + GS+L
Sbjct: 1200 TS-WVNSVVFNPDGSTLASGSSDQTVRLWEINSSKCL-CTFQGHTSWVNSVVFNPDGSML 1257
Query: 234 VSGGTEKVVRVWDTRSGSKILKLKGHTDNIRALLLDSTGRVLFI----RIIRFYDKGQQR 289
SG ++K VR+WD S + +GHT+ + ++ + G +L + +R ++ +
Sbjct: 1258 ASGSSDKTVRLWDISSSKCLHTFQGHTNWVNSVAFNPDGSMLASGSGDQTVRLWEISSSK 1317
Query: 290 CVHSYAVHTDSVWALASTPTFSHVYSGGRDFSLYLTDLQTRESSLLCTGE-HPIRQLALH 348
C+H++ HT V ++ +P + + SG D ++ L + + E G + + +
Sbjct: 1318 CLHTFQGHTSWVSSVTFSPDGTMLASGSDDQTVRLWSISSGECLYTFLGHTNWVGSVIFS 1377
Query: 349 NDSIWVA--STDSSVHRWPAEGCDPQKIFQRGNSFLAGNLSFSRARVSLEGST--PCF*I 404
D +A S D +V W Q N+++ G++ FS L + +
Sbjct: 1378 PDGAILASGSGDQTVRLWSISSGKCLYTLQGHNNWV-GSIVFSPDGTLLASGSDDQTVRL 1436
Query: 405 MNF**VPIYKEPTLTIRGTPGIVQHEVLNNKRHVLTKDTSD-SVKLWEITKGVVVEDYGK 463
N E T+ G V+ ++ +L + D ++KLW++ G + K
Sbjct: 1437 WNI----SSGECLYTLHGHINSVRSVAFSSDGLILASGSDDETIKLWDVKTGECI----K 1488
Query: 464 VSFKEKKEELFEMVSVPGWFTVD 486
EK E + SV G V+
Sbjct: 1489 TLKSEKIYEGMNITSVRGLTEVE 1511
Score = 83.2 bits (204), Expect = 2e-15
Identities = 60/227 (26%), Positives = 108/227 (47%), Gaps = 13/227 (5%)
Query: 50 DGSDYLFTGSRDGRLKRWALAEDAATCSATFESHVDWVNDAVLVGDNT-LVSCSSDTTLK 108
DGS L +GS D ++ W ++ ++ C TF+ H +WVN D + L S S D T++
Sbjct: 1253 DGS-MLASGSSDKTVRLWDIS--SSKCLHTFQGHTNWVNSVAFNPDGSMLASGSGDQTVR 1309
Query: 109 TWNALSAGTCVRTHRQHTDYVTCLAAAGKNSNIVASGGLGGEVFVWDIEAALAPVSKCND 168
W +S+ C+ T + HT +V+ + + + ++ASG V +W I + +C
Sbjct: 1310 LWE-ISSSKCLHTFQGHTSWVSSVTFS-PDGTMLASGSDDQTVRLWSISSG-----ECLY 1362
Query: 169 AMVDESSNGINGSANLLPSTNLRTISSSHSISLHTAQTQGYIPIAAKGHKESVYALAMDE 228
+ + N + GS P + S + + G +GH V ++
Sbjct: 1363 TFLGHT-NWV-GSVIFSPDGAILASGSGDQTVRLWSISSGKCLYTLQGHNNWVGSIVFSP 1420
Query: 229 GGSILVSGGTEKVVRVWDTRSGSKILKLKGHTDNIRALLLDSTGRVL 275
G++L SG ++ VR+W+ SG + L GH +++R++ S G +L
Sbjct: 1421 DGTLLASGSDDQTVRLWNISSGECLYTLHGHINSVRSVAFSSDGLIL 1467
Score = 77.8 bits (190), Expect = 1e-13
Identities = 61/209 (29%), Positives = 96/209 (45%), Gaps = 13/209 (6%)
Query: 50 DGSDYLFTGSRDGRLKRWALAEDAATCSATFESHVDWVNDAVLVGDNT-LVSCSSDTTLK 108
DGS L +GS D ++ W ++ ++ C TF+ H WV+ D T L S S D T++
Sbjct: 1295 DGS-MLASGSGDQTVRLWEIS--SSKCLHTFQGHTSWVSSVTFSPDGTMLASGSDDQTVR 1351
Query: 109 TWNALSAGTCVRTHRQHTDYVTCLAAAGKNSNIVASGGLGGEVFVWDIEAALAPVSKCND 168
W+ +S+G C+ T HT++V + + + I+ASG V +W I + KC
Sbjct: 1352 LWS-ISSGECLYTFLGHTNWVGSVIFSPDGA-ILASGSGDQTVRLWSISSG-----KCLY 1404
Query: 169 AMVDESSNGINGSANLLPSTNLRTISSSHSISLHTAQTQGYIPIAAKGHKESVYALAMDE 228
+ + N GS P L S + G GH SV ++A
Sbjct: 1405 TL--QGHNNWVGSIVFSPDGTLLASGSDDQTVRLWNISSGECLYTLHGHINSVRSVAFSS 1462
Query: 229 GGSILVSGGTEKVVRVWDTRSGSKILKLK 257
G IL SG ++ +++WD ++G I LK
Sbjct: 1463 DGLILASGSDDETIKLWDVKTGECIKTLK 1491
>YS00_ANASP (Q8YTC2) Hypothetical WD-repeat protein alr2800
Length = 1258
Score = 99.4 bits (246), Expect = 3e-20
Identities = 93/365 (25%), Positives = 158/365 (42%), Gaps = 27/365 (7%)
Query: 49 SDGSDYLFTGSRDGRLKRWALAEDAATCSATFESHVDWVNDAVLVGDN-TLVSCSSDTTL 107
S + L + D +K W++ + C T H V D TL S S D T+
Sbjct: 693 SPDGEILASCGADENVKLWSVRD--GVCIKTLTGHEHEVFSVAFHPDGETLASASGDKTI 750
Query: 108 KTWNALSAGTCVRTHRQHTDYVTCLAAAGKNSNIVASGGLGGEVFVWDIEAALAPVSKCN 167
K W+ + GTC++T HTD+V C+A + + N +AS + +WD+ KC
Sbjct: 751 KLWD-IQDGTCLQTLTGHTDWVRCVAFS-PDGNTLASSAADHTIKLWDVSQG-----KCL 803
Query: 168 DAMVDESSNGINGSANLLPSTNLRTISSSHSISL---HTAQT-QGYIPIAAKGHKESVYA 223
+ + + A L + S +I + HT + + YI GH SVY+
Sbjct: 804 RTLKSHTG-WVRSVAFSADGQTLASGSGDRTIKIWNYHTGECLKTYI-----GHTNSVYS 857
Query: 224 LAMDEGGSILVSGGTEKVVRVWDTRSGSKILKLKGHTDNIRALLLDSTGRVL----FIRI 279
+A ILVSG ++ +++WD ++ I L GHT+ + ++ G+ L +
Sbjct: 858 IAYSPDSKILVSGSGDRTIKLWDCQTHICIKTLHGHTNEVCSVAFSPDGQTLACVSLDQS 917
Query: 280 IRFYDKGQQRCVHSYAVHTDSVWALASTPTFSHVYSGGRDFSLYLTDLQTRESSLLCTGE 339
+R ++ +C+ ++ +TD +A +P + SG D ++ L D QT + G
Sbjct: 918 VRLWNCRTGQCLKAWYGNTDWALPVAFSPDRQILASGSNDKTVKLWDWQTGKYISSLEGH 977
Query: 340 HP-IRQLALHNDS--IWVASTDSSVHRWPAEGCDPQKIFQRGNSFLAGNLSFSRARVSLE 396
I +A DS + ASTDSSV W +I ++ + + ++
Sbjct: 978 TDFIYGIAFSPDSQTLASASTDSSVRLWNISTGQCFQILLEHTDWVYAVVFHPQGKIIAT 1037
Query: 397 GSTPC 401
GS C
Sbjct: 1038 GSADC 1042
Score = 95.9 bits (237), Expect = 3e-19
Identities = 76/293 (25%), Positives = 134/293 (44%), Gaps = 16/293 (5%)
Query: 44 LTSTISDGSDYLFTGSRDGRLKRWALAEDAATCSATFESHVDWVNDAVLVGDN-TLVSCS 102
L S L +GS D +K W ++ E H D++ D+ TL S S
Sbjct: 940 LPVAFSPDRQILASGSNDKTVKLWDW--QTGKYISSLEGHTDFIYGIAFSPDSQTLASAS 997
Query: 103 SDTTLKTWNALSAGTCVRTHRQHTDYVTCLAAAGKNSNIVASGGLGGEVFVWDIEAALAP 162
+D++++ WN +S G C + +HTD+V + + I+A+G V +W+I
Sbjct: 998 TDSSVRLWN-ISTGQCFQILLEHTDWVYAVVFHPQGK-IIATGSADCTVKLWNISTG--- 1052
Query: 163 VSKCNDAMVDESSNGINGSANLLPSTNLRTISSSHSISLHTAQTQGYIPIAAKGHKESVY 222
+C + E S+ I G A L + S+ S+ L T + I +GH VY
Sbjct: 1053 --QCLKTL-SEHSDKILGMAWSPDGQLLASASADQSVRLWDCCTGRCVGIL-RGHSNRVY 1108
Query: 223 ALAMDEGGSILVSGGTEKVVRVWDTRSGSKILKLKGHTDNIRALLLDSTGRVL----FIR 278
+ G I+ + T++ V++WD + G + L GHT+ + + G++L +
Sbjct: 1109 SAIFSPNGEIIATCSTDQTVKIWDWQQGKCLKTLTGHTNWVFDIAFSPDGKILASASHDQ 1168
Query: 279 IIRFYDKGQQRCVHSYAVHTDSVWALASTPTFSHVYSGGRDFSLYLTDLQTRE 331
+R +D +C H HT V ++A +P V SG +D ++ + +++T E
Sbjct: 1169 TVRIWDVNTGKCHHICIGHTHLVSSVAFSPDGEVVASGSQDQTVRIWNVKTGE 1221
Score = 50.8 bits (120), Expect = 1e-05
Identities = 32/124 (25%), Positives = 55/124 (43%), Gaps = 4/124 (3%)
Query: 208 GYIPIAAKGHKESVYALAMDEGGSILVSGGTEKVVRVWDTRSGSKILKLKGHTDNIRALL 267
G + + +GH V + G IL S G ++ V++W R G I L GH + ++
Sbjct: 674 GKLLLICRGHSNWVRFVVFSPDGEILASCGADENVKLWSVRDGVCIKTLTGHEHEVFSVA 733
Query: 268 LDSTGRVLFI----RIIRFYDKGQQRCVHSYAVHTDSVWALASTPTFSHVYSGGRDFSLY 323
G L + I+ +D C+ + HTD V +A +P + + S D ++
Sbjct: 734 FHPDGETLASASGDKTIKLWDIQDGTCLQTLTGHTDWVRCVAFSPDGNTLASSAADHTIK 793
Query: 324 LTDL 327
L D+
Sbjct: 794 LWDV 797
Score = 47.0 bits (110), Expect = 2e-04
Identities = 35/152 (23%), Positives = 69/152 (45%), Gaps = 7/152 (4%)
Query: 220 SVYALAMDEGGSILVSGGTEKVVRVWDTRSGSKILKLKGHTDNIRALLLDSTGRVLFI-- 277
++ + A G +L + T+ VRVW+ +SG +L +GH++ +R ++ G +L
Sbjct: 644 NILSAAFSPEGQLLATCDTDCHVRVWEVKSGKLLLICRGHSNWVRFVVFSPDGEILASCG 703
Query: 278 --RIIRFYDKGQQRCVHSYAVHTDSVWALASTPTFSHVYSGGRDFSLYLTDLQTRESSLL 335
++ + C+ + H V+++A P + S D ++ L D+Q
Sbjct: 704 ADENVKLWSVRDGVCIKTLTGHEHEVFSVAFHPDGETLASASGDKTIKLWDIQDGTCLQT 763
Query: 336 CTGEHP-IRQLALHNDSIWVAST--DSSVHRW 364
TG +R +A D +AS+ D ++ W
Sbjct: 764 LTGHTDWVRCVAFSPDGNTLASSAADHTIKLW 795
>HET1_PODAN (Q00808) Vegetatible incompatibility protein HET-E-1
Length = 1356
Score = 93.6 bits (231), Expect = 2e-18
Identities = 104/429 (24%), Positives = 174/429 (40%), Gaps = 27/429 (6%)
Query: 42 ALLTSTISDGSDYLFTGSRDGRLKRWALAEDAATCSATFESHVDWVNDAVLVGDNTLV-S 100
++L+ S + +GS D +K W A T T E H V D V S
Sbjct: 843 SVLSVAFSADGQRVASGSDDKTIKIWDTASGTGT--QTLEGHGGSVWSVAFSPDRERVAS 900
Query: 101 CSSDTTLKTWNALSAGTCVRTHRQHTDYVTCLAAAGKNSNIVASGGLGGEVFVWDIEAAL 160
S D T+K W+A S GTC +T H V +A + VASG + +WD
Sbjct: 901 GSDDKTIKIWDAAS-GTCTQTLEGHGGRVQSVAFSPDGQR-VASGSDDHTIKIWD----- 953
Query: 161 APVSKCNDAMVDESSNGINGSANLLPSTNLRTISSSHSISLHTAQT-QGYIPIAAKGHKE 219
A C + S+ ++ + + R S S ++ T G +GH
Sbjct: 954 AASGTCTQTLEGHGSSVLSVAFS---PDGQRVASGSGDKTIKIWDTASGTCTQTLEGHGG 1010
Query: 220 SVYALAMDEGGSILVSGGTEKVVRVWDTRSGSKILKLKGHTDNIRALLLDSTGRVLFI-- 277
SV+++A G + SG +K +++WDT SG+ L+GH +++++ G+ +
Sbjct: 1011 SVWSVAFSPDGQRVASGSDDKTIKIWDTASGTCTQTLEGHGGWVQSVVFSPDGQRVASGS 1070
Query: 278 --RIIRFYDKGQQRCVHSYAVHTDSVWALASTPTFSHVYSGGRDFSLYLTDLQTRESSLL 335
I+ +D C + H DSVW++A +P V SG D ++ + D + +
Sbjct: 1071 DDHTIKIWDAVSGTCTQTLEGHGDSVWSVAFSPDGQRVASGSIDGTIKIWDAASGTCTQT 1130
Query: 336 CTGEHP-IRQLALHNDSIWVA--STDSSVHRW-PAEGCDPQKIFQRGNSFLAGNLSFSRA 391
G + +A D VA S D ++ W A G Q + G + S
Sbjct: 1131 LEGHGGWVHSVAFSPDGQRVASGSIDGTIKIWDAASGTCTQTLEGHGGWVQSVAFSPDGQ 1190
Query: 392 RVSLEGSTPCF*IMNF**VPIYKEPTLTIRGTPGIVQHEVLN-NKRHVLTKDTSDSVKLW 450
RV+ S I + T T+ G G VQ + + + V + + +++K+W
Sbjct: 1191 RVASGSSDKTIKIWD----TASGTCTQTLEGHGGWVQSVAFSPDGQRVASGSSDNTIKIW 1246
Query: 451 EITKGVVVE 459
+ G +
Sbjct: 1247 DTASGTCTQ 1255
Score = 57.8 bits (138), Expect = 1e-07
Identities = 57/254 (22%), Positives = 102/254 (39%), Gaps = 13/254 (5%)
Query: 215 KGHKESVYALAMDEGGSILVSGGTEKVVRVWDTRSGSKILKLKGHTDNIRALLLDSTGRV 274
+GH SV ++A G + SG +K +++WDT SG+ L+GH ++ ++
Sbjct: 838 EGHGSSVLSVAFSADGQRVASGSDDKTIKIWDTASGTGTQTLEGHGGSVWSVAFSPDRER 897
Query: 275 LFI----RIIRFYDKGQQRCVHSYAVHTDSVWALASTPTFSHVYSGGRDFSLYLTDLQTR 330
+ + I+ +D C + H V ++A +P V SG D ++ + D +
Sbjct: 898 VASGSDDKTIKIWDAASGTCTQTLEGHGGRVQSVAFSPDGQRVASGSDDHTIKIWDAASG 957
Query: 331 ESSLLCTGE-HPIRQLALHNDSIWVA--STDSSVHRW-PAEGCDPQKIFQRGNSFLAGNL 386
+ G + +A D VA S D ++ W A G Q + G S +
Sbjct: 958 TCTQTLEGHGSSVLSVAFSPDGQRVASGSGDKTIKIWDTASGTCTQTLEGHGGSVWSVAF 1017
Query: 387 SFSRARVSLEGSTPCF*IMNF**VPIYKEPTLTIRGTPGIVQHEVLN-NKRHVLTKDTSD 445
S RV+ I + T T+ G G VQ V + + + V +
Sbjct: 1018 SPDGQRVASGSDDKTIKIWD----TASGTCTQTLEGHGGWVQSVVFSPDGQRVASGSDDH 1073
Query: 446 SVKLWEITKGVVVE 459
++K+W+ G +
Sbjct: 1074 TIKIWDAVSGTCTQ 1087
>PKWA_THECU (P49695) Probable serine/threonine-protein kinase pkwA
(EC 2.7.1.37)
Length = 742
Score = 90.5 bits (223), Expect = 1e-17
Identities = 78/324 (24%), Positives = 129/324 (39%), Gaps = 60/324 (18%)
Query: 49 SDGSDYLFTGSRDGRLKRWALAEDAATCSATFESHVDWVNDAVLVGDNTLV-SCSSDTTL 107
S G L GS D + W +A T E H DWV D L+ S S D T+
Sbjct: 468 SPGGSLLAGGSGDKLIHVWDVASGDEL--HTLEGHTDWVRAVAFSPDGALLASGSDDATV 525
Query: 108 KTWNALSAGTCVRTHRQHTDYVTCLAAAGKNSNIVASGGLGGEVFVWDIEAALAPVSKCN 167
+ W+ +A HT YV +A + + ++VASG G +W++
Sbjct: 526 RLWDVAAAEERA-VFEGHTHYVLDIAFS-PDGSMVASGSRDGTARLWNV----------- 572
Query: 168 DAMVDESSNGINGSANLLPSTNLRTISSSHSISLHTAQTQGYIPIAAKGHKESVYALAMD 227
+ H++ KGH + VYA+A
Sbjct: 573 ------------------------ATGTEHAV--------------LKGHTDYVYAVAFS 594
Query: 228 EGGSILVSGGTEKVVRVWDTRSGSKILKLKGHTDNIRALLLDSTGRVLF---IRIIRFYD 284
GS++ SG + +R+WD +G + L+ +N+ +L G +L + +D
Sbjct: 595 PDGSMVASGSRDGTIRLWDVATGKERDVLQAPAENVVSLAFSPDGSMLVHGSDSTVHLWD 654
Query: 285 KGQQRCVHSYAVHTDSVWALASTPTFSHVYSGGRDFSLYLTDLQTRESSLLCTGE-HPIR 343
+H++ HTD V A+A +P + + SG D ++ L D+ +E G P+
Sbjct: 655 VASGEALHTFEGHTDWVRAVAFSPDGALLASGSDDRTIRLWDVAAQEEHTTLEGHTEPVH 714
Query: 344 QLALHNDSIWVAST--DSSVHRWP 365
+A H + +AS D ++ WP
Sbjct: 715 SVAFHPEGTTLASASEDGTIRIWP 738
Score = 69.3 bits (168), Expect = 3e-11
Identities = 69/231 (29%), Positives = 100/231 (42%), Gaps = 56/231 (24%)
Query: 185 LPSTNLRTISS---SHSISLHTAQTQGYIPIAAKGHKESVYALAMDEGGSILVSGGTEKV 241
LP T LR SS S S H P +E+V A+A GGS+L G +K+
Sbjct: 428 LPETPLRPDSSTAPSESADPHELNE----PRILTTDREAV-AVAFSPGGSLLAGGSGDKL 482
Query: 242 VRVWDTRSGSKILKLKGHTDNIRALLLDSTGRVLFI----RIIRFYD--KGQQRCV---H 292
+ VWD SG ++ L+GHTD +RA+ G +L +R +D ++R V H
Sbjct: 483 IHVWDVASGDELHTLEGHTDWVRAVAFSPDGALLASGSDDATVRLWDVAAAEERAVFEGH 542
Query: 293 SYAV-------------------------------------HTDSVWALASTPTFSHVYS 315
++ V HTD V+A+A +P S V S
Sbjct: 543 THYVLDIAFSPDGSMVASGSRDGTARLWNVATGTEHAVLKGHTDYVYAVAFSPDGSMVAS 602
Query: 316 GGRDFSLYLTDLQT-RESSLLCTGEHPIRQLALHND-SIWVASTDSSVHRW 364
G RD ++ L D+ T +E +L + LA D S+ V +DS+VH W
Sbjct: 603 GSRDGTIRLWDVATGKERDVLQAPAENVVSLAFSPDGSMLVHGSDSTVHLW 653
Score = 37.7 bits (86), Expect = 0.11
Identities = 32/108 (29%), Positives = 48/108 (43%), Gaps = 7/108 (6%)
Query: 50 DGSDYLFTGSRDGRLKRWALAEDAATCSATFESHVDWVNDAVLVGDNTLV-SCSSDTTLK 108
DGS + D + W +A A TFE H DWV D L+ S S D T++
Sbjct: 638 DGS--MLVHGSDSTVHLWDVASGEAL--HTFEGHTDWVRAVAFSPDGALLASGSDDRTIR 693
Query: 109 TWNALSAGTCVRTHRQHTDYVTCLAAAGKNSNIVASGGLGGEVFVWDI 156
W+ ++A T HT+ V +A + + + AS G + +W I
Sbjct: 694 LWD-VAAQEEHTTLEGHTEPVHSVAFHPEGTTL-ASASEDGTIRIWPI 739
>YH92_CAEEL (Q23256) Hypothetical WD-repeat protein ZC302.2 in
chromosome V
Length = 501
Score = 79.0 bits (193), Expect = 4e-14
Identities = 85/328 (25%), Positives = 136/328 (40%), Gaps = 38/328 (11%)
Query: 4 VASTGNANNSTRPRKEKRLTYVLNDSDDTKHCAGINCLALLTSTISDGSDYLFTGSRDGR 63
VA T +++ T+ + + V S TK + I S YL TGS D +
Sbjct: 185 VAPTTSSSGITKKPENGEFSLVKTISGHTKSVSVIK--------FSYCGKYLGTGSADKQ 236
Query: 64 LKRWALAEDAATCSATFESHVDWVNDAVLVGDNTLV-SCSSDTTLKTWNALSAGTCVRTH 122
+K W + T T SH +ND ++ + S S DTT+K ++ +S G C+RT
Sbjct: 237 IKVWNTVD--MTYLQTLASHQLGINDFSWSSNSQFIASASDDTTVKIFDVIS-GACLRTM 293
Query: 123 RQHTDYVTCLAAAGKNSNIVASGGLGGEVFVWDIEAALAPVSKCNDAMVDE-SSNGINGS 181
R HT+YV C + S+++AS G V VWD + L KC A D +S N
Sbjct: 294 RGHTNYVFC-CSFNPQSSLIASAGFDETVRVWDFKTGLC--VKCIPAHSDPITSISYNHD 350
Query: 182 ANLLPSTN----LRTISSSHSISLHTAQTQGYIPIAAKGHKESVYALAMDEGGSILVSGG 237
N + +++ +R ++ L T + P+ + G L+S
Sbjct: 351 GNTMATSSYDGCIRVWDAASGSCLKTLVDTDHAPVT---------FVCFSPNGKYLLSAQ 401
Query: 238 TEKVVRVWDTRSGSKILKLKGHTDNIRALLLDSTGRVLFIRIIRFYDKGQ--------QR 289
+ +++WD + + GH N + L + L II + G+ ++
Sbjct: 402 LDSSLKLWDPKKAKPLKYYNGH-KNKKYCLFANMSVPLGKHIISGSEDGRILVWSIQTKQ 460
Query: 290 CVHSYAVHTDSVWALASTPTFSHVYSGG 317
V HT V A S PT + + SGG
Sbjct: 461 IVQILEGHTTPVLATDSHPTLNIIASGG 488
Score = 45.4 bits (106), Expect = 5e-04
Identities = 31/118 (26%), Positives = 56/118 (47%), Gaps = 7/118 (5%)
Query: 215 KGHKESVYALAMDEGGSILVSGGTEKVVRVWDTRSGSKILKLKGHTDNIRALLLDSTGRV 274
+GH V+ + + S++ S G ++ VRVWD ++G + + H+D I ++ + G
Sbjct: 294 RGHTNYVFCCSFNPQSSLIASAGFDETVRVWDFKTGLCVKCIPAHSDPITSISYNHDGNT 353
Query: 275 L----FIRIIRFYDKGQQRCVHSYAVHTD--SVWALASTPTFSHVYSGGRDFSLYLTD 326
+ + IR +D C+ + V TD V + +P ++ S D SL L D
Sbjct: 354 MATSSYDGCIRVWDAASGSCLKT-LVDTDHAPVTFVCFSPNGKYLLSAQLDSSLKLWD 410
>FBW7_MOUSE (Q8VBV4) F-box/WD-repeat protein 7 (F-box and WD-40
domain protein 7) (F-box protein FBW7) (F-box protein
Fbxw6) (F-box-WD40 repeat protein 6) (SEL-10)
Length = 629
Score = 77.8 bits (190), Expect = 1e-13
Identities = 87/322 (27%), Positives = 131/322 (40%), Gaps = 40/322 (12%)
Query: 17 RKEKRLTYVLNDSDDTKHCAGINCLALLTSTISDGSDYLFTGSRDGRLKRWALAEDAATC 76
R E + VL DD I CL + I GSD D LK W+ C
Sbjct: 289 RGELKSPKVLKGHDDHV----ITCLQFCGNRIVSGSD-------DNTLKVWSAV--TGKC 335
Query: 77 SATFESHVDWVNDAVLVGDNTLVSCSSDTTLKTWNALSAGTCVRTHRQHTDYVTCLAAAG 136
T H V + + DN ++S S+D TLK WNA G C+ T HT V C+
Sbjct: 336 LRTLVGHTGGVWSSQM-RDNIIISGSTDRTLKVWNA-ETGECIHTLYGHTSTVRCMHLHE 393
Query: 137 KNSNIVASGGLGGEVFVWDIEAALAPVSKCNDAMVDESSNGINGSANLLPSTNLRTISSS 196
K V SG + VWDIE +C ++ G + + R +S +
Sbjct: 394 KR---VVSGSRDATLRVWDIETG-----QCLHVLM-----GHVAAVRCVQYDGRRVVSGA 440
Query: 197 HS--ISLHTAQTQGYIPIAAKGHKESVYALAMDEGGSILVSGGTEKVVRVWDTRSGSKIL 254
+ + + +T+ + +GH VY+L D G +VSG + +RVWD +G+ I
Sbjct: 441 YDFMVKVWDPETETCLH-TLQGHTNRVYSLQFD--GIHVVSGSLDTSIRVWDVETGNCIH 497
Query: 255 KLKGHTDNIRALLLDSTGRVL--FIRIIRFYDKGQQRCVHSY---AVHTDSVWALASTPT 309
L GH + L V ++ +D +C+ + + H +V L
Sbjct: 498 TLTGHQSLTSGMELKDNILVSGNADSTVKIWDIKTGQCLQTLQGPSKHQSAVTCLQFNKN 557
Query: 310 FSHVYSGGRDFSLYLTDLQTRE 331
F V + D ++ L DL+T E
Sbjct: 558 F--VITSSDDGTVKLWDLKTGE 577
Score = 70.9 bits (172), Expect = 1e-11
Identities = 72/287 (25%), Positives = 116/287 (40%), Gaps = 60/287 (20%)
Query: 81 ESHVDWVNDAVLVGDNTLVSCSSDTTLKTWNALSAGTCVRTHRQHTDYVTCLAAAGKNSN 140
+ H D V + N +VS S D TLK W+A++ G C+RT HT V ++ N
Sbjct: 299 KGHDDHVITCLQFCGNRIVSGSDDNTLKVWSAVT-GKCLRTLVGHTGGVW---SSQMRDN 354
Query: 141 IVASGGLGGEVFVWDIEAALAPVSKCNDAMVDESSNGINGSANLLPSTNLRTISSSHSIS 200
I+ SG + VW+ E +C
Sbjct: 355 IIISGSTDRTLKVWNAETG-----EC---------------------------------- 375
Query: 201 LHTAQTQGYIPIAAKGHKESVYALAMDEGGSILVSGGTEKVVRVWDTRSGSKILKLKGHT 260
+HT GH +V + + E +VSG + +RVWD +G + L GH
Sbjct: 376 IHTLY----------GHTSTVRCMHLHE--KRVVSGSRDATLRVWDIETGQCLHVLMGHV 423
Query: 261 DNIRALLLDSTGRV--LFIRIIRFYDKGQQRCVHSYAVHTDSVWALASTPTFSHVYSGGR 318
+R + D V + +++ +D + C+H+ HT+ V++L HV SG
Sbjct: 424 AAVRCVQYDGRRVVSGAYDFMVKVWDPETETCLHTLQGHTNRVYSLQFDGI--HVVSGSL 481
Query: 319 DFSLYLTDLQTRESSLLCTGEHPIRQLALHNDSIWVA-STDSSVHRW 364
D S+ + D++T TG + D+I V+ + DS+V W
Sbjct: 482 DTSIRVWDVETGNCIHTLTGHQSLTSGMELKDNILVSGNADSTVKIW 528
Score = 56.6 bits (135), Expect = 2e-07
Identities = 64/255 (25%), Positives = 106/255 (41%), Gaps = 24/255 (9%)
Query: 216 GHKESVYALAMDEGGSILVSGGTEKVVRVWDTRSGSKILKLKGHTDNIRALLLDSTGRVL 275
GH V++ M + +I++SG T++ ++VW+ +G I L GHT +R + L V
Sbjct: 341 GHTGGVWSSQMRD--NIIISGSTDRTLKVWNAETGECIHTLYGHTSTVRCMHLHEKRVVS 398
Query: 276 FIR--IIRFYDKGQQRCVHSYAVHTDSVWALASTPTFSHVYSGGRDFSLYLTDLQTRESS 333
R +R +D +C+H H +V + V SG DF + + D +T E+
Sbjct: 399 GSRDATLRVWDIETGQCLHVLMGHVAAVRCVQYDG--RRVVSGAYDFMVKVWDPET-ETC 455
Query: 334 LLCTGEHPIRQLALHNDSIWV--ASTDSSVHRWPAEGCDPQKIFQRGNSFLAGNLSFSRA 391
L H R +L D I V S D+S+ W E + S +G
Sbjct: 456 LHTLQGHTNRVYSLQFDGIHVVSGSLDTSIRVWDVETGNCIHTLTGHQSLTSGMELKDNI 515
Query: 392 RVSLEGSTPCF*IMNF**VPIYKEPT-LTIRGTPGIVQHE-----VLNNKRHVLTKDTSD 445
VS + V I+ T ++ G +H+ + NK V+T
Sbjct: 516 LVSGNADST---------VKIWDIKTGQCLQTLQGPSKHQSAVTCLQFNKNFVITSSDDG 566
Query: 446 SVKLWEITKGVVVED 460
+VKLW++ G + +
Sbjct: 567 TVKLWDLKTGEFIRN 581
>FBW7_HUMAN (Q969H0) F-box/WD-repeat protein 7 (F-box and WD-40
domain protein 7) (F-box protein FBX30) (hCdc4)
(Archipelago homolog) (hAgo) (SEL-10)
Length = 707
Score = 77.0 bits (188), Expect = 2e-13
Identities = 72/245 (29%), Positives = 103/245 (41%), Gaps = 33/245 (13%)
Query: 17 RKEKRLTYVLNDSDDTKHCAGINCLALLTSTISDGSDYLFTGSRDGRLKRWALAEDAATC 76
R E + VL DD I CL + I GSD D LK W+ C
Sbjct: 367 RGELKSPKVLKGHDDHV----ITCLQFCGNRIVSGSD-------DNTLKVWSAV--TGKC 413
Query: 77 SATFESHVDWVNDAVLVGDNTLVSCSSDTTLKTWNALSAGTCVRTHRQHTDYVTCLAAAG 136
T H V + + DN ++S S+D TLK WNA G C+ T HT V C+
Sbjct: 414 LRTLVGHTGGVWSSQM-RDNIIISGSTDRTLKVWNA-ETGECIHTLYGHTSTVRCMHLHE 471
Query: 137 KNSNIVASGGLGGEVFVWDIEAALAPVSKCNDAMVDESSNGINGSANLLPSTNLRTISSS 196
K V SG + VWDIE +C ++ G + + R +S +
Sbjct: 472 KR---VVSGSRDATLRVWDIETG-----QCLHVLM-----GHVAAVRCVQYDGRRVVSGA 518
Query: 197 HS--ISLHTAQTQGYIPIAAKGHKESVYALAMDEGGSILVSGGTEKVVRVWDTRSGSKIL 254
+ + + +T+ + +GH VY+L D G +VSG + +RVWD +G+ I
Sbjct: 519 YDFMVKVWDPETETCLH-TLQGHTNRVYSLQFD--GIHVVSGSLDTSIRVWDVETGNCIH 575
Query: 255 KLKGH 259
L GH
Sbjct: 576 TLTGH 580
Score = 70.9 bits (172), Expect = 1e-11
Identities = 72/287 (25%), Positives = 116/287 (40%), Gaps = 60/287 (20%)
Query: 81 ESHVDWVNDAVLVGDNTLVSCSSDTTLKTWNALSAGTCVRTHRQHTDYVTCLAAAGKNSN 140
+ H D V + N +VS S D TLK W+A++ G C+RT HT V ++ N
Sbjct: 377 KGHDDHVITCLQFCGNRIVSGSDDNTLKVWSAVT-GKCLRTLVGHTGGVW---SSQMRDN 432
Query: 141 IVASGGLGGEVFVWDIEAALAPVSKCNDAMVDESSNGINGSANLLPSTNLRTISSSHSIS 200
I+ SG + VW+ E +C
Sbjct: 433 IIISGSTDRTLKVWNAETG-----EC---------------------------------- 453
Query: 201 LHTAQTQGYIPIAAKGHKESVYALAMDEGGSILVSGGTEKVVRVWDTRSGSKILKLKGHT 260
+HT GH +V + + E +VSG + +RVWD +G + L GH
Sbjct: 454 IHTLY----------GHTSTVRCMHLHE--KRVVSGSRDATLRVWDIETGQCLHVLMGHV 501
Query: 261 DNIRALLLDSTGRV--LFIRIIRFYDKGQQRCVHSYAVHTDSVWALASTPTFSHVYSGGR 318
+R + D V + +++ +D + C+H+ HT+ V++L HV SG
Sbjct: 502 AAVRCVQYDGRRVVSGAYDFMVKVWDPETETCLHTLQGHTNRVYSLQFDGI--HVVSGSL 559
Query: 319 DFSLYLTDLQTRESSLLCTGEHPIRQLALHNDSIWVA-STDSSVHRW 364
D S+ + D++T TG + D+I V+ + DS+V W
Sbjct: 560 DTSIRVWDVETGNCIHTLTGHQSLTSGMELKDNILVSGNADSTVKIW 606
Score = 56.2 bits (134), Expect = 3e-07
Identities = 64/255 (25%), Positives = 106/255 (41%), Gaps = 24/255 (9%)
Query: 216 GHKESVYALAMDEGGSILVSGGTEKVVRVWDTRSGSKILKLKGHTDNIRALLLDSTGRVL 275
GH V++ M + +I++SG T++ ++VW+ +G I L GHT +R + L V
Sbjct: 419 GHTGGVWSSQMRD--NIIISGSTDRTLKVWNAETGECIHTLYGHTSTVRCMHLHEKRVVS 476
Query: 276 FIR--IIRFYDKGQQRCVHSYAVHTDSVWALASTPTFSHVYSGGRDFSLYLTDLQTRESS 333
R +R +D +C+H H +V + V SG DF + + D +T E+
Sbjct: 477 GSRDATLRVWDIETGQCLHVLMGHVAAVRCVQYDG--RRVVSGAYDFMVKVWDPET-ETC 533
Query: 334 LLCTGEHPIRQLALHNDSIWV--ASTDSSVHRWPAEGCDPQKIFQRGNSFLAGNLSFSRA 391
L H R +L D I V S D+S+ W E + S +G
Sbjct: 534 LHTLQGHTNRVYSLQFDGIHVVSGSLDTSIRVWDVETGNCIHTLTGHQSLTSGMELKDNI 593
Query: 392 RVSLEGSTPCF*IMNF**VPIYKEPT-LTIRGTPGIVQHE-----VLNNKRHVLTKDTSD 445
VS + V I+ T ++ G +H+ + NK V+T
Sbjct: 594 LVSGNADST---------VKIWDIKTGQCLQTLQGPNKHQSAVTCLQFNKNFVITSSDDG 644
Query: 446 SVKLWEITKGVVVED 460
+VKLW++ G + +
Sbjct: 645 TVKLWDLKTGEFIRN 659
Score = 47.0 bits (110), Expect = 2e-04
Identities = 31/107 (28%), Positives = 55/107 (50%), Gaps = 10/107 (9%)
Query: 54 YLFTGSRDGRLKRWALAEDAATCSATFESHVDWVNDAVLVGDNTLVSCSSDTTLKTWNAL 113
++ +GS D ++ W + + C T H + + + DN LVS ++D+T+K W+ +
Sbjct: 553 HVVSGSLDTSIRVWDV--ETGNCIHTLTGHQS-LTSGMELKDNILVSGNADSTVKIWD-I 608
Query: 114 SAGTCVRTHR---QHTDYVTCLAAAGKNSNIVASGGLGGEVFVWDIE 157
G C++T + +H VTCL N N V + G V +WD++
Sbjct: 609 KTGQCLQTLQGPNKHQSAVTCLQF---NKNFVITSSDDGTVKLWDLK 652
Score = 44.7 bits (104), Expect = 0.001
Identities = 35/140 (25%), Positives = 61/140 (43%), Gaps = 15/140 (10%)
Query: 197 HSISLHTAQTQGYIPIAAKGHKESVYALAMDEGGSILVSGGTEKVVRVWDTRSGSKILKL 256
H I + + + P KGH + V + G+ +VSG + ++VW +G + L
Sbjct: 359 HRIDTNWRRGELKSPKVLKGHDDHVIT-CLQFCGNRIVSGSDDNTLKVWSAVTGKCLRTL 417
Query: 257 KGHTDNI-------RALLLDSTGRVLFIRIIRFYDKGQQRCVHSYAVHTDSVWALASTPT 309
GHT + ++ ST R L + ++ C+H+ HT +V +
Sbjct: 418 VGHTGGVWSSQMRDNIIISGSTDRTL-----KVWNAETGECIHTLYGHTSTVRCMHLHE- 471
Query: 310 FSHVYSGGRDFSLYLTDLQT 329
V SG RD +L + D++T
Sbjct: 472 -KRVVSGSRDATLRVWDIET 490
>SE10_CAEEL (Q93794) F-box/WD-repeat protein sel-10
(Suppressor/enhancer of lin-12) (Sel-10 protein)
Length = 587
Score = 74.7 bits (182), Expect = 8e-13
Identities = 69/264 (26%), Positives = 117/264 (44%), Gaps = 33/264 (12%)
Query: 53 DYLFTGSRDGRLKRWALAEDAATCSATFESHVDWVNDAVLVGDNTLVSCSSDTTLKTWNA 112
D L TGS D LK W + + + + W + G +VS S+D T+K W+
Sbjct: 267 DVLVTGSDDNTLKVWCIDKGEVMYTLVGHTGGVWTSQISQCG-RYIVSGSTDRTVKVWST 325
Query: 113 LSAGTCVRTHRQHTDYVTCLAAAGKNSNIVASGGLGGEVFVWDIEAA--LAPVSKCNDAM 170
+ G+ + T + HT V C+A AG +I+ +G + VWD+E+ LA +
Sbjct: 326 VD-GSLLHTLQGHTSTVRCMAMAG---SILVTGSRDTTLRVWDVESGRHLATL------- 374
Query: 171 VDESSNGINGSANLLPSTNLRTISSSH--SISLHTAQTQGYIPIAAKGHKESVYALAMDE 228
+G + + + +S + ++ + A T G GH VY+L +
Sbjct: 375 -----HGHHAAVRCVQFDGTTVVSGGYDFTVKIWNAHT-GRCIRTLTGHNNRVYSLLFES 428
Query: 229 GGSILVSGGTEKVVRVWD-TRSGSK--ILKLKGHTD-----NIRALLLDSTGRVLFIRII 280
SI+ SG + +RVWD TR + + L+GHT +R +L S +R+
Sbjct: 429 ERSIVCSGSLDTSIRVWDFTRPEGQECVALLQGHTSLTSGMQLRGNILVSCNADSHVRV- 487
Query: 281 RFYDKGQQRCVHSYAVHTDSVWAL 304
+D + CVH + H ++ +L
Sbjct: 488 --WDIHEGTCVHMLSGHRSAITSL 509
Score = 70.5 bits (171), Expect = 2e-11
Identities = 75/294 (25%), Positives = 117/294 (39%), Gaps = 59/294 (20%)
Query: 77 SATFESHVDWVNDAVLVGDNTLVSCSSDTTLKTWNALSAGTCVRTHRQHTDYVTCLAAAG 136
SA H D V + + D+ LV+ S D TLK W + G + T HT
Sbjct: 248 SAVLRGHEDHVITCMQIHDDVLVTGSDDNTLKVW-CIDKGEVMYTLVGHT---------- 296
Query: 137 KNSNIVASGGLGGEVFVWDIEAALAPVSKCNDAMVDESSNGINGSANLLPSTNLRTISSS 196
GG VW + +S+C +V S++ ++ S+
Sbjct: 297 --------GG------VW-----TSQISQCGRYIVSGSTD-----------RTVKVWSTV 326
Query: 197 HSISLHTAQTQGYIPIAAKGHKESVYALAMDEGGSILVSGGTEKVVRVWDTRSGSKILKL 256
LHT Q GH +V +AM GSILV+G + +RVWD SG + L
Sbjct: 327 DGSLLHTLQ----------GHTSTVRCMAM--AGSILVTGSRDTTLRVWDVESGRHLATL 374
Query: 257 KGHTDNIRALLLDSTGRVL--FIRIIRFYDKGQQRCVHSYAVHTDSVWALASTPTFSHVY 314
GH +R + D T V + ++ ++ RC+ + H + V++L S V
Sbjct: 375 HGHHAAVRCVQFDGTTVVSGGYDFTVKIWNAHTGRCIRTLTGHNNRVYSLLFESERSIVC 434
Query: 315 SGGRDFSLYLTDL---QTRESSLLCTGEHPIRQ-LALHNDSIWVASTDSSVHRW 364
SG D S+ + D + +E L G + + L + + + DS V W
Sbjct: 435 SGSLDTSIRVWDFTRPEGQECVALLQGHTSLTSGMQLRGNILVSCNADSHVRVW 488
Score = 61.2 bits (147), Expect = 9e-09
Identities = 63/228 (27%), Positives = 94/228 (40%), Gaps = 25/228 (10%)
Query: 45 TSTISDGSDYLFTGSRDGRLKRWALAEDAATCSATFESHVDWVNDAVLVGDNTLVSCSSD 104
TS IS Y+ +GS D +K W+ + + T + H V + G + LV+ S D
Sbjct: 301 TSQISQCGRYIVSGSTDRTVKVWSTVDGSLL--HTLQGHTSTVRCMAMAG-SILVTGSRD 357
Query: 105 TTLKTWNALSAGTCVRTHRQHTDYVTCLAAAGKNSNIVASGGLGGEVFVWDIEAALAPVS 164
TTL+ W+ S H H V C+ G V SGG V +W+ A
Sbjct: 358 TTLRVWDVESGRHLATLHGHHAA-VRCVQFDGTT---VVSGGYDFTVKIWN-----AHTG 408
Query: 165 KCNDAMVDESSNGINGSANLLPSTNLRTISSSHSISLHT-----AQTQGYIPIAA-KGHK 218
+C + G N L + R+I S S+ + +G +A +GH
Sbjct: 409 RCIRTLT-----GHNNRVYSLLFESERSIVCSGSLDTSIRVWDFTRPEGQECVALLQGHT 463
Query: 219 ESVYALAMDEGGSILVSGGTEKVVRVWDTRSGSKILKLKGHTDNIRAL 266
+ + G+ILVS + VRVWD G+ + L GH I +L
Sbjct: 464 SLTSGMQLR--GNILVSCNADSHVRVWDIHEGTCVHMLSGHRSAITSL 509
Score = 57.8 bits (138), Expect = 1e-07
Identities = 65/265 (24%), Positives = 107/265 (39%), Gaps = 21/265 (7%)
Query: 207 QGYIPIAAKGHKESVYALAMDEGGSILVSGGTEKVVRVWDTRSGSKILKLKGHTDNIRAL 266
+G + GH V+ + + G +VSG T++ V+VW T GS + L+GHT +R +
Sbjct: 285 KGEVMYTLVGHTGGVWTSQISQCGRYIVSGSTDRTVKVWSTVDGSLLHTLQGHTSTVRCM 344
Query: 267 LLDSTGRVLFIR--IIRFYDKGQQRCVHSYAVHTDSVWALASTPTFSHVYSGGRDFSLYL 324
+ + V R +R +D R + + H +V + T V SGG DF++ +
Sbjct: 345 AMAGSILVTGSRDTTLRVWDVESGRHLATLHGHHAAVRCVQFDGT--TVVSGGYDFTVKI 402
Query: 325 TDLQTRESSLLCTGEHPIRQLALHNDS----IWVASTDSSVHRWP---AEGCDPQKIFQR 377
+ T TG H R +L +S + S D+S+ W EG + + Q
Sbjct: 403 WNAHTGRCIRTLTG-HNNRVYSLLFESERSIVCSGSLDTSIRVWDFTRPEGQECVALLQG 461
Query: 378 GNSFLAGNLSFSRARVSLEGSTPCF*IMNF**VPIYKEPTLT--IRGTPGIVQHEVLNNK 435
S +G VS + V E T + G + +
Sbjct: 462 HTSLTSGMQLRGNILVSCNADSHVR-------VWDIHEGTCVHMLSGHRSAITSLQWFGR 514
Query: 436 RHVLTKDTSDSVKLWEITKGVVVED 460
V T +VKLW+I +G ++ D
Sbjct: 515 NMVATSSDDGTVKLWDIERGALIRD 539
Score = 51.6 bits (122), Expect = 7e-06
Identities = 33/102 (32%), Positives = 51/102 (49%), Gaps = 5/102 (4%)
Query: 57 TGSRDGRLKRWALAE-DAATCSATFESHVDWVNDAVLVGDNTLVSCSSDTTLKTWNALSA 115
+GS D ++ W + C A + H + L G N LVSC++D+ ++ W+ +
Sbjct: 435 SGSLDTSIRVWDFTRPEGQECVALLQGHTSLTSGMQLRG-NILVSCNADSHVRVWD-IHE 492
Query: 116 GTCVRTHRQHTDYVTCLAAAGKNSNIVASGGLGGEVFVWDIE 157
GTCV H +T L G+N +VA+ G V +WDIE
Sbjct: 493 GTCVHMLSGHRSAITSLQWFGRN--MVATSSDDGTVKLWDIE 532
>YL24_ANASP (Q8YV57) Hypothetical WD-repeat protein all2124
Length = 1683
Score = 73.9 bits (180), Expect = 1e-12
Identities = 79/367 (21%), Positives = 142/367 (38%), Gaps = 71/367 (19%)
Query: 32 TKHCAGINCLALLTSTISDGSDYLFTGSRDGRLKRWALAEDAATCSATFESHVDWVNDAV 91
T H AG+ +T S + GS D +K W + T H DWVN
Sbjct: 1194 TGHSAGV-----ITVRFSPDGQTIAAGSEDKTVKLWHRQDGKLL--KTLNGHQDWVNSLS 1246
Query: 92 LVGDN-TLVSCSSDTTLKTWNALSAGTCVRTHRQHTDYV---------TCLAAAGKN--- 138
D TL S S+D T+K W ++ G V+T + H D V +A+A ++
Sbjct: 1247 FSPDGKTLASASADKTIKLWR-IADGKLVKTLKGHNDSVWDVNFSSDGKAIASASRDNTI 1305
Query: 139 ----------------------------SNIVASGGLGGEVFVWDIEAALAPVSKCNDAM 170
SNI+AS L + +W ++P+
Sbjct: 1306 KLWNRHGIELETFTGHSGGVYAVNFLPDSNIIASASLDNTIRLWQ-RPLISPLE------ 1358
Query: 171 VDESSNGINGSANLLPSTNLRTISSSHSISLHTAQTQGYIPIAAKGHKESVYALAMDEGG 230
V ++G+ + L + + T + +I L +Q G + G+K ++Y ++ G
Sbjct: 1359 VLAGNSGVYAVSFLHDGSIIATAGADGNIQLWHSQ-DGSLLKTLPGNK-AIYGISFTPQG 1416
Query: 231 SILVSGGTEKVVRVWDTRSGSKILKLKGHTDNIRALLLDSTGRVLFI----RIIRFYDKG 286
++ S +K V++W R G + L GH + + + G+ L ++ ++
Sbjct: 1417 DLIASANADKTVKIWRVRDGKALKTLIGHDNEVNKVNFSPDGKTLASASRDNTVKLWNVS 1476
Query: 287 QQRCVHSYAVHTDSVWALASTPTFSHVYSGGRDFSLYLTDLQTRESSLLCTGEHPIRQLA 346
+ + HTD V+ ++ +P + S D ++ L D + I+ L
Sbjct: 1477 DGKFKKTLKGHTDEVFWVSFSPDGKIIASASADKTIRLWD---------SFSGNLIKSLP 1527
Query: 347 LHNDSIW 353
HND ++
Sbjct: 1528 AHNDLVY 1534
Score = 59.3 bits (142), Expect = 4e-08
Identities = 66/332 (19%), Positives = 131/332 (38%), Gaps = 20/332 (6%)
Query: 43 LLTSTISDGSDYLFTGSRDGRLKRWALAEDAATCSATFESHVDWVNDAVLVGDN-TLVSC 101
+++ +IS + +GS D +K W+ T H D V D T+ S
Sbjct: 1075 VISISISRDGQTIASGSLDKTIKLWSRD---GRLFRTLNGHEDAVYSVSFSPDGQTIASG 1131
Query: 102 SSDTTLKTWNALSAGTCVRTHRQHTDYVTCLAAAGKNSNIVASGGLGGEVFVWDIEAALA 161
SD T+K W S GT ++T H V + + N+ AS + +WD +
Sbjct: 1132 GSDKTIKLWQT-SDGTLLKTITGHEQTVNNVYFSPDGKNL-ASASSDHSIKLWDTTSGQL 1189
Query: 162 PVSKCNDAMVDESSNGINGSANLLPSTNLRTISSSHSISLHTAQTQGYIPIAAKGHKESV 221
++ + + + + +T+ H + G + GH++ V
Sbjct: 1190 LMTLTGHSAGVITVRFSPDGQTIAAGSEDKTVKLWH-------RQDGKLLKTLNGHQDWV 1242
Query: 222 YALAMDEGGSILVSGGTEKVVRVWDTRSGSKILKLKGHTDNIRALLLDSTGRVLFI---- 277
+L+ G L S +K +++W G + LKGH D++ + S G+ +
Sbjct: 1243 NSLSFSPDGKTLASASADKTIKLWRIADGKLVKTLKGHNDSVWDVNFSSDGKAIASASRD 1302
Query: 278 RIIRFYDKGQQRCVHSYAVHTDSVWALASTPTFSHVYSGGRDFSLYLTDLQTRESSLLCT 337
I+ +++ + ++ H+ V+A+ P + + S D ++ L +
Sbjct: 1303 NTIKLWNRHGIE-LETFTGHSGGVYAVNFLPDSNIIASASLDNTIRLWQRPLISPLEVLA 1361
Query: 338 GEHPIRQLA-LHNDS-IWVASTDSSVHRWPAE 367
G + ++ LH+ S I A D ++ W ++
Sbjct: 1362 GNSGVYAVSFLHDGSIIATAGADGNIQLWHSQ 1393
Score = 57.8 bits (138), Expect = 1e-07
Identities = 55/226 (24%), Positives = 96/226 (42%), Gaps = 17/226 (7%)
Query: 53 DYLFTGSRDGRLKRWALAEDAATCSATFESHVDWVNDAVLVGDN-TLVSCSSDTTLKTWN 111
D + + + D +K W + + A T H + VN D TL S S D T+K WN
Sbjct: 1417 DLIASANADKTVKIWRVRDGKAL--KTLIGHDNEVNKVNFSPDGKTLASASRDNTVKLWN 1474
Query: 112 ALSAGTCVRTHRQHTDYVTCLAAAGKNSNIVASGGLGGEVFVWDIEAA--LAPVSKCNDA 169
+S G +T + HTD V ++ + + I+AS + +WD + + + ND
Sbjct: 1475 -VSDGKFKKTLKGHTDEVFWVSFS-PDGKIIASASADKTIRLWDSFSGNLIKSLPAHNDL 1532
Query: 170 MVDESSNGINGSANLLPSTNLRTISSSHSISLHTAQTQGYIPIAAKGHKESVYALAMDEG 229
+ S N ++L ST S+ ++ L + G++ GH VY+ +
Sbjct: 1533 VY---SVNFNPDGSMLAST-----SADKTVKLWRSH-DGHLLHTFSGHSNVVYSSSFSPD 1583
Query: 230 GSILVSGGTEKVVRVWDTRSGSKILKLKGHTDNIRALLLDSTGRVL 275
G + S +K V++W G + L H + + + G+ L
Sbjct: 1584 GRYIASASEDKTVKIWQI-DGHLLTTLPQHQAGVMSAIFSPDGKTL 1628
Score = 55.8 bits (133), Expect = 4e-07
Identities = 38/184 (20%), Positives = 82/184 (43%), Gaps = 11/184 (5%)
Query: 140 NIVASGGLGGEVFVWDIEAALAPVSKCNDAMVDESSNGINGSANLLPSTNLRTISSSHSI 199
+++AS V +W + A + D N +N S + L + S +++
Sbjct: 1417 DLIASANADKTVKIWRVRDGKALKTLIGH---DNEVNKVNFSPD---GKTLASASRDNTV 1470
Query: 200 SLHTAQTQGYIPIAAKGHKESVYALAMDEGGSILVSGGTEKVVRVWDTRSGSKILKLKGH 259
L + G KGH + V+ ++ G I+ S +K +R+WD+ SG+ I L H
Sbjct: 1471 KLWNV-SDGKFKKTLKGHTDEVFWVSFSPDGKIIASASADKTIRLWDSFSGNLIKSLPAH 1529
Query: 260 TDNIRALLLDSTGRVLFI----RIIRFYDKGQQRCVHSYAVHTDSVWALASTPTFSHVYS 315
D + ++ + G +L + ++ + +H+++ H++ V++ + +P ++ S
Sbjct: 1530 NDLVYSVNFNPDGSMLASTSADKTVKLWRSHDGHLLHTFSGHSNVVYSSSFSPDGRYIAS 1589
Query: 316 GGRD 319
D
Sbjct: 1590 ASED 1593
Score = 51.6 bits (122), Expect = 7e-06
Identities = 68/396 (17%), Positives = 147/396 (36%), Gaps = 73/396 (18%)
Query: 121 THRQHTDYVTCLAAAGKNSNIVASGGLGGEVFVWDIEAALAPVSKCNDAMVDESSNGING 180
+H+Q + L AA + ++++A V ++ AL + + N +G
Sbjct: 1020 SHQQLAALIASLKAAQQVNHVIAVPNNLKLATVTTLQQALFEMQERN-----RLEGHKDG 1074
Query: 181 SANLLPSTNLRTISS-SHSISLHTAQTQGYIPIAAKGHKESVYALAMDEGGSILVSGGTE 239
++ S + +TI+S S ++ G + GH+++VY+++ G + SGG++
Sbjct: 1075 VISISISRDGQTIASGSLDKTIKLWSRDGRLFRTLNGHEDAVYSVSFSPDGQTIASGGSD 1134
Query: 240 KVVRVWDTRSGSKILKLKGHTDNIRALLLDSTGRVLFI----RIIRFYDKGQQRCVHSYA 295
K +++W T G+ + + GH + + G+ L I+ +D + + +
Sbjct: 1135 KTIKLWQTSDGTLLKTITGHEQTVNNVYFSPDGKNLASASSDHSIKLWDTTSGQLLMTLT 1194
Query: 296 VHTDSVWALASTPTFSHVYSGGRDFSLYLTDLQTRESSLLCTGEHP-------------- 341
H+ V + +P + +G D ++ L Q + G
Sbjct: 1195 GHSAGVITVRFSPDGQTIAAGSEDKTVKLWHRQDGKLLKTLNGHQDWVNSLSFSPDGKTL 1254
Query: 342 -------------------IRQLALHNDSIW------------VASTDSSVHRWPAEGCD 370
++ L HNDS+W AS D+++ W G +
Sbjct: 1255 ASASADKTIKLWRIADGKLVKTLKGHNDSVWDVNFSSDGKAIASASRDNTIKLWNRHGIE 1314
Query: 371 PQKIFQRGNSFLAGNL---SFSRARVSLEGSTPCF*IMNF**VPIYKEPTLT----IRGT 423
+ A N S A SL+ + + +++ P ++ + G
Sbjct: 1315 LETFTGHSGGVYAVNFLPDSNIIASASLDNT-----------IRLWQRPLISPLEVLAGN 1363
Query: 424 PGIVQHEVLNNKRHVLTKDTSDSVKLWEITKGVVVE 459
G+ L++ + T +++LW G +++
Sbjct: 1364 SGVYAVSFLHDGSIIATAGADGNIQLWHSQDGSLLK 1399
>KMHA_DICDI (P42527) Myosin heavy chain kinase A (EC 2.7.1.129) (MHCK
A)
Length = 1146
Score = 72.4 bits (176), Expect = 4e-12
Identities = 71/300 (23%), Positives = 127/300 (41%), Gaps = 30/300 (10%)
Query: 76 CSATFESHVDWVNDAVLVGDNTLVSCSS--DTTLKTWNALSAGTCVRT---HRQHTDYVT 130
C +T +S + VN ++ DN + C+ D T + ++ C+ T HR+ + +
Sbjct: 861 CVSTIQSFRERVN-SIAFFDNQKLLCAGYGDGTYRVFDVNDNWKCLYTVNGHRKSIESIA 919
Query: 131 CLAAAGKNSNIVASGGLGGEVFVWDIEAALAPVSKCNDAMVDESSNGINGSANLLPSTNL 190
C NSN + + + V I + +KC + +V G G N + +
Sbjct: 920 C------NSNYIFTSSPDNTIKVHIIRSGN---TKCIETLV-----GHTGEVNCVVANEK 965
Query: 191 RTISSSHSISLHTAQTQGYIPIAAKGHKESVYALAMDEGGSILVSGGTEKVVRVWDTRSG 250
S S+ ++ + I + + Y + G L SGG ++++ VWDT +
Sbjct: 966 YLFSCSYDKTIKVWDLSTFKEIKSFEGVHTKYIKTLALSGRYLFSGGNDQIIYVWDTETL 1025
Query: 251 SKILKLKGHTDNIRALLLDSTGRVLFI----RIIRFYDKGQQRCVHSYAVHTDSVWALAS 306
S + ++GH D + L L T LF +I+ +D C+ + H +SV +
Sbjct: 1026 SMLFNMQGHEDWV--LSLHCTASYLFSTSKDNVIKIWDLSNFSCIDTLKGHWNSVSSCVV 1083
Query: 307 TPTFSHVYSGGRDFSLYLTDLQTRESSLLCTGEHP--IRQLALHNDSIWVASTDSSVHRW 364
+ +YSG D S+ + DL T E H ++ L + N+ I A+ D S+ W
Sbjct: 1084 KDRY--LYSGSEDNSIKVWDLDTLECVYTIPKSHSLGVKCLMVFNNQIISAAFDGSIKVW 1141
Score = 45.1 bits (105), Expect = 7e-04
Identities = 34/105 (32%), Positives = 52/105 (49%), Gaps = 8/105 (7%)
Query: 52 SDYLFTGSRDGRLKRWALAEDAATCSATFESHVDWVNDAVLVGDNTLVSCSSDTTLKTWN 111
+ YLF+ S+D +K W L+ +C T + H + V+ V V D L S S D ++K W+
Sbjct: 1045 ASYLFSTSKDNVIKIWDLSN--FSCIDTLKGHWNSVSSCV-VKDRYLYSGSEDNSIKVWD 1101
Query: 112 ALSAGTCVRT-HRQHTDYVTCLAAAGKNSNIVASGGLGGEVFVWD 155
L CV T + H+ V CL +N + S G + VW+
Sbjct: 1102 -LDTLECVYTIPKSHSLGVKCLMVF---NNQIISAAFDGSIKVWE 1142
>PWP2_SCHPO (Q9C1X1) Periodic tryptophan protein 2 homolog
Length = 854
Score = 70.5 bits (171), Expect = 2e-11
Identities = 63/244 (25%), Positives = 111/244 (44%), Gaps = 21/244 (8%)
Query: 45 TSTISDGSDYLFTGSRD-GRLKRWALAEDAATCSATFESHVDWVNDAVLVGDNT-LVSCS 102
T T++ D++ GS G+L W ++ +SH D ++ D +++ +
Sbjct: 301 TLTVNSTGDWIAIGSSKLGQLLVWEWQSESYVLKQ--QSHYDALSTLQYSSDGQRIITGA 358
Query: 103 SDTTLKTWNALSAGTCVRTHRQHTDYVTCLAAAGKNSNIVASGGLGGEVFVWDIEAALAP 162
D +K W+ +++G C+ T QHT V+ L + K N++ S L G V WD+
Sbjct: 359 DDGKIKVWD-MNSGFCIVTFTQHTSAVSGLCFS-KRGNVLFSSSLDGSVRAWDLIRYRNF 416
Query: 163 VSKCNDAMVDESSNGINGSANLLPSTNLRTISSSHSISLHTAQTQGYIPIAAKGHKESVY 222
+ + V S ++ S ++ + + S I + + QT G + GH+ V
Sbjct: 417 RTFTAPSRVQFSCIAVDPSGEIVCAGS----QDSFEIFMWSVQT-GQLLETLAGHEGPVS 471
Query: 223 ALAMDEGGSILVSGGTEKVVRVWD--TRSG--------SKILKLKGHTDNIRALLLDSTG 272
+L+ + GS+L SG +K VR+WD +RSG S +L L H D + G
Sbjct: 472 SLSFNSSGSLLASGSWDKTVRIWDIFSRSGIVEPLPIPSDVLSLAFHPDGKEVCVASLDG 531
Query: 273 RVLF 276
++ F
Sbjct: 532 QLTF 535
Score = 48.5 bits (114), Expect = 6e-05
Identities = 39/169 (23%), Positives = 78/169 (46%), Gaps = 10/169 (5%)
Query: 205 QTQGYIPIAAKGHKESVYALAMDEGGSILVSGGTEKVVRVWDTRSGSKILKLKGHTDNIR 264
Q++ Y+ + + H +++ L G +++G + ++VWD SG I+ HT +
Sbjct: 327 QSESYV-LKQQSHYDALSTLQYSSDGQRIITGADDGKIKVWDMNSGFCIVTFTQHTSAVS 385
Query: 265 ALLLDSTGRVLFIR----IIRFYDKGQQRCVHSYAVHTDSVWA-LASTPTFSHVYSGGRD 319
L G VLF +R +D + R ++ + ++ +A P+ V +G +D
Sbjct: 386 GLCFSKRGNVLFSSSLDGSVRAWDLIRYRNFRTFTAPSRVQFSCIAVDPSGEIVCAGSQD 445
Query: 320 -FSLYLTDLQTRE-SSLLCTGEHPIRQLALHNDSIWVA--STDSSVHRW 364
F +++ +QT + L E P+ L+ ++ +A S D +V W
Sbjct: 446 SFEIFMWSVQTGQLLETLAGHEGPVSSLSFNSSGSLLASGSWDKTVRIW 494
>TRA7_HUMAN (Q6Q0C0) E3 ubiquitin protein ligase TRAF7 (EC 6.3.2.-)
(TNF receptor associated factor 7) (Ring finger and WD
repeat domain 1) (RING finger protein 119)
Length = 670
Score = 68.9 bits (167), Expect = 4e-11
Identities = 78/283 (27%), Positives = 120/283 (41%), Gaps = 45/283 (15%)
Query: 53 DYLFTGSRDGRLKRWALAEDAAT---CSATFESHVDWVNDAVLVGDNTLVSCSSDTTLKT 109
D LF+GS D +K W D T C T E H D + A+ + L S S+D T+
Sbjct: 409 DLLFSGSSDKTIKVW----DTCTTYKCQKTLEGH-DGIVLALCIQGCKLYSGSADCTIIV 463
Query: 110 WNALSAGTCVRTHRQHTDYVTCLAAAGKNSNIVASGGLGGEVFVWDI-------EAALAP 162
W+ + V T R H + V L ++ N++ SG L + VWDI + L
Sbjct: 464 WDIQNLQK-VNTIRAHDNPVCTLVSS---HNVLFSGSLKA-IKVWDIVGTELKLKKELTG 518
Query: 163 VSKCNDAMVDESSNGINGSANLLPSTNLRTISSSHSISLHTAQTQGYIPIAAKGHKESVY 222
++ A+V S +GS + ++RT+ +H QT G SVY
Sbjct: 519 LNHWVRALVAAQSYLYSGSYQTIKIWDIRTLDC-----IHVLQTSG----------GSVY 563
Query: 223 ALAMDEGGSILVSGGTEKVVRVWDTRSGSKILKLKGHTDNIRALLLDSTG------RVLF 276
++A+ +V G E ++ VWD S ++ L GH + AL + ST +
Sbjct: 564 SIAVTNHH--IVCGTYENLIHVWDIESKEQVRTLTGHVGTVYALAVISTPDQTKVFSASY 621
Query: 277 IRIIRFYDKGQQRCVHSYAVHTDSVWALASTPTFSHVYSGGRD 319
R +R + C + H SV ALA + ++SG D
Sbjct: 622 DRSLRVWSMDNMICTQTLLRHQGSVTALAVSR--GRLFSGAVD 662
Score = 39.3 bits (90), Expect = 0.038
Identities = 20/54 (37%), Positives = 32/54 (59%), Gaps = 1/54 (1%)
Query: 216 GHKESVYALAMDEGGSILVSGGTEKVVRVWDTRSGSKILK-LKGHTDNIRALLL 268
GH+ V+ L + G +L SG ++K ++VWDT + K K L+GH + AL +
Sbjct: 394 GHQGPVWCLCVYSMGDLLFSGSSDKTIKVWDTCTTYKCQKTLEGHDGIVLALCI 447
>TRA7_MOUSE (Q922B6) E3 ubiquitin protein ligase TRAF7 (EC 6.3.2.-)
(TNF receptor associated factor 7)
Length = 594
Score = 68.2 bits (165), Expect = 8e-11
Identities = 78/283 (27%), Positives = 120/283 (41%), Gaps = 45/283 (15%)
Query: 53 DYLFTGSRDGRLKRWALAEDAAT---CSATFESHVDWVNDAVLVGDNTLVSCSSDTTLKT 109
D LF+GS D +K W D T C T E H D + A+ + L S S+D T+
Sbjct: 333 DLLFSGSSDKTIKVW----DTCTTYKCQKTLEGH-DGIVLALCIQGCKLYSGSADCTIIV 387
Query: 110 WNALSAGTCVRTHRQHTDYVTCLAAAGKNSNIVASGGLGGEVFVWDI-------EAALAP 162
W+ + V T R H + V L ++ N++ SG L + VWDI + L
Sbjct: 388 WDIQNLQK-VNTIRAHDNPVCTLVSS---HNMLFSGSLKA-IKVWDIVGTELKLKKELTG 442
Query: 163 VSKCNDAMVDESSNGINGSANLLPSTNLRTISSSHSISLHTAQTQGYIPIAAKGHKESVY 222
++ A+V S +GS + ++RT+ +H QT G SVY
Sbjct: 443 LNHWVRALVAAQSYLYSGSYQTIKIWDIRTLDC-----IHVLQTSG----------GSVY 487
Query: 223 ALAMDEGGSILVSGGTEKVVRVWDTRSGSKILKLKGHTDNIRALLLDSTG------RVLF 276
++A+ +V G E ++ VWD S ++ L GH + AL + ST +
Sbjct: 488 SIAVTNHH--IVCGTYENLIHVWDIESKEQVRTLTGHVGTVYALAVISTPDQTKVFSASY 545
Query: 277 IRIIRFYDKGQQRCVHSYAVHTDSVWALASTPTFSHVYSGGRD 319
R +R + C + H SV ALA + ++SG D
Sbjct: 546 DRSLRVWSMDNMICTQTLLRHQGSVTALAVSR--GRLFSGAVD 586
Score = 39.3 bits (90), Expect = 0.038
Identities = 20/54 (37%), Positives = 32/54 (59%), Gaps = 1/54 (1%)
Query: 216 GHKESVYALAMDEGGSILVSGGTEKVVRVWDTRSGSKILK-LKGHTDNIRALLL 268
GH+ V+ L + G +L SG ++K ++VWDT + K K L+GH + AL +
Sbjct: 318 GHQGPVWCLCVYSMGDLLFSGSSDKTIKVWDTCTTYKCQKTLEGHDGIVLALCI 371
>PF20_CHLRE (P93107) Flagellar WD-repeat protein PF20
Length = 606
Score = 68.2 bits (165), Expect = 8e-11
Identities = 71/271 (26%), Positives = 110/271 (40%), Gaps = 18/271 (6%)
Query: 55 LFTGSRDGRLKRWAL-AEDAATCSATFESHVDWVNDAVLVGDNT-LVSCSSDTTLKTWNA 112
L T S D K W + D C E H DWV T L S D+ +K W+
Sbjct: 341 LVTASDDKTWKMWHMPGGDLIMCG---EGHKDWVAGVDFHPAGTCLASGGGDSAVKIWD- 396
Query: 113 LSAGTCVRTHRQHTDYVTCLAAAGKNSNIVASGGLGGEVFVWDIEAALAPVSKCNDAMVD 172
CV T H + + +VASG L V +WD+ P KC A+
Sbjct: 397 FEKQRCVTTFTDHKQAIWSVRFHHLGE-VVASGSLDHTVRLWDL-----PAGKCRMALRG 450
Query: 173 ESSNGINGSANLLPSTNLRTISSSHSISLHTAQTQGYIPIAAKGHKESVYALAMDEGGSI 232
+ +N A S++L T SS ++S+ A+ G GH+ S ++ + G+
Sbjct: 451 HVDS-VNDLAWQPFSSSLATASSDKTVSVWDARA-GLCTQTYYGHQNSCNGVSFNILGTQ 508
Query: 233 LVSGGTEKVVRVWDTRSGSKILKLKGHTDNIRALLLDSTGRVLFIRI----IRFYDKGQQ 288
L S + VV++WDTR +++ + D +G+VL + ++ Y
Sbjct: 509 LASTDADGVVKLWDTRMTAEVATINTGKHPANKSCFDRSGQVLAVACDDGKVKAYSTTDG 568
Query: 289 RCVHSYAVHTDSVWALASTPTFSHVYSGGRD 319
A H D+V A+ P ++ S G D
Sbjct: 569 VLQAELAGHEDAVQAVLFDPAGQYLVSCGSD 599
Score = 62.4 bits (150), Expect = 4e-09
Identities = 54/194 (27%), Positives = 85/194 (42%), Gaps = 20/194 (10%)
Query: 57 TGSRDGRLKRWALAEDAATCSATFESHVDWVND-AVLVGDNTLVSCSSDTTLKTWNALSA 115
+GS D ++ W L A C HVD VND A ++L + SSD T+ W+A A
Sbjct: 427 SGSLDHTVRLWDLP--AGKCRMALRGHVDSVNDLAWQPFSSSLATASSDKTVSVWDA-RA 483
Query: 116 GTCVRTHRQHTDYVTCLAAAGKNSNIV----ASGGLGGEVFVWDIEAALAPVSKCNDAMV 171
G C +T+ H + + G + NI+ AS G V +WD A V+ N
Sbjct: 484 GLCTQTYYGHQN-----SCNGVSFNILGTQLASTDADGVVKLWDTRMT-AEVATINTGKH 537
Query: 172 DESSNGINGSANLLPSTNLRTISSSHSISLHTAQTQGYIPIAAKGHKESVYALAMDEGGS 231
+ + + S +L ++ + T G + GH+++V A+ D G
Sbjct: 538 PANKSCFDRSGQVL------AVACDDGKVKAYSTTDGVLQAELAGHEDAVQAVLFDPAGQ 591
Query: 232 ILVSGGTEKVVRVW 245
LVS G++ R+W
Sbjct: 592 YLVSCGSDNTFRLW 605
Score = 57.0 bits (136), Expect = 2e-07
Identities = 47/174 (27%), Positives = 76/174 (43%), Gaps = 8/174 (4%)
Query: 215 KGHKESVYALAMDEGGSILVSGGTEKVVRVWDTRSGSKILKLKGHTDNIRALLLDSTGRV 274
KGH SV LA+ ILV+ +K ++W G I+ +GH D + + G
Sbjct: 323 KGHLLSVANLALHPTKPILVTASDDKTWKMWHMPGGDLIMCGEGHKDWVAGVDFHPAGTC 382
Query: 275 LFI----RIIRFYDKGQQRCVHSYAVHTDSVWALASTPTFSHVYSGGRDFSLYLTDLQTR 330
L ++ +D +QRCV ++ H ++W++ V SG D ++ L DL
Sbjct: 383 LASGGGDSAVKIWDFEKQRCVTTFTDHKQAIWSVRFHHLGEVVASGSLDHTVRLWDLPAG 442
Query: 331 ESSLLCTGE-HPIRQLALH--NDSIWVASTDSSVHRWPAE-GCDPQKIFQRGNS 380
+ + G + LA + S+ AS+D +V W A G Q + NS
Sbjct: 443 KCRMALRGHVDSVNDLAWQPFSSSLATASSDKTVSVWDARAGLCTQTYYGHQNS 496
Score = 52.0 bits (123), Expect = 6e-06
Identities = 58/269 (21%), Positives = 100/269 (36%), Gaps = 57/269 (21%)
Query: 79 TFESHVDWV-NDAVLVGDNTLVSCSSDTTLKTWNALSAGTCVRTHRQHTDYVTCLAAAGK 137
TF+ H+ V N A+ LV+ S D T K W+ + G + H D+V +
Sbjct: 321 TFKGHLLSVANLALHPTKPILVTASDDKTWKMWH-MPGGDLIMCGEGHKDWVAGVDFHPA 379
Query: 138 NSNIVASGGLGGEVFVWDIEAALAPVSKCNDAMVDESSNGINGSANLLPSTNLRTISSSH 197
+ +ASGG V +WD E +C D
Sbjct: 380 GT-CLASGGGDSAVKIWDFEK-----QRCVTTFTD------------------------- 408
Query: 198 SISLHTAQTQGYIPIAAKGHKESVYALAMDEGGSILVSGGTEKVVRVWDTRSGSKILKLK 257
HK++++++ G ++ SG + VR+WD +G + L+
Sbjct: 409 -------------------HKQAIWSVRFHHLGEVVASGSLDHTVRLWDLPAGKCRMALR 449
Query: 258 GHTDNIRAL----LLDSTGRVLFIRIIRFYDKGQQRCVHSYAVHTDSVWALASTPTFSHV 313
GH D++ L S + + +D C +Y H +S ++ + +
Sbjct: 450 GHVDSVNDLAWQPFSSSLATASSDKTVSVWDARAGLCTQTYYGHQNSCNGVSFNILGTQL 509
Query: 314 YSGGRDFSLYLTDLQ-TRESSLLCTGEHP 341
S D + L D + T E + + TG+HP
Sbjct: 510 ASTDADGVVKLWDTRMTAEVATINTGKHP 538
>YIRF_SCHPO (Q9P7I3) Hypothetical WD-repeat protein C664.15 in
chromosome I
Length = 651
Score = 66.2 bits (160), Expect = 3e-10
Identities = 73/305 (23%), Positives = 135/305 (43%), Gaps = 58/305 (19%)
Query: 80 FESHVDWVNDAVLVGD-NTLVSCSSDTTLKTWNALSAGTCVRTHRQHTDYVTCLAAAGKN 138
F++H +++ TL + S+D T+K W+ ++ + + + H+DYV+CLA
Sbjct: 328 FQAHSEYITSLDFSHPFGTLATASTDKTVKVWD-MAGVVYLGSLKGHSDYVSCLAI---Q 383
Query: 139 SNIVASGGLGGEVFVWDIEAALAPVSKCNDAMVDESSNGINGSANLLPSTNLRTISSSHS 198
+ +A+G + V +W+++ ND + N + S N P + ++++
Sbjct: 384 DSFIATGSMDTTVRLWNLD---------NDVL--HKDNPVEESLNSPPDQPVD--NATNV 430
Query: 199 ISLHTAQTQGYIPIAAKGHKESVYALAMDEGGSILVSGGTEKVVRVWDTRSGSKILKLK- 257
++ HTA P+ A AL+ D+ +L++G +K VR WD +G I L
Sbjct: 431 LTSHTA------PVTA-------LALSSDD---VLITGADDKTVRQWDIVTGRCIQTLDF 474
Query: 258 -----GHTDNIRALLLDSTGRVLFIR----------------IIRFYDKGQQRCVHSYAV 296
T + L+ D+ + FIR +IR +D V S+
Sbjct: 475 VWAETHDTSTSQILVSDTYKQEPFIRALDCLDAAVASGTVDGLIRIWDLRIGLPVRSFIG 534
Query: 297 HTDSVWALASTPTFSHVYSGGRDFSLYLTDLQTRESSLLCTGEHPIRQLALHNDSIWVAS 356
HT + +L +H+YSG D S+ + DL++ + E + L L+ + VAS
Sbjct: 535 HTAPISSLQFDS--NHLYSGSYDNSVRIWDLRSGSPINIIPMEKKVNSLRLYQGRLAVAS 592
Query: 357 TDSSV 361
+ +V
Sbjct: 593 DEPNV 597
Score = 45.1 bits (105), Expect = 7e-04
Identities = 59/239 (24%), Positives = 93/239 (38%), Gaps = 61/239 (25%)
Query: 34 HCAGINCLALLTSTISDGSDYLFTGSRDGRLKRWALAEDAA------------------- 74
H ++CLA+ S I+ TGS D ++ W L D
Sbjct: 373 HSDYVSCLAIQDSFIA-------TGSMDTTVRLWNLDNDVLHKDNPVEESLNSPPDQPVD 425
Query: 75 TCSATFESHVDWVNDAVLVGDNTLVSCSSDTTLKTWNALSAGTCVRTHRQHTDYVTCLAA 134
+ SH V L D+ L++ + D T++ W+ ++ G C++T D+V
Sbjct: 426 NATNVLTSHTAPVTALALSSDDVLITGADDKTVRQWDIVT-GRCIQT----LDFVWAETH 480
Query: 135 AGKNSNIVASGGLGGEVFVWDIEAALAPVSKCNDAMVDESSNGINGSANLLPSTNLRTIS 194
S I+ S E F+ ++ C DA V +S ++G L+ +LR
Sbjct: 481 DTSTSQILVSDTYKQEPFIRALD--------CLDAAV--ASGTVDG---LIRIWDLRIGL 527
Query: 195 SSHSISLHTAQTQGYIPIAAKGHKESVYALAMDEGGSILVSGGTEKVVRVWDTRSGSKI 253
S HTA PI++ + + L SG + VR+WD RSGS I
Sbjct: 528 PVRSFIGHTA------PISS-----------LQFDSNHLYSGSYDNSVRIWDLRSGSPI 569
>LI23_CAEEL (Q09990) F-box/WD-repeat protein lin-23 (Abnormal cell
lineage protein 23)
Length = 665
Score = 66.2 bits (160), Expect = 3e-10
Identities = 64/264 (24%), Positives = 119/264 (44%), Gaps = 32/264 (12%)
Query: 9 NANNSTRPRKEKRLTYVLNDSDDTKHCAGINCLALLTSTISDGSDYLFTGSRDGRLKRWA 68
N +N+ + R ++T + S+++K G+ CL D + +G RD +K W
Sbjct: 200 NIDNNWK-RGNYKMTRINCQSENSK---GVYCLQY-------DDDKIVSGLRDNTIKIWD 248
Query: 69 LAEDAATCSATFESHVDWVNDAVLVGDN-TLVSCSSDTTLKTWNALSAGTCVRTHRQHTD 127
+ +CS H V L DN ++S SSD T++ W+ + G C++T H +
Sbjct: 249 RKD--YSCSRILSGHTGSV--LCLQYDNRVIISGSSDATVRVWD-VETGECIKTLIHHCE 303
Query: 128 YVTCLAAAGKNSNIVASGGLGGEVFVWDIEAALAPVSKCNDAMVDESSNGINGSANLLPS 187
V L A + I+ + + VWD+ + D + G + N++
Sbjct: 304 AVLHLRFA---NGIMVTCSKDRSIAVWDMVSP-------RDITIRRVLVGHRAAVNVVDF 353
Query: 188 TNLRTISSS--HSISLHTAQTQGYIPIAAKGHKESVYALAMDEGGSILVSGGTEKVVRVW 245
+ +S+S +I + + T ++ A GH+ + L G ++VSG ++ +R+W
Sbjct: 354 DDRYIVSASGDRTIKVWSMDTLEFVRTLA-GHRRGIACLQYR--GRLVVSGSSDNTIRLW 410
Query: 246 DTRSGSKILKLKGHTDNIRALLLD 269
D SG + L+GH + +R + D
Sbjct: 411 DIHSGVCLRVLEGHEELVRCIRFD 434
Score = 50.4 bits (119), Expect = 2e-05
Identities = 33/109 (30%), Positives = 53/109 (48%), Gaps = 7/109 (6%)
Query: 54 YLFTGSRDGRLKRWALAEDAATCSATFESHVDWVNDAVLVGDNTLVSCSSDTTLKTWNAL 113
Y+ + S D +K W++ D T H + G +VS SSD T++ W+ +
Sbjct: 357 YIVSASGDRTIKVWSM--DTLEFVRTLAGHRRGIACLQYRG-RLVVSGSSDNTIRLWD-I 412
Query: 114 SAGTCVRTHRQHTDYVTCLAAAGKNSNIVASGGLGGEVFVWDIEAALAP 162
+G C+R H + V C+ K + SG G++ VWD++AAL P
Sbjct: 413 HSGVCLRVLEGHEELVRCIRFDEKR---IVSGAYDGKIKVWDLQAALDP 458
Score = 34.3 bits (77), Expect = 1.2
Identities = 23/89 (25%), Positives = 42/89 (46%), Gaps = 8/89 (8%)
Query: 217 HKESVYALAMDEGGSILVSGGTEKVVRVWDTRSGSKILKLKGHTDNIRALLLDSTGRVLF 276
+ + VY L D+ +VSG + +++WD + S L GHT ++ L D+ RV+
Sbjct: 221 NSKGVYCLQYDDDK--IVSGLRDNTIKIWDRKDYSCSRILSGHTGSVLCLQYDN--RVII 276
Query: 277 I----RIIRFYDKGQQRCVHSYAVHTDSV 301
+R +D C+ + H ++V
Sbjct: 277 SGSSDATVRVWDVETGECIKTLIHHCEAV 305
>LIS1_RAT (P63004) Platelet-activating factor acetylhydrolase IB
alpha subunit (PAF acetylhydrolase 45 kDa subunit)
(PAF-AH 45 kDa subunit) (PAF-AH alpha) (PAFAH alpha)
(Lissencephaly-1 protein) (LIS-1)
Length = 409
Score = 65.9 bits (159), Expect = 4e-10
Identities = 42/159 (26%), Positives = 75/159 (46%), Gaps = 7/159 (4%)
Query: 213 AAKGHKESVYALAMDEGGSILVSGGTEKVVRVWDTRSGSKILKLKGHTDNIRALLLDSTG 272
A GH+ V + S++VS + ++VWD +G LKGHTD+++ + D +G
Sbjct: 102 ALSGHRSPVTRVIFHPVFSVMVSASEDATIKVWDYETGDFERTLKGHTDSVQDISFDHSG 161
Query: 273 RVLFI----RIIRFYDKGQQRCVHSYAVHTDSVWALASTPTFSHVYSGGRDFSLYLTDLQ 328
++L I+ +D C+ + H +V ++A P H+ S RD ++ + ++Q
Sbjct: 162 KLLASCSADMTIKLWDFQGFECIRTMHGHDHNVSSVAIMPNGDHIVSASRDKTIKMWEVQ 221
Query: 329 TRESSLLCTGEHP-IRQLALHNDSIWVA--STDSSVHRW 364
T TG +R + + D +A S D +V W
Sbjct: 222 TGYCVKTFTGHREWVRMVRPNQDGTLIASCSNDQTVRVW 260
Score = 64.7 bits (156), Expect = 8e-10
Identities = 59/283 (20%), Positives = 115/283 (39%), Gaps = 22/283 (7%)
Query: 55 LFTGSRDGRLKRWALAEDAATCSATFESHVDWVNDAVLVGDNTLV-SCSSDTTLKTWNAL 113
+ + S D +K W + T + H D V D L+ SCS+D T+K W+
Sbjct: 122 MVSASEDATIKVWDY--ETGDFERTLKGHTDSVQDISFDHSGKLLASCSADMTIKLWD-F 178
Query: 114 SAGTCVRTHRQHTDYVTCLAAAGKNSNIVASGGLGGEVFVWDIEAALAPVSKCNDA---- 169
C+RT H V+ +A +IV S + +W+++ +
Sbjct: 179 QGFECIRTMHGHDHNVSSVAIMPNGDHIV-SASRDKTIKMWEVQTGYCVKTFTGHREWVR 237
Query: 170 MVDESSNGINGSANLLPSTNLRTISSSHSISLHTAQTQGYIPIAAKGHKESVYALAMDEG 229
MV + +G ++ T + ++ + + + + + S +++ G
Sbjct: 238 MVRPNQDGTLIASCSNDQTVRVWVVATKECKAELREHEHVVECISWAPESSYSSISEATG 297
Query: 230 ---------GSILVSGGTEKVVRVWDTRSGSKILKLKGHTDNIRALLLDSTGRVLFI--- 277
G L+SG +K +++WD +G ++ L GH + +R +L S G+ +
Sbjct: 298 SETKKSGKPGPFLLSGSRDKTIKMWDVSTGMCLMTLVGHDNWVRGVLFHSGGKFILSCAD 357
Query: 278 -RIIRFYDKGQQRCVHSYAVHTDSVWALASTPTFSHVYSGGRD 319
+ +R +D +RC+ + H V +L T +V +G D
Sbjct: 358 DKTLRVWDYKNKRCMKTLNAHEHFVTSLDFHKTAPYVVTGSVD 400
Score = 63.2 bits (152), Expect = 2e-09
Identities = 51/220 (23%), Positives = 87/220 (39%), Gaps = 34/220 (15%)
Query: 48 ISDGSDYLFTGSRDGRLKRWALAEDAATCSATFESHVDWVNDAVLVGDNTLV-SCSSDTT 106
I D++ + SRD +K W + C TF H +WV D TL+ SCS+D T
Sbjct: 199 IMPNGDHIVSASRDKTIKMWEV--QTGYCVKTFTGHREWVRMVRPNQDGTLIASCSNDQT 256
Query: 107 LKTWNALSAGTCVRTHRQHTDYVTCLAAAGKNSNIVASGGLGGEVFVWDIEAALAPVSKC 166
++ W ++ C R+H V C++ W E++ + +S+
Sbjct: 257 VRVW-VVATKECKAELREHEHVVECIS--------------------WAPESSYSSISEA 295
Query: 167 NDAMVDESSNGINGSANLLPSTNLRTISSSHSISLHTAQTQGYIPIAAKGHKESVYALAM 226
+ +S P L + S +I + T G + GH V +
Sbjct: 296 TGSETKKSGK---------PGPFLLSGSRDKTIKMWDVST-GMCLMTLVGHDNWVRGVLF 345
Query: 227 DEGGSILVSGGTEKVVRVWDTRSGSKILKLKGHTDNIRAL 266
GG ++S +K +RVWD ++ + L H + +L
Sbjct: 346 HSGGKFILSCADDKTLRVWDYKNKRCMKTLNAHEHFVTSL 385
Score = 49.3 bits (116), Expect = 4e-05
Identities = 62/296 (20%), Positives = 118/296 (38%), Gaps = 36/296 (12%)
Query: 96 NTLVSCSSDTTLKTWNALSAGTCVRTHRQHTDYVTCLAAAGKNSNIVASGGLGGEVFVWD 155
+ +VS S D T+K W+ G RT + HTD V + + + ++AS + +WD
Sbjct: 120 SVMVSASEDATIKVWD-YETGDFERTLKGHTDSVQDI-SFDHSGKLLASCSADMTIKLWD 177
Query: 156 IEAALAPVSKCNDAMVDESSNGINGSANLLPSTNLRTISSSHSISLHTAQTQGYIPIAAK 215
+ +C M N ++ A + ++ + S +I + QT GY
Sbjct: 178 FQG-----FECIRTMHGHDHN-VSSVAIMPNGDHIVSASRDKTIKMWEVQT-GYCVKTFT 230
Query: 216 GHKESVYALAMDEGGSILVSGGTEKVVRVWDTRSGSKILKLKGHTDNIRAL--------- 266
GH+E V + ++ G+++ S ++ VRVW + +L+ H + +
Sbjct: 231 GHREWVRMVRPNQDGTLIASCSNDQTVRVWVVATKECKAELREHEHVVECISWAPESSYS 290
Query: 267 -LLDSTGRVL--------FI------RIIRFYDKGQQRCVHSYAVHTDSVWALASTPTFS 311
+ ++TG F+ + I+ +D C+ + H + V +
Sbjct: 291 SISEATGSETKKSGKPGPFLLSGSRDKTIKMWDVSTGMCLMTLVGHDNWVRGVLFHSGGK 350
Query: 312 HVYSGGRDFSLYLTDLQTRE-SSLLCTGEHPIRQLALHNDSIWV--ASTDSSVHRW 364
+ S D +L + D + + L EH + L H + +V S D +V W
Sbjct: 351 FILSCADDKTLRVWDYKNKRCMKTLNAHEHFVTSLDFHKTAPYVVTGSVDQTVKVW 406
>LIS1_MOUSE (P63005) Platelet-activating factor acetylhydrolase IB
alpha subunit (PAF acetylhydrolase 45 kDa subunit)
(PAF-AH 45 kDa subunit) (PAF-AH alpha) (PAFAH alpha)
(Lissencephaly-1 protein) (LIS-1)
Length = 409
Score = 65.9 bits (159), Expect = 4e-10
Identities = 42/159 (26%), Positives = 75/159 (46%), Gaps = 7/159 (4%)
Query: 213 AAKGHKESVYALAMDEGGSILVSGGTEKVVRVWDTRSGSKILKLKGHTDNIRALLLDSTG 272
A GH+ V + S++VS + ++VWD +G LKGHTD+++ + D +G
Sbjct: 102 ALSGHRSPVTRVIFHPVFSVMVSASEDATIKVWDYETGDFERTLKGHTDSVQDISFDHSG 161
Query: 273 RVLFI----RIIRFYDKGQQRCVHSYAVHTDSVWALASTPTFSHVYSGGRDFSLYLTDLQ 328
++L I+ +D C+ + H +V ++A P H+ S RD ++ + ++Q
Sbjct: 162 KLLASCSADMTIKLWDFQGFECIRTMHGHDHNVSSVAIMPNGDHIVSASRDKTIKMWEVQ 221
Query: 329 TRESSLLCTGEHP-IRQLALHNDSIWVA--STDSSVHRW 364
T TG +R + + D +A S D +V W
Sbjct: 222 TGYCVKTFTGHREWVRMVRPNQDGTLIASCSNDQTVRVW 260
Score = 64.7 bits (156), Expect = 8e-10
Identities = 59/283 (20%), Positives = 115/283 (39%), Gaps = 22/283 (7%)
Query: 55 LFTGSRDGRLKRWALAEDAATCSATFESHVDWVNDAVLVGDNTLV-SCSSDTTLKTWNAL 113
+ + S D +K W + T + H D V D L+ SCS+D T+K W+
Sbjct: 122 MVSASEDATIKVWDY--ETGDFERTLKGHTDSVQDISFDHSGKLLASCSADMTIKLWD-F 178
Query: 114 SAGTCVRTHRQHTDYVTCLAAAGKNSNIVASGGLGGEVFVWDIEAALAPVSKCNDA---- 169
C+RT H V+ +A +IV S + +W+++ +
Sbjct: 179 QGFECIRTMHGHDHNVSSVAIMPNGDHIV-SASRDKTIKMWEVQTGYCVKTFTGHREWVR 237
Query: 170 MVDESSNGINGSANLLPSTNLRTISSSHSISLHTAQTQGYIPIAAKGHKESVYALAMDEG 229
MV + +G ++ T + ++ + + + + + S +++ G
Sbjct: 238 MVRPNQDGTLIASCSNDQTVRVWVVATKECKAELREHEHVVECISWAPESSYSSISEATG 297
Query: 230 ---------GSILVSGGTEKVVRVWDTRSGSKILKLKGHTDNIRALLLDSTGRVLFI--- 277
G L+SG +K +++WD +G ++ L GH + +R +L S G+ +
Sbjct: 298 SETKKSGKPGPFLLSGSRDKTIKMWDVSTGMCLMTLVGHDNWVRGVLFHSGGKFILSCAD 357
Query: 278 -RIIRFYDKGQQRCVHSYAVHTDSVWALASTPTFSHVYSGGRD 319
+ +R +D +RC+ + H V +L T +V +G D
Sbjct: 358 DKTLRVWDYKNKRCMKTLNAHEHFVTSLDFHKTAPYVVTGSVD 400
Score = 63.2 bits (152), Expect = 2e-09
Identities = 51/220 (23%), Positives = 87/220 (39%), Gaps = 34/220 (15%)
Query: 48 ISDGSDYLFTGSRDGRLKRWALAEDAATCSATFESHVDWVNDAVLVGDNTLV-SCSSDTT 106
I D++ + SRD +K W + C TF H +WV D TL+ SCS+D T
Sbjct: 199 IMPNGDHIVSASRDKTIKMWEV--QTGYCVKTFTGHREWVRMVRPNQDGTLIASCSNDQT 256
Query: 107 LKTWNALSAGTCVRTHRQHTDYVTCLAAAGKNSNIVASGGLGGEVFVWDIEAALAPVSKC 166
++ W ++ C R+H V C++ W E++ + +S+
Sbjct: 257 VRVW-VVATKECKAELREHEHVVECIS--------------------WAPESSYSSISEA 295
Query: 167 NDAMVDESSNGINGSANLLPSTNLRTISSSHSISLHTAQTQGYIPIAAKGHKESVYALAM 226
+ +S P L + S +I + T G + GH V +
Sbjct: 296 TGSETKKSGK---------PGPFLLSGSRDKTIKMWDVST-GMCLMTLVGHDNWVRGVLF 345
Query: 227 DEGGSILVSGGTEKVVRVWDTRSGSKILKLKGHTDNIRAL 266
GG ++S +K +RVWD ++ + L H + +L
Sbjct: 346 HSGGKFILSCADDKTLRVWDYKNKRCMKTLNAHEHFVTSL 385
Score = 49.3 bits (116), Expect = 4e-05
Identities = 62/296 (20%), Positives = 118/296 (38%), Gaps = 36/296 (12%)
Query: 96 NTLVSCSSDTTLKTWNALSAGTCVRTHRQHTDYVTCLAAAGKNSNIVASGGLGGEVFVWD 155
+ +VS S D T+K W+ G RT + HTD V + + + ++AS + +WD
Sbjct: 120 SVMVSASEDATIKVWD-YETGDFERTLKGHTDSVQDI-SFDHSGKLLASCSADMTIKLWD 177
Query: 156 IEAALAPVSKCNDAMVDESSNGINGSANLLPSTNLRTISSSHSISLHTAQTQGYIPIAAK 215
+ +C M N ++ A + ++ + S +I + QT GY
Sbjct: 178 FQG-----FECIRTMHGHDHN-VSSVAIMPNGDHIVSASRDKTIKMWEVQT-GYCVKTFT 230
Query: 216 GHKESVYALAMDEGGSILVSGGTEKVVRVWDTRSGSKILKLKGHTDNIRAL--------- 266
GH+E V + ++ G+++ S ++ VRVW + +L+ H + +
Sbjct: 231 GHREWVRMVRPNQDGTLIASCSNDQTVRVWVVATKECKAELREHEHVVECISWAPESSYS 290
Query: 267 -LLDSTGRVL--------FI------RIIRFYDKGQQRCVHSYAVHTDSVWALASTPTFS 311
+ ++TG F+ + I+ +D C+ + H + V +
Sbjct: 291 SISEATGSETKKSGKPGPFLLSGSRDKTIKMWDVSTGMCLMTLVGHDNWVRGVLFHSGGK 350
Query: 312 HVYSGGRDFSLYLTDLQTRE-SSLLCTGEHPIRQLALHNDSIWV--ASTDSSVHRW 364
+ S D +L + D + + L EH + L H + +V S D +V W
Sbjct: 351 FILSCADDKTLRVWDYKNKRCMKTLNAHEHFVTSLDFHKTAPYVVTGSVDQTVKVW 406
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.319 0.135 0.412
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 90,733,837
Number of Sequences: 164201
Number of extensions: 3976559
Number of successful extensions: 11868
Number of sequences better than 10.0: 383
Number of HSP's better than 10.0 without gapping: 242
Number of HSP's successfully gapped in prelim test: 142
Number of HSP's that attempted gapping in prelim test: 9177
Number of HSP's gapped (non-prelim): 1655
length of query: 744
length of database: 59,974,054
effective HSP length: 118
effective length of query: 626
effective length of database: 40,598,336
effective search space: 25414558336
effective search space used: 25414558336
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 70 (31.6 bits)
Medicago: description of AC148345.14


